CD4: Difference between revisions
Gerald Chi (talk | contribs) mNo edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
{{other uses|CD-4 (disambiguation)}} | |||
{{Infobox_gene}} | |||
| | {{Infobox protein family | ||
| Symbol = CD4-extrcel | |||
| Name = CD4, Cluster of differentiation 4, extracellular | |||
| image = PDB 1wip EBI.jpg | |||
| width = | |||
| caption = structure of t-cell surface glycoprotein cd4, monoclinic crystal form | |||
| Pfam = PF09191 | |||
| Pfam_clan = | |||
| InterPro = IPR015274 | |||
| SMART = | |||
| PROSITE = | |||
| MEROPS = | |||
| SCOP = 1cid | |||
| TCDB = | |||
| OPM family = 230 | |||
| OPM protein = 2klu | |||
| CAZy = | |||
| CDD = cd07695 | |||
}} | |||
[[File:CD4 correceptor.png|thumb|Image of CD4 co-receptor binding to MHC (Major Histocompatibility Complex) non-polymorphic region.]] | |||
In [[molecular biology]], '''CD4''' ([[cluster of differentiation]] 4) is a [[glycoprotein]] found on the surface of immune cells such as [[T helper cell]]s, [[monocytes]], [[macrophages]], and [[dendritic cells]]. It was discovered in the late 1970s and was originally known as leu-3 and T4 (after the OKT4 [[Monoclonal antibodies|monoclonal antibody]] that reacted with it) before being named CD4 in 1984.<ref name="isbn0-387-12056-4">{{cite book |vauthors=Bernard A, Boumsell L, Hill C | chapter = Joint Report of the First International Workshop on Human Leucocyte Differentiation Antigens by the Investigators of the Participating Laboratories |veditors=Bernard A, Boumsell L, Dausset J, Milstein C, Schlossman SF | title = Leucocyte typing: human leucocyte differentiation antigens detected by monoclonal antibodies: specification, classification, nomenclature | quote = Report on the first international references workshop sponsored by INSERM, WHO and IUIS | publisher = Springer | location = Berlin | year = 1984 | pages = 45–48 | isbn = 0-387-12056-4 | oclc = | doi = 10.1007/978-3-642-68857-7_3 }}</ref> In humans, the CD4 protein is encoded by the ''CD4'' [[gene]].<ref name="pmid3086883">{{cite journal | vauthors = Isobe M, Huebner K, Maddon PJ, Littman DR, Axel R, Croce CM | title = The gene encoding the T-cell surface protein T4 is located on human chromosome 12 | journal = Proc. Natl. Acad. Sci. U.S.A. | volume = 83 | issue = 12 | pages = 4399–402 | date = June 1986 | pmid = 3086883 | pmc = 323740 | doi = 10.1073/pnas.83.12.4399 | url = http://www.pnas.org/cgi/pmidlookup?view=long&pmid=3086883 | bibcode = 1986PNAS...83.4399I }}</ref><ref name="pmid8723724">{{cite journal | vauthors = Ansari-Lari MA, Muzny DM, Lu J, Lu F, Lilley CE, Spanos S, Malley T, Gibbs RA | title = A gene-rich cluster between the CD4 and triosephosphate isomerase genes at human chromosome 12p13 | journal = Genome Res. | volume = 6 | issue = 4 | pages = 314–26 | date = April 1996 | pmid = 8723724 | doi = 10.1101/gr.6.4.314 | url = http://www.genome.org/cgi/pmidlookup?view=long&pmid=8723724 }}</ref> | |||
'''CD4''' ([[cluster of differentiation]] 4) is a [[glycoprotein]] | |||
[[T helper cell|CD4+ T helper cells]] are [[leukocytes|white blood cells]] that are an essential part of the human immune system. They are often referred to as CD4 cells, T-helper cells or T4 cells. They are called helper cells because one of their main roles is to send signals to other types of immune cells, including CD8 killer cells, which then destroy the infectious particle. If CD4 cells become depleted, for example in untreated HIV infection, or following immune suppression prior to a transplant, the body is left vulnerable to a wide range of infections that it would otherwise have been able to fight. | |||
==Structure== | ==Structure== | ||
Like many cell surface receptors/markers, CD4 is a member of the [[immunoglobulin superfamily]]. | [[File:CD4 receptor.png|right|thumb|Schematic representation of CD4 [[Receptor (biochemistry)|receptor]].]] | ||
Like many cell surface receptors/markers, CD4 is a member of the [[immunoglobulin superfamily]]. | |||
It has four immunoglobulin domains (D<sub>1</sub> to D<sub>4</sub>) that are exposed on the extracellular surface of the cell: | |||
* D<sub>1</sub> and D<sub>3</sub> resemble [[immunoglobulin]] variable (IgV) domains. | |||
* D<sub>2</sub> and D<sub>4</sub> resemble [[immunoglobulin]] constant (IgC) domains. | |||
The [[immunoglobulin]] variable (IgV) domain of D<sub>1</sub> adopts an immunoglobulin-like β-sandwich fold with seven β-strands in 2 β-sheets, in a [[Beta sheet#Greek key motif|Greek key topology]].<ref name="pmid8493535">{{cite journal | vauthors = Brady RL, Dodson EJ, Dodson GG, Lange G, Davis SJ, Williams AF, Barclay AN | title = Crystal structure of domains 3 and 4 of rat CD4: relation to the NH2-terminal domains | journal = Science | volume = 260 | issue = 5110 | pages = 979–83 | date = May 1993 | pmid = 8493535 | doi = 10.1126/science.8493535 | bibcode = 1993Sci...260..979B }}</ref> | |||
CD4 interacts with the β<sub>2</sub>-domain of [[MHC class II]] molecules through its D<sub>1</sub> domain. T cells displaying CD4 molecules (and not [[CD8]]) on their surface, therefore, are specific for antigens presented by MHC II and not by [[MHC class I]] (they are ''MHC class II-restricted''). MHC class I contains [[Beta-2 microglobulin]]. | |||
The short [[cytoplasmic]]/[[intracellular]] tail (C) of CD4 contains a special sequence of [[amino acids]] that allow it to recruit and interact with the [[tyrosine kinase]] [[Lck]]. | |||
== Function == | |||
CD4 is a [[co-receptor]] of the [[T cell receptor]] (TCR) and assists the latter in communicating with [[antigen-presenting cell|antigen-presenting cells]]. The TCR complex and CD4 each bind to distinct regions of the antigen-presenting MHCII molecule - α1/β1 and β2, respectively. In CD4 the interaction involves its [[extracellular]] D<sub>1</sub> domain. The resulting close proximity between the TCR complex and CD4 (extracellular and intracellular) allows the [[tyrosine kinase]] Lck bound to the cytoplasmic tail of CD4 to tyrosine-phosphorylate the [[Immunoreceptor tyrosine activation motif]]s (ITAM) on the cytoplasmic domains of CD3 to amplify the signal generated by the TCR. Lck is essential for the activation of many molecular components of the signaling cascade of an activated T cell. Depending on the signal, different types of [[T helper cells]] result. Phosphorylated ITAM motifs on CD3 recruit and activate SH2 domain-containing protein tyrosine kinases (PTK) such as Zap70 to further mediate downstream signalling through tyrosine phosphorylation, leading to transcription factor activation including NF-κB and consequent T cell activation.{{citation needed|date=August 2016}} | |||
=== Other interactions === | |||
CD4 has also been shown to [[Protein-protein interaction|interact]] with [[SPG21]],<ref name=pmid11113139>{{cite journal | vauthors = Zeitlmann L, Sirim P, Kremmer E, Kolanus W | title = Cloning of ACP33 as a novel intracellular ligand of CD4 | journal = J. Biol. Chem. | volume = 276 | issue = 12 | pages = 9123–32 | date = Mar 2001 | pmid = 11113139 | doi = 10.1074/jbc.M009270200 }}</ref> [[Lck]]<ref name="pmid20724730">{{cite journal | vauthors = Rudd CE, Trevillyan JM, Dasgupta JD, Wong LL, Schlossman SF | title = Pillars article: the CD4 receptor is complexed in detergent lysates to a protein-tyrosine kinase (pp58) from human T lymphocytes. 1988 | journal = J. Immunol. | volume = 185 | issue = 5 | pages = 2645–9 | date = September 2010 | pmid = 20724730 | pmc = 3791413 | doi = }}</ref><ref name="pmid2455897">{{cite journal | vauthors = Rudd CE, Trevillyan JM, Dasgupta JD, Wong LL, Schlossman SF | title = The CD4 receptor is complexed in detergent lysates to a protein-tyrosine kinase (pp58) from human T lymphocytes | journal = Proc. Natl. Acad. Sci. U.S.A. | volume = 85 | issue = 14 | pages = 5190–4 | date = July 1988 | pmid = 2455897 | pmc = 281714 | doi = 10.1073/pnas.85.14.5190 | bibcode = 1988PNAS...85.5190R }}</ref><ref name="pmid2470098">{{cite journal | vauthors = Barber EK, Dasgupta JD, Schlossman SF, Trevillyan JM, Rudd CE | title = The CD4 and CD8 antigens are coupled to a protein-tyrosine kinase (p56lck) that phosphorylates the CD3 complex | journal = Proc. Natl. Acad. Sci. U.S.A. | volume = 86 | issue = 9 | pages = 3277–81 | date = May 1989 | pmid = 2470098 | pmc = 287114 | doi = 10.1073/pnas.86.9.3277 | bibcode = 1989PNAS...86.3277B }}</ref><ref name="pmid12007789">{{cite journal | vauthors = Hawash IY, Hu XE, Adal A, Cassady JM, Geahlen RL, Harrison ML | title = The oxygen-substituted palmitic acid analogue, 13-oxypalmitic acid, inhibits Lck localization to lipid rafts and T cell signaling | journal = Biochim. Biophys. Acta | volume = 1589 | issue = 2 | pages = 140–50 | date = April 2002 | pmid = 12007789 | doi = 10.1016/S0167-4889(02)00165-9 }}</ref><ref name="pmid11854499">{{cite journal | vauthors = Foti M, Phelouzat MA, Holm A, Rasmusson BJ, Carpentier JL | title = p56Lck anchors CD4 to distinct microdomains on microvilli | journal = Proc. Natl. Acad. Sci. U.S.A. | volume = 99 | issue = 4 | pages = 2008–13 | date = February 2002 | pmid = 11854499 | pmc = 122310 | doi = 10.1073/pnas.042689099 | bibcode = 2002PNAS...99.2008F }}</ref> and [[Protein unc-119 homolog]].<ref name="pmid14757743">{{cite journal | vauthors = Gorska MM, Stafford SJ, Cen O, Sur S, Alam R | title = Unc119, a Novel Activator of Lck/Fyn, Is Essential for T Cell Activation | journal = J. Exp. Med. | volume = 199 | issue = 3 | pages = 369–79 | date = February 2004 | pmid = 14757743 | pmc = 2211793 | doi = 10.1084/jem.20030589 }}</ref> | |||
== Disease == | |||
{{anchor|count}} | |||
=== HIV infection === | |||
[[HIV-1]] uses CD4 to gain entry into host T-cells and achieves this through its [[viral envelope]] protein known as [[gp120]].<ref name="pmid9641677">{{cite journal | vauthors = Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA | title = Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody | journal = Nature | volume = 393 | issue = 6686 | pages = 648–59 | date = June 1998 | pmid = 9641677 | doi = 10.1038/31405 | bibcode = 1998Natur.393..648K }}</ref> The binding to CD4 creates a shift in the conformation of gp120 allowing HIV-1 to bind to a co-receptor expressed on the host cell. These co-receptors are [[chemokine receptors]] [[CCR5]] or [[CXCR4]]. Following a structural change in another viral protein ([[gp41]]), HIV inserts a [[fusion peptide]] into the host cell that allows the outer membrane of the virus to fuse with the [[cell membrane]]. | |||
=== HIV pathology === | |||
HIV infection leads to a progressive reduction in the number of [[T helper cells|T cells expressing CD4]]. Medical professionals refer to the CD4 count to decide when to begin treatment during HIV infection, although recent medical guidelines have changed to recommend treatment at all CD4 counts as soon as HIV is diagnosed. A CD4 count measures the number of T cells expressing CD4. While CD4 counts are not a direct [[HIV]] test—e.g. they do not check the presence of viral DNA, or specific antibodies against HIV—they are used to assess the immune system of a patient.{{citation needed|date=August 2016}} | |||
[[National Institutes of Health]] guidelines recommend treatment of any HIV-positive individuals, regardless of CD4 count<ref name="urlaidsinfo.nih.gov">{{cite web | url = http://aidsinfo.nih.gov/contentfiles/lvguidelines/glchunk/glchunk_37.pdf | title = Guidelines for the Use of Antiretroviral Agents in HIV-1-Infected Adults and Adolescents| work = AIDSinfo | publisher = U.S. Department of Health & Human Services | date = 2013-02-13 }}</ref> Normal [[blood values]] are usually expressed as the number of cells per microliter (μL, or equivalently, cubic millimeter, mm<sup>3</sup>) of blood, with normal values for CD4 cells being 500–1200 cells/mm<sup>3</sup>.<ref name="pmid1349272">{{cite journal | vauthors = Bofill M, Janossy G, Lee CA, MacDonald-Burns D, Phillips AN, Sabin C, Timms A, Johnson MA, Kernoff PB | title = Laboratory control values for CD4 and CD8 T lymphocytes. Implications for HIV-1 diagnosis | journal = Clin. Exp. Immunol. | volume = 88 | issue = 2 | pages = 243–52 | date = May 1992 | pmid = 1349272 | pmc = 1554313 | doi = 10.1111/j.1365-2249.1992.tb03068.x}}</ref> Patients often undergo treatments when the CD4 counts reach a level of 350 cells per microliter in Europe but usually around 500/μL in the US; people with less than 200 cells per microliter are at high risk of contracting AIDS defined illnesses. Medical professionals also refer to CD4 tests to determine efficacy of treatment.{{citation needed|date=August 2016}} | |||
[[Viral load]] testing provides more information about the efficacy for therapy than CD4 counts.<ref name="HIV Medicine Association February 2016">{{Citation |author1 = HIV Medicine Association |author1-link = HIV Medicine Association |date = February 2016 |title = Five Things Physicians and Patients Should Question |publisher = HIV Medicine Association |work = [[Choosing Wisely]]: an initiative of the [[ABIM Foundation]] |page = |url = http://www.choosingwisely.org/societies/hiv-medicine-association/ |accessdate = 9 May 2016}}</ref> For the first 2 years of HIV therapy, CD4 counts may be done every 3–6 months.<ref name="HIV Medicine Association February 2016"/> If a patient's viral load becomes undetectable after 2 years then CD4 counts might not be needed if they are consistently above 500/mm<sup>3</sup>.<ref name="HIV Medicine Association February 2016"/> If the count remains at 300–500/mm<sup>3</sup>, then the tests can be done annually.<ref name="HIV Medicine Association February 2016"/> It is not necessary to schedule CD4 counts with viral load tests and the two should be done independently when each is indicated.<ref name="HIV Medicine Association February 2016"/> | |||
[[File:Reference ranges for blood tests - white blood cells.png|thumb|500px|center|[[Reference ranges for blood tests]] of white blood cells, comparing CD4+ cell amount (shown in green-yellow) with other cells.]] | |||
=== Other diseases === | |||
The | CD4 continues to be expressed in most [[neoplasm]]s derived from [[T helper cells]]. It is therefore possible to use CD4 [[immunohistochemistry]] on tissue [[biopsy]] samples to identify most forms of peripheral T cell [[lymphoma]] and related malignant conditions.<ref name="isbn1-84110-100-1">{{cite book |author1=Kumarasen Cooper |author2=Anthony S-Y. Leong | title = Manual of diagnostic antibodies for immunohistology | publisher = Greenwich Medical Media | location = London | year = 2003 | pages = 65 | isbn = 1-84110-100-1 }}</ref> The antigen has also been associated with a number of [[autoimmunity|autoimmune diseases]] such as [[vitiligo]] and [[type I diabetes mellitus]].<ref name="pmid19843086">{{cite journal | vauthors = Zamani M, Tabatabaiefar MA, Mosayyebi S, Mashaghi A, Mansouri P | title = Possible association of the CD4 gene polymorphism with vitiligo in an Iranian population | journal = Clin. Exp. Dermatol. | volume = 35 | issue = 5 | pages = 521–4 | date = July 2010 | pmid = 19843086 | doi = 10.1111/j.1365-2230.2009.03667.x }}</ref> | ||
== | T-cells play a large part in autoinflammatory diseases.<ref name="pmid24164192">{{cite journal | vauthors = Ciccarelli F, De Martinis M, Ginaldi L | title = An update on autoinflammatory diseases | journal = Curr. Med. Chem. | volume = 21 | issue = 3 | pages = 261–9 | year = 2014 | pmid = 24164192 | pmc = 3905709 | doi = 10.2174/09298673113206660303 }}</ref> When testing a drug's efficacy or studying diseases, it is helpful to quantify the amount of T-cells. | ||
[[ | <!-- Deleted image removed: [[File:CD4-R113-5ug-tonsil.png|thumbnail|Immunohistochemistry CD4+ T-cells stained with DAB]] --> | ||
CD4 | on fresh-frozen tissue with CD4+, CD8+, and CD3+ T-cell markers (which stain different markers on a T-cell - giving different results).<ref>{{cite web|title=550280 - BD Biosciences|url=http://www.bdbiosciences.com/us/reagents/research/antibodies-buffers/immunology-reagents/anti-mouse-antibodies/cell-surface-antigens/purified-rat-anti-mouse-cd4-rm4-5/p/550280|website=BD Biosciences|publisher=Becton Dickinson}}</ref> | ||
== See also == | |||
*[[CD4+ T cells and antitumor immunity]] | |||
{{Clear}} | |||
== References == | |||
{{reflist|2}} | |||
{{InterPro content|IPR015274}} | |||
==Further reading== | == Further reading == | ||
{{refbegin | 2}} | {{refbegin | 2}} | ||
* {{cite journal | vauthors = Miceli MC, Parnes JR | title = Role of CD4 and CD8 in T cell activation and differentiation | journal = Adv. Immunol. | volume = 53 | issue = | pages = 59–122 | year = 1993 | pmid = 8512039 | doi = 10.1016/S0065-2776(08)60498-8 | isbn = 978-0-12-022453-1 | series = Advances in Immunology }} | |||
* {{cite journal | vauthors = Geyer M, Fackler OT, Peterlin BM | title = Structure–function relationships in HIV-1 Nef | journal = EMBO Rep. | volume = 2 | issue = 7 | pages = 580–5 | year = 2001 | pmid = 11463741 | pmc = 1083955 | doi = 10.1093/embo-reports/kve141 }} | |||
*{{cite journal | * {{cite journal | vauthors = Greenway AL, Holloway G, McPhee DA, Ellis P, Cornall A, Lidman M | title = HIV-1 Nef control of cell signalling molecules: multiple strategies to promote virus replication | journal = J. Biosci. | volume = 28 | issue = 3 | pages = 323–35 | year = 2004 | pmid = 12734410 | doi = 10.1007/BF02970151 }} | ||
*{{cite journal | * {{cite journal | vauthors = Bénichou S, Benmerah A | title = [The HIV nef and the Kaposi-sarcoma-associated virus K3/K5 proteins: "parasites"of the endocytosis pathway] | journal = Med Sci (Paris) | volume = 19 | issue = 1 | pages = 100–6 | year = 2003 | pmid = 12836198 | doi = 10.1051/medsci/2003191100 }} | ||
*{{cite journal | * {{cite journal | vauthors = Leavitt SA, SchOn A, Klein JC, Manjappara U, Chaiken IM, Freire E | title = Interactions of HIV-1 proteins gp120 and Nef with cellular partners define a novel allosteric paradigm | journal = Curr. Protein Pept. Sci. | volume = 5 | issue = 1 | pages = 1–8 | year = 2004 | pmid = 14965316 | doi = 10.2174/1389203043486955 }} | ||
*{{cite journal | * {{cite journal | vauthors = Tolstrup M, Ostergaard L, Laursen AL, Pedersen SF, Duch M | title = HIV/SIV escape from immune surveillance: focus on Nef | journal = Curr. HIV Res. | volume = 2 | issue = 2 | pages = 141–51 | year = 2004 | pmid = 15078178 | doi = 10.2174/1570162043484924 }} | ||
*{{cite journal | * {{cite journal | vauthors = Hout DR, Mulcahy ER, Pacyniak E, Gomez LM, Gomez ML, Stephens EB | title = Vpu: a multifunctional protein that enhances the pathogenesis of human immunodeficiency virus type 1 | journal = Curr. HIV Res. | volume = 2 | issue = 3 | pages = 255–70 | year = 2005 | pmid = 15279589 | doi = 10.2174/1570162043351246 }} | ||
*{{cite journal | * {{cite journal | vauthors = Joseph AM, Kumar M, Mitra D | title = Nef: "necessary and enforcing factor" in HIV infection | journal = Curr. HIV Res. | volume = 3 | issue = 1 | pages = 87–94 | year = 2005 | pmid = 15638726 | doi = 10.2174/1570162052773013 }} | ||
*{{cite journal | * {{cite journal | vauthors = Anderson JL, Hope TJ | title = HIV accessory proteins and surviving the host cell | journal = Current HIV/AIDS reports | volume = 1 | issue = 1 | pages = 47–53 | year = 2005 | pmid = 16091223 | doi = 10.1007/s11904-004-0007-x }} | ||
*{{cite journal | * {{cite journal | vauthors = Li L, Li HS, Pauza CD, Bukrinsky M, Zhao RY | title = Roles of HIV-1 auxiliary proteins in viral pathogenesis and host-pathogen interactions | journal = Cell Res. | volume = 15 | issue = 11–12 | pages = 923–34 | year = 2006 | pmid = 16354571 | doi = 10.1038/sj.cr.7290370 }} | ||
*{{cite journal | * {{cite journal | vauthors = Stove V, Verhasselt B | title = Modelling thymic HIV-1 Nef effects | journal = Curr. HIV Res. | volume = 4 | issue = 1 | pages = 57–64 | year = 2006 | pmid = 16454711 | doi = 10.2174/157016206775197583 }} | ||
*{{cite journal | |||
*{{cite journal | |||
}} | |||
{{refend}} | {{refend}} | ||
== External links == | |||
* {{MeshName|CD1+Antigen}} | |||
* [http://www.ebioscience.com/ebioscience/whatsnew/mousecdchart.htm Mouse CD Antigen Chart] | |||
* [http://www.ebioscience.com/ebioscience/whatsnew/humancdchart.htm Human CD Antigen Chart] | |||
* *[http://www.bio.davidson.edu/Courses/Molbio/MolStudents/spring2005/Greendyke/gp120.htm Human Immunodeficiency Virus Glycoprotein 120] | |||
* {{UCSC gene info|CD4}} | |||
{{PDB Gallery|geneid=920}} | |||
{{T cell receptor}} | {{T cell receptor}} | ||
{{Clusters of differentiation}} | {{Clusters of differentiation}} | ||
{{Clusters of differentiation by lineage}} | |||
{{Interleukin receptor modulators}} | |||
{{DEFAULTSORT:Cd4}} | |||
[[Category:Clusters of differentiation]] | |||
[[Category:Glycoproteins]] | [[Category:Glycoproteins]] | ||
[[Category: | [[Category:Immunology]] | ||
[[Category:Protein domains]] | |||
[[Category:T cells]] | |||
Revision as of 10:36, 13 September 2017
| VALUE_ERROR (nil) | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Aliases | |||||||
| External IDs | GeneCards: [1] | ||||||
| Orthologs | |||||||
| Species | Human | Mouse | |||||
| Entrez |
|
| |||||
| Ensembl |
|
| |||||
| UniProt |
|
| |||||
| RefSeq (mRNA) |
|
| |||||
| RefSeq (protein) |
|
| |||||
| Location (UCSC) | n/a | n/a | |||||
| PubMed search | n/a | n/a | |||||
| Wikidata | |||||||
| |||||||
| CD4, Cluster of differentiation 4, extracellular | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| File:PDB 1wip EBI.jpg structure of t-cell surface glycoprotein cd4, monoclinic crystal form | |||||||||
| Identifiers | |||||||||
| Symbol | CD4-extrcel | ||||||||
| Pfam | PF09191 | ||||||||
| InterPro | IPR015274 | ||||||||
| SCOP | 1cid | ||||||||
| SUPERFAMILY | 1cid | ||||||||
| OPM superfamily | 230 | ||||||||
| OPM protein | 2klu | ||||||||
| CDD | cd07695 | ||||||||
| |||||||||
In molecular biology, CD4 (cluster of differentiation 4) is a glycoprotein found on the surface of immune cells such as T helper cells, monocytes, macrophages, and dendritic cells. It was discovered in the late 1970s and was originally known as leu-3 and T4 (after the OKT4 monoclonal antibody that reacted with it) before being named CD4 in 1984.[1] In humans, the CD4 protein is encoded by the CD4 gene.[2][3]
CD4+ T helper cells are white blood cells that are an essential part of the human immune system. They are often referred to as CD4 cells, T-helper cells or T4 cells. They are called helper cells because one of their main roles is to send signals to other types of immune cells, including CD8 killer cells, which then destroy the infectious particle. If CD4 cells become depleted, for example in untreated HIV infection, or following immune suppression prior to a transplant, the body is left vulnerable to a wide range of infections that it would otherwise have been able to fight.
Structure
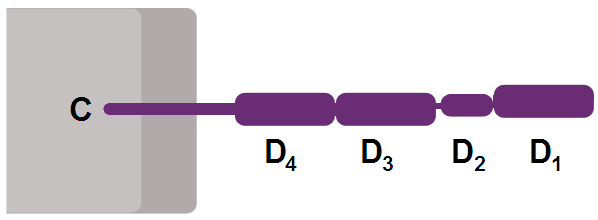
Like many cell surface receptors/markers, CD4 is a member of the immunoglobulin superfamily.
It has four immunoglobulin domains (D1 to D4) that are exposed on the extracellular surface of the cell:
- D1 and D3 resemble immunoglobulin variable (IgV) domains.
- D2 and D4 resemble immunoglobulin constant (IgC) domains.
The immunoglobulin variable (IgV) domain of D1 adopts an immunoglobulin-like β-sandwich fold with seven β-strands in 2 β-sheets, in a Greek key topology.[4]
CD4 interacts with the β2-domain of MHC class II molecules through its D1 domain. T cells displaying CD4 molecules (and not CD8) on their surface, therefore, are specific for antigens presented by MHC II and not by MHC class I (they are MHC class II-restricted). MHC class I contains Beta-2 microglobulin.
The short cytoplasmic/intracellular tail (C) of CD4 contains a special sequence of amino acids that allow it to recruit and interact with the tyrosine kinase Lck.
Function
CD4 is a co-receptor of the T cell receptor (TCR) and assists the latter in communicating with antigen-presenting cells. The TCR complex and CD4 each bind to distinct regions of the antigen-presenting MHCII molecule - α1/β1 and β2, respectively. In CD4 the interaction involves its extracellular D1 domain. The resulting close proximity between the TCR complex and CD4 (extracellular and intracellular) allows the tyrosine kinase Lck bound to the cytoplasmic tail of CD4 to tyrosine-phosphorylate the Immunoreceptor tyrosine activation motifs (ITAM) on the cytoplasmic domains of CD3 to amplify the signal generated by the TCR. Lck is essential for the activation of many molecular components of the signaling cascade of an activated T cell. Depending on the signal, different types of T helper cells result. Phosphorylated ITAM motifs on CD3 recruit and activate SH2 domain-containing protein tyrosine kinases (PTK) such as Zap70 to further mediate downstream signalling through tyrosine phosphorylation, leading to transcription factor activation including NF-κB and consequent T cell activation.[citation needed]
Other interactions
CD4 has also been shown to interact with SPG21,[5] Lck[6][7][8][9][10] and Protein unc-119 homolog.[11]
Disease
HIV infection
HIV-1 uses CD4 to gain entry into host T-cells and achieves this through its viral envelope protein known as gp120.[12] The binding to CD4 creates a shift in the conformation of gp120 allowing HIV-1 to bind to a co-receptor expressed on the host cell. These co-receptors are chemokine receptors CCR5 or CXCR4. Following a structural change in another viral protein (gp41), HIV inserts a fusion peptide into the host cell that allows the outer membrane of the virus to fuse with the cell membrane.
HIV pathology
HIV infection leads to a progressive reduction in the number of T cells expressing CD4. Medical professionals refer to the CD4 count to decide when to begin treatment during HIV infection, although recent medical guidelines have changed to recommend treatment at all CD4 counts as soon as HIV is diagnosed. A CD4 count measures the number of T cells expressing CD4. While CD4 counts are not a direct HIV test—e.g. they do not check the presence of viral DNA, or specific antibodies against HIV—they are used to assess the immune system of a patient.[citation needed]
National Institutes of Health guidelines recommend treatment of any HIV-positive individuals, regardless of CD4 count[13] Normal blood values are usually expressed as the number of cells per microliter (μL, or equivalently, cubic millimeter, mm3) of blood, with normal values for CD4 cells being 500–1200 cells/mm3.[14] Patients often undergo treatments when the CD4 counts reach a level of 350 cells per microliter in Europe but usually around 500/μL in the US; people with less than 200 cells per microliter are at high risk of contracting AIDS defined illnesses. Medical professionals also refer to CD4 tests to determine efficacy of treatment.[citation needed]
Viral load testing provides more information about the efficacy for therapy than CD4 counts.[15] For the first 2 years of HIV therapy, CD4 counts may be done every 3–6 months.[15] If a patient's viral load becomes undetectable after 2 years then CD4 counts might not be needed if they are consistently above 500/mm3.[15] If the count remains at 300–500/mm3, then the tests can be done annually.[15] It is not necessary to schedule CD4 counts with viral load tests and the two should be done independently when each is indicated.[15]
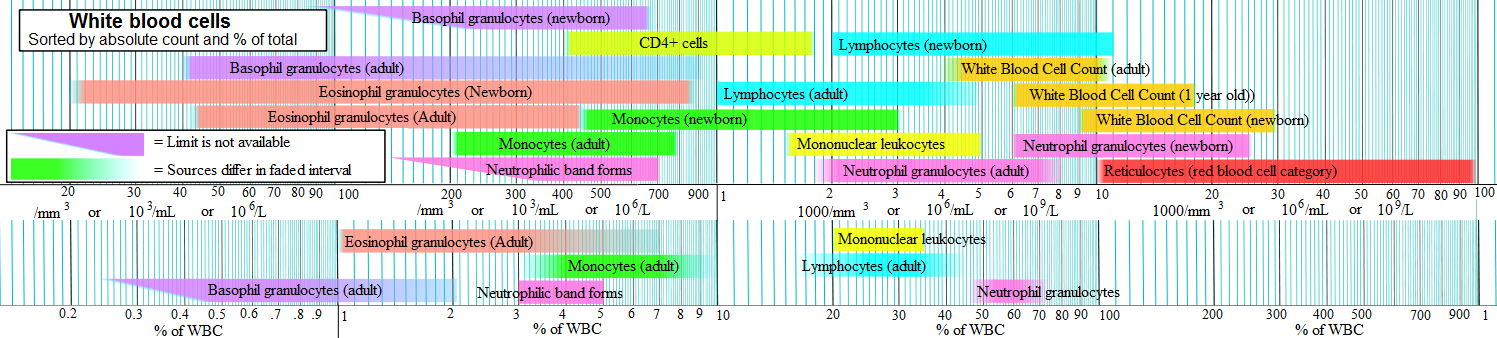
Other diseases
CD4 continues to be expressed in most neoplasms derived from T helper cells. It is therefore possible to use CD4 immunohistochemistry on tissue biopsy samples to identify most forms of peripheral T cell lymphoma and related malignant conditions.[16] The antigen has also been associated with a number of autoimmune diseases such as vitiligo and type I diabetes mellitus.[17]
T-cells play a large part in autoinflammatory diseases.[18] When testing a drug's efficacy or studying diseases, it is helpful to quantify the amount of T-cells. on fresh-frozen tissue with CD4+, CD8+, and CD3+ T-cell markers (which stain different markers on a T-cell - giving different results).[19]
See also
References
- ↑ Bernard A, Boumsell L, Hill C (1984). "Joint Report of the First International Workshop on Human Leucocyte Differentiation Antigens by the Investigators of the Participating Laboratories". In Bernard A, Boumsell L, Dausset J, Milstein C, Schlossman SF. Leucocyte typing: human leucocyte differentiation antigens detected by monoclonal antibodies: specification, classification, nomenclature. Berlin: Springer. pp. 45–48. doi:10.1007/978-3-642-68857-7_3. ISBN 0-387-12056-4.
Report on the first international references workshop sponsored by INSERM, WHO and IUIS
- ↑ Isobe M, Huebner K, Maddon PJ, Littman DR, Axel R, Croce CM (June 1986). "The gene encoding the T-cell surface protein T4 is located on human chromosome 12". Proc. Natl. Acad. Sci. U.S.A. 83 (12): 4399–402. Bibcode:1986PNAS...83.4399I. doi:10.1073/pnas.83.12.4399. PMC 323740. PMID 3086883.
- ↑ Ansari-Lari MA, Muzny DM, Lu J, Lu F, Lilley CE, Spanos S, Malley T, Gibbs RA (April 1996). "A gene-rich cluster between the CD4 and triosephosphate isomerase genes at human chromosome 12p13". Genome Res. 6 (4): 314–26. doi:10.1101/gr.6.4.314. PMID 8723724.
- ↑ Brady RL, Dodson EJ, Dodson GG, Lange G, Davis SJ, Williams AF, Barclay AN (May 1993). "Crystal structure of domains 3 and 4 of rat CD4: relation to the NH2-terminal domains". Science. 260 (5110): 979–83. Bibcode:1993Sci...260..979B. doi:10.1126/science.8493535. PMID 8493535.
- ↑ Zeitlmann L, Sirim P, Kremmer E, Kolanus W (Mar 2001). "Cloning of ACP33 as a novel intracellular ligand of CD4". J. Biol. Chem. 276 (12): 9123–32. doi:10.1074/jbc.M009270200. PMID 11113139.
- ↑ Rudd CE, Trevillyan JM, Dasgupta JD, Wong LL, Schlossman SF (September 2010). "Pillars article: the CD4 receptor is complexed in detergent lysates to a protein-tyrosine kinase (pp58) from human T lymphocytes. 1988". J. Immunol. 185 (5): 2645–9. PMC 3791413. PMID 20724730.
- ↑ Rudd CE, Trevillyan JM, Dasgupta JD, Wong LL, Schlossman SF (July 1988). "The CD4 receptor is complexed in detergent lysates to a protein-tyrosine kinase (pp58) from human T lymphocytes". Proc. Natl. Acad. Sci. U.S.A. 85 (14): 5190–4. Bibcode:1988PNAS...85.5190R. doi:10.1073/pnas.85.14.5190. PMC 281714. PMID 2455897.
- ↑ Barber EK, Dasgupta JD, Schlossman SF, Trevillyan JM, Rudd CE (May 1989). "The CD4 and CD8 antigens are coupled to a protein-tyrosine kinase (p56lck) that phosphorylates the CD3 complex". Proc. Natl. Acad. Sci. U.S.A. 86 (9): 3277–81. Bibcode:1989PNAS...86.3277B. doi:10.1073/pnas.86.9.3277. PMC 287114. PMID 2470098.
- ↑ Hawash IY, Hu XE, Adal A, Cassady JM, Geahlen RL, Harrison ML (April 2002). "The oxygen-substituted palmitic acid analogue, 13-oxypalmitic acid, inhibits Lck localization to lipid rafts and T cell signaling". Biochim. Biophys. Acta. 1589 (2): 140–50. doi:10.1016/S0167-4889(02)00165-9. PMID 12007789.
- ↑ Foti M, Phelouzat MA, Holm A, Rasmusson BJ, Carpentier JL (February 2002). "p56Lck anchors CD4 to distinct microdomains on microvilli". Proc. Natl. Acad. Sci. U.S.A. 99 (4): 2008–13. Bibcode:2002PNAS...99.2008F. doi:10.1073/pnas.042689099. PMC 122310. PMID 11854499.
- ↑ Gorska MM, Stafford SJ, Cen O, Sur S, Alam R (February 2004). "Unc119, a Novel Activator of Lck/Fyn, Is Essential for T Cell Activation". J. Exp. Med. 199 (3): 369–79. doi:10.1084/jem.20030589. PMC 2211793. PMID 14757743.
- ↑ Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA (June 1998). "Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody". Nature. 393 (6686): 648–59. Bibcode:1998Natur.393..648K. doi:10.1038/31405. PMID 9641677.
- ↑ "Guidelines for the Use of Antiretroviral Agents in HIV-1-Infected Adults and Adolescents" (PDF). AIDSinfo. U.S. Department of Health & Human Services. 2013-02-13.
- ↑ Bofill M, Janossy G, Lee CA, MacDonald-Burns D, Phillips AN, Sabin C, Timms A, Johnson MA, Kernoff PB (May 1992). "Laboratory control values for CD4 and CD8 T lymphocytes. Implications for HIV-1 diagnosis". Clin. Exp. Immunol. 88 (2): 243–52. doi:10.1111/j.1365-2249.1992.tb03068.x. PMC 1554313. PMID 1349272.
- ↑ 15.0 15.1 15.2 15.3 15.4 HIV Medicine Association (February 2016), "Five Things Physicians and Patients Should Question", Choosing Wisely: an initiative of the ABIM Foundation, HIV Medicine Association, retrieved 9 May 2016
- ↑ Kumarasen Cooper; Anthony S-Y. Leong (2003). Manual of diagnostic antibodies for immunohistology. London: Greenwich Medical Media. p. 65. ISBN 1-84110-100-1.
- ↑ Zamani M, Tabatabaiefar MA, Mosayyebi S, Mashaghi A, Mansouri P (July 2010). "Possible association of the CD4 gene polymorphism with vitiligo in an Iranian population". Clin. Exp. Dermatol. 35 (5): 521–4. doi:10.1111/j.1365-2230.2009.03667.x. PMID 19843086.
- ↑ Ciccarelli F, De Martinis M, Ginaldi L (2014). "An update on autoinflammatory diseases". Curr. Med. Chem. 21 (3): 261–9. doi:10.2174/09298673113206660303. PMC 3905709. PMID 24164192.
- ↑ "550280 - BD Biosciences". BD Biosciences. Becton Dickinson.
Further reading
- Miceli MC, Parnes JR (1993). "Role of CD4 and CD8 in T cell activation and differentiation". Adv. Immunol. Advances in Immunology. 53: 59–122. doi:10.1016/S0065-2776(08)60498-8. ISBN 978-0-12-022453-1. PMID 8512039.
- Geyer M, Fackler OT, Peterlin BM (2001). "Structure–function relationships in HIV-1 Nef". EMBO Rep. 2 (7): 580–5. doi:10.1093/embo-reports/kve141. PMC 1083955. PMID 11463741.
- Greenway AL, Holloway G, McPhee DA, Ellis P, Cornall A, Lidman M (2004). "HIV-1 Nef control of cell signalling molecules: multiple strategies to promote virus replication". J. Biosci. 28 (3): 323–35. doi:10.1007/BF02970151. PMID 12734410.
- Bénichou S, Benmerah A (2003). "[The HIV nef and the Kaposi-sarcoma-associated virus K3/K5 proteins: "parasites"of the endocytosis pathway]". Med Sci (Paris). 19 (1): 100–6. doi:10.1051/medsci/2003191100. PMID 12836198.
- Leavitt SA, SchOn A, Klein JC, Manjappara U, Chaiken IM, Freire E (2004). "Interactions of HIV-1 proteins gp120 and Nef with cellular partners define a novel allosteric paradigm". Curr. Protein Pept. Sci. 5 (1): 1–8. doi:10.2174/1389203043486955. PMID 14965316.
- Tolstrup M, Ostergaard L, Laursen AL, Pedersen SF, Duch M (2004). "HIV/SIV escape from immune surveillance: focus on Nef". Curr. HIV Res. 2 (2): 141–51. doi:10.2174/1570162043484924. PMID 15078178.
- Hout DR, Mulcahy ER, Pacyniak E, Gomez LM, Gomez ML, Stephens EB (2005). "Vpu: a multifunctional protein that enhances the pathogenesis of human immunodeficiency virus type 1". Curr. HIV Res. 2 (3): 255–70. doi:10.2174/1570162043351246. PMID 15279589.
- Joseph AM, Kumar M, Mitra D (2005). "Nef: "necessary and enforcing factor" in HIV infection". Curr. HIV Res. 3 (1): 87–94. doi:10.2174/1570162052773013. PMID 15638726.
- Anderson JL, Hope TJ (2005). "HIV accessory proteins and surviving the host cell". Current HIV/AIDS reports. 1 (1): 47–53. doi:10.1007/s11904-004-0007-x. PMID 16091223.
- Li L, Li HS, Pauza CD, Bukrinsky M, Zhao RY (2006). "Roles of HIV-1 auxiliary proteins in viral pathogenesis and host-pathogen interactions". Cell Res. 15 (11–12): 923–34. doi:10.1038/sj.cr.7290370. PMID 16354571.
- Stove V, Verhasselt B (2006). "Modelling thymic HIV-1 Nef effects". Curr. HIV Res. 4 (1): 57–64. doi:10.2174/157016206775197583. PMID 16454711.
External links
- CD1+Antigen at the US National Library of Medicine Medical Subject Headings (MeSH)
- Mouse CD Antigen Chart
- Human CD Antigen Chart
- *Human Immunodeficiency Virus Glycoprotein 120
- Human CD4 genome location and CD4 gene details page in the UCSC Genome Browser.