Immunoglobulin supergene family: Difference between revisions
m (→Class III) |
m (→Class IV) |
||
| Line 646: | Line 646: | ||
"The 1C7 gene [...] is located immediately adjacent to the B144 gene. RNA for B144 and 1C7 are transcribed in convergent directions such that there is a slight overlap between the 3' ends of the two mRNAs. [Human] 1C7 also shows multiple splice forms with 9 forms of the human mRNA reported so far.(21) The major forms encode proteins containing a leader sequence, a probable trans-membrane segment, an external sequence including an immunoglobulin-like domain, and at least three alternative forms of the putative intracellular segment of the protein. One alternative splice modifies the structure of the immunoglobulin-like domain, changing it from a sequence more closely resembling those of the V regions of Ig molecules to one that is more similar to IgC2 regions. Of the three alternative putative intracellular domains, one encodes multiple proline repeats suggestive of SH3 binding domains."<ref name="gruen01"/> | "The 1C7 gene [...] is located immediately adjacent to the B144 gene. RNA for B144 and 1C7 are transcribed in convergent directions such that there is a slight overlap between the 3' ends of the two mRNAs. [Human] 1C7 also shows multiple splice forms with 9 forms of the human mRNA reported so far.(21) The major forms encode proteins containing a leader sequence, a probable trans-membrane segment, an external sequence including an immunoglobulin-like domain, and at least three alternative forms of the putative intracellular segment of the protein. One alternative splice modifies the structure of the immunoglobulin-like domain, changing it from a sequence more closely resembling those of the V regions of Ig molecules to one that is more similar to IgC2 regions. Of the three alternative putative intracellular domains, one encodes multiple proline repeats suggestive of SH3 binding domains."<ref name="gruen01"/> | ||
" | "The existence of the G1 gene was initially noted as a part of a screen of MHC cosmids for embedded genes. The G1 and AIF1 transcripts appear to be derived by alternative splicing from partially overlapping genomic templates. A third human interferon gamma-responsive transcript, IRT-1, has been noted that shares some internal sequences with both G1 and AIF1, but on the basis of the predicted open reading frame it shares only limited amino acid sequences with G1."<ref name="gruen01"/> | ||
" | Gene ID: 199 is AIF1 allograft inflammatory factor 1 on 6p21.33: "This gene encodes a protein that binds actin and calcium. This gene is induced by cytokines and interferon and may promote macrophage activation and growth of vascular smooth muscle cells and T-lymphocytes. Polymorphisms in this gene may be associated with systemic sclerosis. Alternative splicing results in multiple transcript variants, but the full-length and coding nature of some of these variants is not certain."<ref name=RefSeq199>{{ cite web | ||
|author=RefSeq | |||
|title=AIF1 allograft inflammatory factor 1 [ Homo sapiens (human) ] | |||
|publisher=National Center for Biotechnology Information, U.S. National Library of Medicine | |||
|location=8600 Rockville Pike, Bethesda MD, 20894 USA | |||
|date=January 2016 | |||
|url=https://www.ncbi.nlm.nih.gov/gene/199 | |||
|accessdate=7 April 2020 }}</ref> | |||
# NP_001305899.1 allograft inflammatory factor 1 isoform 1: "Transcript Variant: This variant (4) uses an alternate splice site in the 5' region and initiates translation at a downstream start codon compared to variant 3. The encoded isoform (1) has a shorter N-terminus than isoform 3. Variants 1 and 4 encode the same isoform (1)."<ref name=RefSeq199/> | |||
# NP_001614.3 allograft inflammatory factor 1 isoform 3: "Transcript Variant: This variant (3) encodes the longest isoform (3)."<ref name=RefSeq199/> | |||
# NP_116573.1 allograft inflammatory factor 1 isoform 1: "Transcript Variant: This variant (1, also known as G1) differs in the 5' UTR, lacks a portion of the 5' coding region, and initiates translation at a downstream start codon compared to variant 3. The encoded isoform (1) has a shorter N-terminus than isoform 3. Variants 1 and 4 encode the same isoform (1)."<ref name=RefSeq199/> | |||
"AIF-1 (allograft inflammatory factor-1) is a Ca<sup>2+</sup> binding protein predominantly expressed by activated monocytes, originally identified in rat cardiac allografts with chronic rejection.(22) The human cDNA homologue is 86% identical to the rat (90% identical to the amino acid sequence) and was identified by reverse transcriptase-PCR of endomyocardial biopsy specimens from human heart transplants and in macrophage cell lines.(23)"<ref name="gruen01"/> | |||
Gene ID: 3303 is [[HSPA1A]] heat shock protein family A (Hsp70) member 1A on 6p21.33: "This intronless gene encodes a 70kDa heat shock protein which is a member of the heat shock protein 70 family. In conjuction with other heat shock proteins, this protein stabilizes existing proteins against aggregation and mediates the folding of newly translated proteins in the cytosol and in organelles. It is also involved in the ubiquitin-proteasome pathway through interaction with the AU-rich element RNA-binding protein 1. The gene is located in the major histocompatibility complex class III region, in a cluster with two closely related genes which encode similar proteins."<ref name=RefSeq3303>{{ cite web | Gene ID: 3303 is [[HSPA1A]] heat shock protein family A (Hsp70) member 1A on 6p21.33: "This intronless gene encodes a 70kDa heat shock protein which is a member of the heat shock protein 70 family. In conjuction with other heat shock proteins, this protein stabilizes existing proteins against aggregation and mediates the folding of newly translated proteins in the cytosol and in organelles. It is also involved in the ubiquitin-proteasome pathway through interaction with the AU-rich element RNA-binding protein 1. The gene is located in the major histocompatibility complex class III region, in a cluster with two closely related genes which encode similar proteins."<ref name=RefSeq3303>{{ cite web | ||
Revision as of 03:12, 8 April 2020
Associate Editor(s)-in-Chief: Henry A. Hoff
The immunoglobulin supergene family is "the group of proteins that have immunoglobulin-like domains, including histocompatibility antigens, the T-cell antigen receptor, poly-IgR, and other proteins involved in the vertebrate immune response (17)."[1]
𝛂1B-glycoprotein
"𝛂1B-glycoprotein(𝛂1B) [...] consists of a single polypeptide chain N-linked to four glucosamine oligosaccharides. The polypeptide has five intrachain disulfide bonds and contains 474 amino acid residues. [...] 𝛂1B exhibits internal duplication and consists of five repeating structural domains, each containing about 95 amino acids and one disulfide bond. [...] several domains of 𝛂1B, especially the third, show statistically significant homology to variable regions of certain immunoglobulin light and heavy chains. 𝛂1B [...] exhibits sequence similarity to other members of the immunoglobulin supergene family such as the receptor for transepithelial transport of IgA and IgM and the secretory component of human IgA."[1]
"Some of the domains of 𝛂1B show significant homology to variable (V) and constant (C) regions of certain immunoglobulins. Likewise, there is statistically significant homology between 𝛂1B and the secretory component (SC) of human IgA (15) and also with the extracellular portion of the rabbit receptor for transepithelial transport of polymeric immunoglobulins (IgA and IgM). Mostov et al. (16) have called the later protein the poly-Ig receptor or poly-IgR and have shown that it is the precursor of SC."[1]
Human genes
Carcinoembryonic antigen genes
Gene ID: 634 is CEACAM1 CEA cell adhesion molecule 1 on 19q13.2: "This gene encodes a member of the carcinoembryonic antigen (CEA) gene family, which belongs to the immunoglobulin superfamily. Two subgroups of the CEA family, the CEA cell adhesion molecules and the pregnancy-specific glycoproteins, are located within a 1.2 Mb cluster on the long arm of chromosome 19. Eleven pseudogenes of the CEA cell adhesion molecule subgroup are also found in the cluster. The encoded protein was originally described in bile ducts of liver as biliary glycoprotein. Subsequently, it was found to be a cell-cell adhesion molecule detected on leukocytes, epithelia, and endothelia. The encoded protein mediates cell adhesion via homophilic as well as heterophilic binding to other proteins of the subgroup. Multiple cellular activities have been attributed to the encoded protein, including roles in the differentiation and arrangement of tissue three-dimensional structure, angiogenesis, apoptosis, tumor suppression, metastasis, and the modulation of innate and adaptive immune responses. Multiple transcript variants encoding different isoforms have been reported, but the full-length nature of all variants has not been defined."[2]
- NP_001020083.1 carcinoembryonic antigen-related cell adhesion molecule 1 isoform 2 precursor: "Transcript Variant: This variant (2) lacks an exon in the 3' coding region that results in a frameshift and an early stop codon, compared to variant 1. The resulting protein (isoform 2) has a distinct C-terminus and is shorter than isoform 1. This variant has been referred to by multiple names, including BGPc, transmembrane carcinoembryonic antigen 3, TM3-CEA, and CEACAM1-4S."[2]
- NP_001171742.1 carcinoembryonic antigen-related cell adhesion molecule 1 isoform 4 precursor: "Transcript Variant: This variant (4) lacks an alternate, in-frame, exon, compared to variant 1. The resulting protein (isoform 3) is shorter when it was compared to isoform 1."[2]
- NP_001171744.1 carcinoembryonic antigen-related cell adhesion molecule 1 isoform 3 precursor: "Transcript Variant: This variant (3) has one and lacks a different alternate, in-frame, segment, compared to variant 1. The resulting protein (isoform 3) is shorter when it was compared to isoform 1. This variant has been referred to as 'alternative spliced isoform 3S' and 'short form 3'."[2]
- NP_001171745.1 carcinoembryonic antigen-related cell adhesion molecule 1 isoform 5 precursor: "Transcript Variant: This variant (5) lacks two coding region segments, one of which shifts the reading frame, compared to variant 1. The resulting protein (isoform 5) has a shorter and distinct C-terminus when it is compared to isoform 1."[2]
- NP_001192273.1 carcinoembryonic antigen-related cell adhesion molecule 1 isoform 6 precursor: "Transcript Variant: This variant (6) lacks a segment, which results in a frameshift, compared to variant 1. The resulting protein (isoform 6) has a distinct C-terminus, compared to isoform 1."[2]
- NP_001703.2 carcinoembryonic antigen-related cell adhesion molecule 1 isoform 1 precursor: "Transcript Variant: This variant (1) represents the longest transcript and encodes the longest protein (isoform 1). This variant has been referred to by multiple names, including transmembrane carcinoembryonic antigen BGPa, TM1-CEA, and CEACAM1-4L."[2]
Major histocompatibility complex genes
Class I
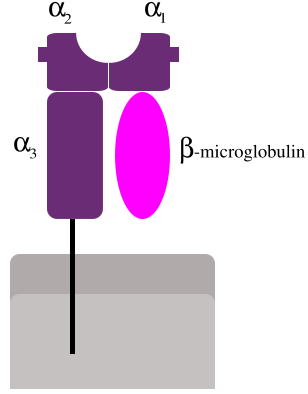
Gene ID: 563 is AZGP1 alpha-2-glycoprotein 1, zinc-binding, on 7q22.1.[3]
- NP_001176.1 zinc-alpha-2-glycoprotein precursor: "IgC_MHC_I_alpha3; Class I major histocompatibility complex (MHC) alpha chain immunoglobulin domain"[3]
Gene ID: 567 is B2M beta-2-microglobulin aka beta chain of MHC class I molecules on 15q21.1: "This gene encodes a serum protein found in association with the major histocompatibility complex (MHC) class I heavy chain on the surface of nearly all nucleated cells. The protein has a predominantly beta-pleated sheet structure that can form amyloid fibrils in some pathological conditions. The encoded antimicrobial protein displays antibacterial activity in amniotic fluid. A mutation in this gene has been shown to result in hypercatabolic hypoproteinemia."[4]
- NP_004039.1 beta-2-microglobulin precursor: "cd05770 Location:24 → 116 IgC_beta2m; Class I major histocompatibility complex (MHC) beta2-microglobulin"[4]
Gene ID: 821 is CANX calnexin aka major histocompatibility complex class I antigen-binding protein p88 on 5q35.3: "This gene encodes a member of the calnexin family of molecular chaperones. The encoded protein is a calcium-binding, endoplasmic reticulum (ER)-associated protein that interacts transiently with newly synthesized N-linked glycoproteins, facilitating protein folding and assembly. It may also play a central role in the quality control of protein folding by retaining incorrectly folded protein subunits within the ER for degradation. Alternatively spliced transcript variants encoding different isoforms have been described."[5]
- NP_001019820.1 calnexin isoform d precursor.[5]
- NP_001350922.1 calnexin isoform a: "Transcript Variant: This variant (3) represents the longest transcript and encodes the longest isoform (a)."[5]
- NP_001350923.1 calnexin isoform b.[5]
- NP_001350924.1 calnexin isoform c precursor.[5]
- NP_001350925.1 calnexin isoform d precursor.[5]
- NP_001350926.1 calnexin isoform d precursor.[5]
- NP_001350927.1 calnexin isoform e precursor.[5]
- NP_001350928.1 calnexin isoform f precursor.[5]
- NP_001350929.1 calnexin isoform g.[5]
- NP_001350930.1 calnexin isoform g.[5]
- NP_001737.1 calnexin isoform d precursor.[5]
- NR_157048.1 RNA Sequence.[5]
Gene ID: 909 is CD1A CD1a molecule on 1q23.1: "This gene encodes a member of the CD1 family of transmembrane glycoproteins, which are structurally related to the major histocompatibility complex (MHC) proteins and form heterodimers with beta-2-microglobulin. The CD1 proteins mediate the presentation of primarily lipid and glycolipid antigens of self or microbial origin to T cells. The human genome contains five CD1 family genes organized in a cluster on chromosome 1. The CD1 family members are thought to differ in their cellular localization and specificity for particular lipid ligands. The protein encoded by this gene localizes to the plasma membrane and to recycling vesicles of the early endocytic system. Alternative splicing results in multiple transcript variants."[6]
- NP_001307581.1 T-cell surface glycoprotein CD1a isoform 2: "Transcript Variant: This variant (2) contains an alternate exon in the 5' UTR, lacks a portion of the 5' coding region, and initiates translation at an alternate start codon, compared to variant 1. The encoded isoform (2) has a distinct N-terminus and is shorter than isoform 1."[6]
- NP_001754.2 T-cell surface glycoprotein CD1a isoform 1 precursor: "Transcript Variant: This variant (1) encodes the longer isoform (1)."[6]
Gene ID: 912 is CD1D CD1d molecule aka HMC class I antigen-like glycoprotein CD1D on 1q23.1: "This gene encodes a divergent member of the CD1 family of transmembrane glycoproteins, which are structurally related to the major histocompatibility complex (MHC) proteins and form heterodimers with beta-2-microglobulin. The CD1 proteins mediate the presentation of primarily lipid and glycolipid antigens of self or microbial origin to T cells. The human genome contains five CD1 family genes organized in a cluster on chromosome 1. The CD1 family members are thought to differ in their cellular localization and specificity for particular lipid ligands. The protein encoded by this gene localizes to late endosomes and lysosomes via a tyrosine-based motif in the cytoplasmic tail. Two transcript variants encoding different isoforms have been found for this gene."[7]
- NP_001306074.1 antigen-presenting glycoprotein CD1d isoform 2 precursor: "Transcript Variant: This variant (2) lacks an alternate in-frame exon and has a shorter 3' UTR compared to variant 1. The resulting isoform (2) has the same N- and C-termini but is shorter compared to isoform 1."[7]
- NP_001358690.1 antigen-presenting glycoprotein CD1d isoform 3.[7]
- NP_001358691.1 antigen-presenting glycoprotein CD1d isoform 1.[7]
- NP_001358692.1 antigen-presenting glycoprotein CD1d isoform 1.[7]
- NP_001757.1 antigen-presenting glycoprotein CD1d isoform 1 precursor: "Transcript Variant: This variant (1) represents the longer transcript and encodes the longer isoform (1)."[7]
Gene ID: 2217 is FCGRT Fc fragment of IgG receptor and transporter aka major histocompatibility complex class I-like Fc receptor on 19q13.33: "This gene encodes a receptor that binds the Fc region of monomeric immunoglobulin G. The encoded protein transfers immunoglobulin G antibodies from mother to fetus across the placenta. This protein also binds immunoglobulin G to protect the antibody from degradation. Alternative splicing results in multiple transcript variants."[8]
- NP_001129491.1 IgG receptor FcRn large subunit p51 precursor: "Transcript Variant: This variant (1) represents the longer transcript. Variants 1 and 2 encode the same protein."[8]
- NP_004098.1 IgG receptor FcRn large subunit p51 precursor: "Transcript Variant: This variant (2) has an alternate 5' UTR, compared to variant 1. Variants 1 and 2 encode the same protein."[8]
Gene ID: 3077 is HFE homeostatic iron regulator on 6p22.2: "The protein encoded by this gene is a membrane protein that is similar to MHC class I-type proteins and associates with beta2-microglobulin (beta2M). It is thought that this protein functions to regulate iron absorption by regulating the interaction of the transferrin receptor with transferrin. The iron storage disorder, hereditary haemochromatosis, is a recessive genetic disorder that results from defects in this gene. At least nine alternatively spliced variants have been described for this gene. Additional variants have been found but their full-length nature has not been determined."[9]
- NP_000401.1 hereditary hemochromatosis protein isoform 1 precursor: "Transcript Variant: This variant (1) encodes the longest isoform."[9]
- NP_001287678.1 hereditary hemochromatosis protein isoform 12 precursor: "Transcript Variant: This variant (12) uses an alternate splice acceptor site at its 3'-terminal exon, compared to variant 1. This variant encodes isoform 12 which has a shorter and distinct C-terminus, compared to isoform 1."[9]
- NP_620572.1 hereditary hemochromatosis protein isoform 3 precursor: "Transcript Variant: This variant (3) lacks an internal in-frame segment of the coding region, as compared to variant 1, resulting in a shorter protein (isoform 3)."[9]
- NP_620573.1 hereditary hemochromatosis protein isoform 4 precursor: "Transcript Variant: This variant (4) lacks an internal in-frame segment of the coding region, compared to variant 1, resulting in a shorter protein (isoform 4)."[9]
- NP_620575.1 hereditary hemochromatosis protein isoform 6 precursor: "Transcript Variant: This variant (6) lacks an internal in-frame segment of the coding region, as compared to variant 1, resulting in a shorter protein (isoform 6)."[9]
- NP_620576.1 hereditary hemochromatosis protein isoform 7 precursor: "Transcript Variant: This variant (7) lacks an internal in-frame segment of the coding region, as compared to variant 1, resulting in a shorter protein isoform (7)."[9]
- NP_620577.1 hereditary hemochromatosis protein isoform 8 precursor: "Transcript Variant: This variant (8) lacks two internal in-frame segments of the coding region, as compared to variant 1, resulting in a shorter protein (isoform 8)."[9]
- NP_620578.1 hereditary hemochromatosis protein isoform 9 precursor: "Transcript Variant: This variant (9) lacks an internal in-frame segment of the coding region through the use of an alternate splice acceptor site, as compared to variant 1, resulting in a shorter protein (isoform 9)."[9]
- NP_620579.1 hereditary hemochromatosis protein isoform 10 precursor: "Transcript Variant: This variant (10) lacks an internal in-frame segment of the coding region, as compared to variant 1, resulting in a shorter protein (isoform 10)."[9]
- NP_620580.1 hereditary hemochromatosis protein isoform 11 precursor: "Transcript Variant: This variant (11) lacks a large internal part of the coding region but the reading frame is maintained, as compared to variant 1. The protein encoded is the shortest isoform (11)."[9]
Gene ID: 3105 is HLA-A major histocompatibility complex, class I, A, on 6p22.1: "HLA-A belongs to the HLA class I heavy chain paralogues. This class I molecule is a heterodimer consisting of a heavy chain and a light chain (beta-2 microglobulin). The heavy chain is anchored in the membrane. Class I molecules play a central role in the immune system by presenting peptides derived from the endoplasmic reticulum lumen. They are expressed in nearly all cells. The heavy chain is approximately 45 kDa and its gene contains 8 exons. Exon 1 encodes the leader peptide, exons 2 and 3 encode the alpha1 and alpha2 domains, which both bind the peptide, exon 4 encodes the alpha3 domain, exon 5 encodes the transmembrane region, and exons 6 and 7 encode the cytoplasmic tail. Polymorphisms within exon 2 and exon 3 are responsible for the peptide binding specificity of each class one molecule. Typing for these polymorphisms is routinely done for bone marrow and kidney transplantation. Hundreds of HLA-A alleles have been described."[10]
- NP_001229687.1 HLA class I histocompatibility antigen, A alpha chain A*01:01:01:01 precursor: "Transcript Variant: This variant (2) represents the A*01:01:01:01 allele of the HLA-A gene, as found in the alternate locus group ALT_REF_LOCI_2 of the reference genome and in the RefSeqGene NG_029217."[10]
- NP_002107.3 HLA class I histocompatibility antigen, A alpha chain A*03:01:0:01 precursor: "Transcript Variant: This variant (1) represents the A*03:01:0:01 allele of the HLA-A gene, as found in the primary assembly of the reference genome."[10]
Gene ID: 3106 is HLA-B major histocompatibility complex, class I, B, on 6p21.33: "HLA-B belongs to the HLA class I heavy chain paralogues. This class I molecule is a heterodimer consisting of a heavy chain and a light chain (beta-2 microglobulin). The heavy chain is anchored in the membrane. Class I molecules play a central role in the immune system by presenting peptides derived from the endoplasmic reticulum lumen. They are expressed in nearly all cells. The heavy chain is approximately 45 kDa and its gene contains 8 exons. Exon 1 encodes the leader peptide, exon 2 and 3 encode the alpha1 and alpha2 domains, which both bind the peptide, exon 4 encodes the alpha3 domain, exon 5 encodes the transmembrane region and exons 6 and 7 encode the cytoplasmic tail. Polymorphisms within exon 2 and exon 3 are responsible for the peptide binding specificity of each class one molecule. Typing for these polymorphisms is routinely done for bone marrow and kidney transplantation. Hundreds of HLA-B alleles have been described."[11]
- NP_005505.2 major histocompatibility complex, class I, B precursor.[11]
Gene ID: 3107 is HLA-C major histocompatibility complex, class I, C, on 6p21.33: "HLA-C belongs to the HLA class I heavy chain paralogues. This class I molecule is a heterodimer consisting of a heavy chain and a light chain (beta-2 microglobulin). The heavy chain is anchored in the membrane. Class I molecules play a central role in the immune system by presenting peptides derived from endoplasmic reticulum lumen. They are expressed in nearly all cells. The heavy chain is approximately 45 kDa and its gene contains 8 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the alpha1 and alpha2 domain, which both bind the peptide, exon 4 encodes the alpha3 domain, exon 5 encodes the transmembrane region, and exons 6 and 7 encode the cytoplasmic tail. Polymorphisms within exon 2 and exon 3 are responsible for the peptide binding specificity of each class one molecule. Typing for these polymorphisms is routinely done for bone marrow and kidney transplantation. Over one hundred HLA-C alleles have been described."[12]
- NP_001229971.1 HLA class I histocompatibility antigen, C alpha chain precursor: "Transcript Variant: This variant (2) represents the C*07:01:01:01 allele of the HLA-C gene, as represented in the alternate locus group ALT_REF_LOCI_2 of the reference genome."[12]
- NP_002108.4 HLA class I histocompatibility antigen, C alpha chain precursor: "Transcript Variant: This variant (1) represents the C*07:02:01 allele of the HLA-C gene, as represented in the assembled chromosome 6 [6p21.33] in the primary assembly of the reference genome."[12]
Gene ID: 3133 is HLA-E major histocompatibility complex, class I, E, on 6p22.1: "HLA-E belongs to the HLA class I heavy chain paralogues. This class I molecule is a heterodimer consisting of a heavy chain and a light chain (beta-2 microglobulin). The heavy chain is anchored in the membrane. HLA-E binds a restricted subset of peptides derived from the leader peptides of other class I molecules. The heavy chain is approximately 45 kDa and its gene contains 8 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the alpha1 and alpha2 domains, which both bind the peptide, exon 4 encodes the alpha3 domain, exon 5 encodes the transmembrane region, and exons 6 and 7 encode the cytoplasmic tail."[13]
- NP_005507.3 HLA class I histocompatibility antigen, alpha chain E precursor.[13]
Gene ID: 3134 is HLA-F major histocompatibility complex, class I, F, on 6p22.1: "This gene belongs to the HLA class I heavy chain paralogues. It encodes a non-classical heavy chain that forms a heterodimer with a beta-2 microglobulin light chain, with the heavy chain anchored in the membrane. Unlike most other HLA heavy chains, this molecule is localized in the endoplasmic reticulum and Golgi apparatus, with a small amount present at the cell surface in some cell types. It contains a divergent peptide-binding groove, and is thought to bind a restricted subset of peptides for immune presentation. This gene exhibits few polymorphisms. Multiple transcript variants encoding different isoforms have been found for this gene. These variants lack a coding exon found in transcripts from other HLA paralogues due to an altered splice acceptor site, resulting in a shorter cytoplasmic domain."[14]
- NP_001091948.1 HLA class I histocompatibility antigen, alpha chain F isoform 3 precursor: "Transcript Variant: This variant (3) lacks an alternate in-frame coding exon and uses an alternate 3' exon, compared to variant 1. The resulting isoform (3) is shorter and has a distinct C-terminus, compared to isoform 1."[14]
- NP_001091949.1 HLA class I histocompatibility antigen, alpha chain F isoform 1 precursor: "Transcript Variant: This variant (1) represents the longest transcript and encodes the longest protein (isoform 1)."[14]
- NP_061823.2 HLA class I histocompatibility antigen, alpha chain F isoform 2 precursor: "Transcript Variant: This variant (2) uses an alternate 3' exon, compared to variant 1. The resulting isoform (2) has a shorter and distinct C-terminus, compared to isoform 1."[14]
Gene ID: 3135 is HLA-G major histocompatibility complex, class I, G, on 6p22.1: "HLA-G belongs to the HLA class I heavy chain paralogues. This class I molecule is a heterodimer consisting of a heavy chain and a light chain (beta-2 microglobulin). The heavy chain is anchored in the membrane. HLA-G is expressed on fetal derived placental cells. The heavy chain is approximately 45 kDa and its gene contains 8 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the alpha1 and alpha2 domain, which both bind the peptide, exon 4 encodes the alpha3 domain, exon 5 encodes the transmembrane region, and exon 6 encodes the cytoplasmic tail."[15]
- NP_001350496.1 HLA class I histocompatibility antigen, alpha chain G isoform 1 precursor: "Transcript Variant: This variant (1) encodes the longer isoform (1)."[15]
- NP_002118.1 HLA class I histocompatibility antigen, alpha chain G isoform 2 precursor: "Transcript Variant: This variant (2) differs in the 5' UTR and coding sequence compared to variant 1. The resulting isoform (2) is shorter at the N-terminus compared to isoform 1."[15]
Gene ID: 3140 is MR1 major histocompatibility complex, class I-related on 1q25.3: "MAIT (mucosal-associated invariant T-cells) lymphocytes represent a small population of T-cells primarily found in the gut. The protein encoded by this gene is an antigen-presenting molecule that presents metabolites of microbial vitamin B to MAITs. This presentation may activate the MAITs to regulate the amounts of specific types of bacteria in the gut. Several transcript variants encoding different isoforms have been found for this gene, and a pseudogene of it has been detected about 36 kbp upstream on the same chromosome."[16]
- NP_001181928.1 major histocompatibility complex class I-related gene protein isoform 2 precursor: "Transcript Variant: This variant (2) uses an alternate in-frame splice site in the coding region, compared to variant 1. This results in a shorter protein (isoform 2), compared to isoform 1."[16]
- NP_001181929.1 major histocompatibility complex class I-related gene protein isoform 3 precursor: "Transcript Variant: This variant (3) lacks an in-frame exon in the coding region, compared to variant 1. This results in a shorter protein (isoform 3), compared to isoform 1."[16]
- NP_001181964.1 major histocompatibility complex class I-related gene protein isoform 4 precursor: "Transcript Variant: This variant (4) lacks two consecutive in-frame exons in the coding region, compared to variant 1. This results in a shorter protein (isoform 4), compared to isoform 1."[16]
- NP_001297142.1 major histocompatibility complex class I-related gene protein isoform 5: "Transcript Variant: This variant (5) contains an alternate exon in the 5' end and lacks an alternate in-frame exon compared to variant 1, which results in translation initiation at a downstream start codon compared to variant 1. The encoded isoform (5) is shorter at the N-terminus and lacks an internal segment compared to isoform 1."[16]
- NP_001522.1 major histocompatibility complex class I-related gene protein isoform 1 precursor: "Transcript Variant: This variant (1) represents the longest transcript and encodes the longest isoform (1)."[16]
Gene ID: 3806 is KIR2DS1 killer cell immunoglobulin like receptor, two Ig domains and short cytoplasmic tail 1 aka MHC class I NK cell receptor Eb6 ActI on 19q13.4: "Killer cell immunoglobulin-like receptors (KIRs) are transmembrane glycoproteins expressed by natural killer cells and subsets of T cells. The KIR genes are polymorphic and highly homologous and they are found in a cluster on chromosome 19q13.4 within the 1 Mb leukocyte receptor complex (LRC). The gene content of the KIR gene cluster varies among haplotypes, although several "framework" genes are found in all haplotypes (KIR3DL3, KIR3DP1, KIR3DL4, KIR3DL2). The KIR proteins are classified by the number of extracellular immunoglobulin domains (2D or 3D) and by whether they have a long (L) or short (S) cytoplasmic domain. KIR proteins with the long cytoplasmic domain transduce inhibitory signals upon ligand binding via an immune tyrosine-based inhibitory motif (ITIM), while KIR proteins with the short cytoplasmic domain lack the ITIM motif and instead associate with the TYRO protein tyrosine kinase binding protein to transduce activating signals. The ligands for several KIR proteins are subsets of HLA class I molecules; thus, KIR proteins are thought to play an important role in regulation of the immune response."[17]
- NP_055327.1 killer cell immunoglobulin-like receptor 2DS1 precursor.[17]
Gene ID: 4277 is MICB MHC class I polypeptide-related sequence B on 6p21.33: "This gene encodes a heavily glycosylated protein which is a ligand for the NKG2D type II receptor. Binding of the ligand activates the cytolytic response of natural killer (NK) cells, CD8 alphabeta T cells, and gammadelta T cells which express the receptor. This protein is stress-induced and is similar to MHC class I molecules; however, it does not associate with beta-2-microglobulin or bind peptides. Alternative splicing results in multiple transcript variants."[18]
- NP_001276089.1 MHC class I polypeptide-related sequence B isoform 2: "Transcript Variant: This variant (2) differs in the 5' UTR, lacks a portion of the 5' coding region and initiates translation at a downstream start codon, compared to variant 1. It encodes isoform 2, which has a shorter N-terminus, compared to isoform 1."[18]
- NP_001276090.1 MHC class I polypeptide-related sequence B isoform 3: "Transcript Variant: This variant (3) contains an alternate in-frame splice site in the 5' coding region, compared to variant 1. It encodes isoform 3, which is shorter than isofom 1."[18]
- NP_005922.2 MHC class I polypeptide-related sequence B isoform 1 precursor: "Transcript Variant: This variant (1) represents the longest transcript and encodes the longest isoform (1)."[18]
Gene ID: 100507436 is MICA MHC class I polypeptide-related sequence A on 6p21.33: "This gene encodes the highly polymorphic major histocompatability complex class I chain-related protein A. The protein product is expressed on the cell surface, although unlike canonical class I molecules it does not seem to associate with beta-2-microglobulin. It is a ligand for the NKG2-D type II integral membrane protein receptor. The protein functions as a stress-induced antigen that is broadly recognized by intestinal epithelial gamma delta T cells. Variations in this gene have been associated with susceptibility to psoriasis 1 and psoriatic arthritis, and the shedding of MICA-related antibodies and ligands is involved in the progression from monoclonal gammopathy of undetermined significance to multiple myeloma. Alternative splicing of this gene results in multiple transcript variants."[19]
- NP_000238.1 MHC class I polypeptide-related sequence A isoform 1 (MICA*001) precursor: "Transcript Variant: This variant (1*001, also known as 1) is derived from the MICA*001 allele. It encodes the longest isoform (1). The MICA*001 allele is found in the c6_QBL (ALT_REF_LOCI_6) alternate assembly."[19]
- NP_001170990.1 MHC class I polypeptide-related sequence A isoform 2 (MICA*00801) precursor: "Transcript Variant: This variant (1*00801, also known as 1) is derived from the MICA*00801 allele. It contains a 4 nt insertion (rs9279200) that results in a frameshift and truncation of the CDS, compared to variant 1 (allele MICA*001). The resulting isoform (2) has a shorter and distinct C-terminus, compared to isoform 1. The MICA*00801 allele is found in the primary, ALT_REF_LOCI_2 and ALT_REF_LOCI_7 assembly units of the GRCh38 reference genome sequence."[19]
- NP_001276081.1 MHC class I polypeptide-related sequence A isoform 3 (MICA*00801): "Transcript Variant: This variant (3) contains an alternate 5' exon and it thus differs in the 5' UTR, lacks a portion of the 5' coding region, and initiates translation from a downstream in-frame start codon, compared to variant 1 (MICA*00801 allele). The encoded isoform (3) is shorter at the N-terminus, compared to isoform 2. This RefSeq represents the MICA*00801 allelic form of variant 3; the MICA*00801 allele is found in the primary, ALT_REF_LOCI_2 and ALT_REF_LOCI_7 assembly units of the GRCh38 reference genome sequence. Both variants 2 and 3 encode the same isoform."[19]
- NP_001276082.1 MHC class I polypeptide-related sequence A isoform 3 (MICA*00801): "Transcript Variant: This variant (2) contains an alternate 5' exon and it thus differs in the 5' UTR, lacks a portion of the 5' coding region, and initiates translation from a downstream in-frame start codon, compared to variant 1 (MICA*00801 allele). The encoded isoform (3) is shorter at the N-terminus, compared to isoform 2. This RefSeq represents the MICA*00801 allelic form of variant 2; the MICA*00801 allele is found in the primary, ALT_REF_LOCI_2 and ALT_REF_LOCI_7 assembly units of the GRCh38 reference genome sequence. Both variants 2 and 3 encode the same isoform."[19]
- NP_001276083.1 MHC class I polypeptide-related sequence A isoform 4 (MICA*00801): "Transcript Variant: This variant (4) contains an alternate 5' exon and uses an alternate splice site in an internal exon, and it thus differs in the 5' UTR, lacks a portion of the 5' coding region, and initiates translation from an alternate start codon, compared to variant 1 (MICA*00801 allele). The encoded isoform (4) has a distinct and shorter N-terminus, compared to isoform 2. This RefSeq represents the MICA*00801 allelic form of variant 4; the MICA*00801 allele is found in the primary, ALT_REF_LOCI_2 and ALT_REF_LOCI_7 assembly units of the GRCh38 reference genome sequence."[19]
Class II
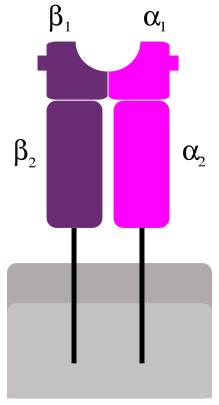
Gene ID: 972 is CD74 CD74 molecule, aka HLA class II histocompatibility antigen gamma chain, on 5q33.1: "The protein encoded by this gene associates with class II major histocompatibility complex (MHC) and is an important chaperone that regulates antigen presentation for immune response. It also serves as cell surface receptor for the cytokine macrophage migration inhibitory factor (MIF) which, when bound to the encoded protein, initiates survival pathways and cell proliferation. This protein also interacts with amyloid precursor protein (APP) and suppresses the production of amyloid beta (Abeta). Multiple alternatively spliced transcript variants encoding different isoforms have been identified."[20]
- NP_001020329.1 HLA class II histocompatibility antigen gamma chain isoform c: "Transcript Variant: This variant (3) lacks three consecutive exons in the 3' coding region, which results in a frame-shift, compared to variant 1. The resulting isoform (c) has a shorter and distinct C-terminus, compared to isoform a."[20]
- NP_001020330.1 HLA class II histocompatibility antigen gamma chain isoform a: "Transcript Variant: This variant (1) encodes the longest isoform (a)."[20]
- NP_001351012.1 HLA class II histocompatibility antigen gamma chain isoform d.[20]
- NP_001351013.1 HLA class II histocompatibility antigen gamma chain isoform e.[20]
- NP_004346.1 HLA class II histocompatibility antigen gamma chain isoform b: "Transcript Variant: This variant (2) lacks an in-frame exon in the 3' coding region, compared to variant 1. The resulting isoform (b) lacks an internal segment in the C-terminal region, compared to isoform a."[20]
- NR_157074.2 RNA Sequence.[20]
Gene ID: 2968 is GTF2H4 general transcription factor IIH subunit 4 on 6p21.33.[21]
Gene ID: 3108 is HLA-DMA major histocompatibility complex, class II, DM alpha, on 6p21.32: "HLA-DMA belongs to the HLA class II alpha chain paralogues. This class II molecule is a heterodimer consisting of an alpha (DMA) and a beta chain (DMB), both anchored in the membrane. It is located in intracellular vesicles. DM plays a central role in the peptide loading of MHC class II molecules by helping to release the CLIP molecule from the peptide binding site. Class II molecules are expressed in antigen presenting cells (APC: B lymphocytes, dendritic cells, macrophages). The alpha chain is approximately 33-35 kDa and its gene contains 5 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, exon 4 encodes the transmembrane domain and the cytoplasmic tail."[22]
- NP_006111.2 HLA class II histocompatibility antigen, DM alpha chain precursor.[22]
Gene ID: 3109 is HLA-DMB major histocompatibility complex, class II, DM beta, on 6p21.32: "HLA-DMB belongs to the HLA class II beta chain paralogues. This class II molecule is a heterodimer consisting of an alpha (DMA) and a beta (DMB) chain, both anchored in the membrane. It is located in intracellular vesicles. DM plays a central role in the peptide loading of MHC class II molecules by helping to release the CLIP (class II-associated invariant chain peptide) molecule from the peptide binding site. Class II molecules are expressed in antigen presenting cells (APC: B lymphocytes, dendritic cells, macrophages). The beta chain is approximately 26-28 kDa and its gene contains 6 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, exon 4 encodes the transmembrane domain and exon 5 encodes the cytoplasmic tail."[23]
- NP_002109.2 HLA class II histocompatibility antigen, DM beta chain precursor.[23]
Gene ID: 3111 is HLA-DOA major histocompatibility complex, class II, DO alpha, on 6p21.32: "HLA-DOA belongs to the HLA class II alpha chain paralogues. HLA-DOA forms a heterodimer with HLA-DOB. The heterodimer, HLA-DO, is found in lysosomes in B cells and regulates HLA-DM-mediated peptide loading on MHC class II molecules. In comparison with classical HLA class II molecules, this gene exhibits very little sequence variation, especially at the protein level."[24]
- NP_002110.1 HLA class II histocompatibility antigen, DO alpha chain precursor.[24]
Gene ID: 3112 is HLA-DOB major histocompatibility complex, class II, DO beta, on 6p21.32: "HLA-DOB belongs to the HLA class II beta chain paralogues. This class II molecule is a heterodimer consisting of an alpha (DOA) and a beta chain (DOB), both anchored in the membrane. It is located in intracellular vesicles. DO suppresses peptide loading of MHC class II molecules by inhibiting HLA-DM. Class II molecules are expressed in antigen presenting cells (APC: B lymphocytes, dendritic cells, macrophages). The beta chain is approximately 26-28 kDa and its gene contains 6 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, exon 4 encodes the transmembrane domain and exon 5 encodes the cytoplasmic tail."[25]
- NP_002111.1 HLA class II histocompatibility antigen, DO beta chain precursor.[25]
Gene ID: 3113 is HLA-DPA1 major histocompatibility complex, class II, DP alpha 1 on 6p21.32: "HLA-DPA1 belongs to the HLA class II alpha chain paralogues. This class II molecule is a heterodimer consisting of an alpha (DPA) and a beta (DPB) chain, both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells (APC: B lymphocytes, dendritic cells, macrophages). The alpha chain is approximately 33-35 kDa and its gene contains 5 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, exon 4 encodes the transmembrane domain and the cytoplasmic tail. Within the DP molecule both the alpha chain and the beta chain contain the polymorphisms specifying the peptide binding specificities, resulting in up to 4 different molecules."[26]
- NP_001229453.1 HLA class II histocompatibility antigen, DP alpha 1 chain precursor: "Transcript Variant: This variant (2) differs in the 5' UTR compared to variant 1. Variants 1, 2 and 3 encode the same protein."[26]
- NP_001229454.1 HLA class II histocompatibility antigen, DP alpha 1 chain precursor: "Transcript Variant: This variant (3) differs in the 5' UTR compared to variant 1. Variants 1, 2 and 3 encode the same protein."[26]
- NP_291032.2 HLA class II histocompatibility antigen, DP alpha 1 chain precursor: "Transcript Variant: This variant (1) represents the shortest transcript. Variants 1, 2 and 3 encode the same protein."[26]
Gene ID: 3115 is HLA-DPB1 major histocompatibility complex, class II, DP beta 1, on 6p21.32: "HLA-DPB belongs to the HLA class II beta chain paralogues. This class II molecule is a heterodimer consisting of an alpha (DPA) and a beta chain (DPB), both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells (APC: B lymphocytes, dendritic cells, macrophages). The beta chain is approximately 26-28 kDa and its gene contains 6 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, exon 4 encodes the transmembrane domain and exon 5 encodes the cytoplasmic tail. Within the DP molecule both the alpha chain and the beta chain contain the polymorphisms specifying the peptide binding specificities, resulting in up to 4 different molecules."[27]
- NP_002112.3 HLA class II histocompatibility antigen, DP beta 1 chain precursor.[27]
Gene ID: 3117 is HLA-DQA1 major histocompatibility complex, class II, DQ alpha 1, on 6p21.32: "HLA-DQA1 belongs to the HLA class II alpha chain paralogues. The class II molecule is a heterodimer consisting of an alpha (DQA) and a beta chain (DQB), both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells (APC: B Lymphocytes, dendritic cells, macrophages). The alpha chain is approximately 33-35 kDa. It is encoded by 5 exons; exon 1 encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, and exon 4 encodes the transmembrane domain and the cytoplasmic tail. Within the DQ molecule both the alpha chain and the beta chain contain the polymorphisms specifying the peptide binding specificities, resulting in up to four different molecules. Typing for these polymorphisms is routinely done for bone marrow transplantation."[28]
- NP_002113.2 HLA class II histocompatibility antigen, DQ alpha 1 chain precursor.[28]
Gene ID: 3118 is HLA-DQA2 major histocompatibility complex, class II, DQ alpha 2, on 6p21.32: "This gene belongs to the HLA class II alpha chain family. The encoded protein forms a heterodimer with a class II beta chain. It is located in intracellular vesicles and plays a central role in the peptide loading of MHC class II molecules by helping to release the CLIP molecule from the peptide binding site. Class II molecules are expressed in antigen presenting cells (B lymphocytes, dendritic cells, macrophages) and are used to present antigenic peptides on the cell surface to be recognized by CD4 T-cells."[29]
- NP_064440.1 HLA class II histocompatibility antigen, DQ alpha 2 chain precursor.[29]
Gene ID: 3119 is HLA-DQB1 major histocompatibility complex, class II, DQ beta 1, on 6p21.32: "HLA-DQB1 belongs to the HLA class II beta chain paralogs. This class II molecule is a heterodimer consisting of an alpha (DQA) and a beta chain (DQB), both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells (APC: B lymphocytes, dendritic cells, macrophages). The beta chain is approximately 26-28 kDa and it contains six exons. Exon 1 encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, exon 4 encodes the transmembrane domain and exon 5 encodes the cytoplasmic tail. Within the DQ molecule both the alpha chain and the beta chain contain the polymorphisms specifying the peptide binding specificities, resulting in up to four different molecules. Typing for these polymorphisms is routinely done for bone marrow transplantation. Alternative splicing results in multiple transcript variants."[30]
- NP_001230890.1 HLA class II histocompatibility antigen, DQ beta 1 chain isoform 2 precursor: "Transcript Variant: This variant (2) includes an alternate in-frame exon in the coding region, compared to variant 1. It encodes isoform 2 which is longer than isoform 1. This transcript represents the DQB1*06:02:01:01 allele of the HLA-DQB1 gene, as represented in the assembled chromosome 6 in the primary assembly of the reference genome."[30]
- NP_001230891.1 HLA class II histocompatibility antigen, DQ beta 1 chain isoform 1 precursor: "Transcript Variant: This variant (3) has the same exon combination as variant 1 but represents the DQB1*02:01:01:01 allele of the HLA-DQB1 gene, as represented in the alternate locus group ALT_REF_LOCI_2 of the reference genome. It encodes isoform 1."[30]
- NP_002114.3 HLA class II histocompatibility antigen, DQ beta 1 chain isoform 1 precursor: "Transcript Variant: This variant (1) is the predominant transcript and encodes isoform 1. This transcript represents the DQB1*06:02:01:01 allele of the HLA-DQB1 gene, as represented in the assembled chromosome 6 in the primary assembly of the reference genome."[30]
Gene ID: 3120 is HLA-DQB2 major histocompatibility complex, class II, DQ beta 2, on 6p21.32: "HLA-DQB2 belongs to the family of HLA class II beta chain paralogs. Class II molecules are heterodimers consisting of an alpha (DQA) and a beta chain (DQB), both anchored in the membrane. They play a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells (APC: B lymphocytes, dendritic cells, macrophages). Polymorphisms in the alpha and beta chains specify the peptide binding specificity, and typing for these polymorphisms is routinely done for bone marrow transplantation. However this gene, HLA-DQB2, is not routinely typed, as it is not thought to have an effect on transplantation. There is conflicting evidence in the literature and public sequence databases for the protein-coding capacity of HLA-DQB2. Because there is evidence of transcription and an intact ORF, HLA-DQB2 is represented in Entrez Gene and in RefSeq as a protein-coding locus."[31]
- NP_001185787.1 HLA class II histocompatibility antigen, DQ beta 2 chain isoform 2 precursor: "Transcript Variant: This variant (2) lacks an in-frame exon in the 3' coding region, compared to variant 1. It encodes a shorter isoform (2), compared to isoform 1."[31]
- NP_001287719.1 HLA class II histocompatibility antigen, DQ beta 2 chain isoform 1 precursor: "Transcript Variant: This variant (1) represents the longer transcript and encodes the longer isoform (1)."[31]
Gene ID: 3121 is HLA-DQB3 major histocompatibility complex, class II, DQ beta 3, on 6p21.3: "not in current annotation release"[32]
Gene ID: 3122 is HLA-DRA major histocompatibility complex, class II, DR alpha, on 6p21.32: "HLA-DRA is one of the HLA class II alpha chain paralogues. This class II molecule is a heterodimer consisting of an alpha and a beta chain, both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells (APC: B lymphocytes, dendritic cells, macrophages). The alpha chain is approximately 33-35 kDa and its gene contains 5 exons. Exon 1 encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, and exon 4 encodes the transmembrane domain and the cytoplasmic tail. DRA does not have polymorphisms in the peptide binding part and acts as the sole alpha chain for DRB1, DRB3, DRB4 and DRB5."[33]
- NP_061984.2 HLA class II histocompatibility antigen, DR alpha chain precursor.[33]
Gene ID: 3123 is HLA-DRB1 major histocompatibility complex, class II, DR beta 1, on 6p21.32: "HLA-DRB1 belongs to the HLA class II beta chain paralogs. The class II molecule is a heterodimer consisting of an alpha (DRA) and a beta chain (DRB), both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells. The beta chain is approximately 26-28 kDa. It is encoded by 6 exons. Exon one encodes the leader peptide; exons 2 and 3 encode the two extracellular domains; exon 4 encodes the transmembrane domain; and exon 5 encodes the cytoplasmic tail. Within the DR molecule the beta chain contains all the polymorphisms specifying the peptide binding specificities. Hundreds of DRB1 alleles have been described and some alleles have increased frequencies associated with certain diseases. There are multiple pseudogenes of this gene."[34]
- NP_001230894.1 major histocompatibility complex, class II, DR beta 1 precursor precursor: "Transcript Variant: This variant (2) represents the DRB1*03:01:01 allele of the HLA-DRB1 gene, as represented in the alternate locus groups ALT_REF_LOCI_2 and ALT_REF_LOCI_6 of the reference genome."[34]
- NP_001346122.1 major histocompatibility complex, class II, DR beta 1 precursor: "Transcript Variant: This variant (3) represents the DRB1*07:01:01 allele of the HLA-DRB1 gene, as represented in the alternate locus groups ALT_REF_LOCI_3 and ALT_REF_LOCI_4 of the reference genome."[34]
- NP_001346123.1 major histocompatibility complex, class II, DR beta 1 precursor: "Transcript Variant: This variant (4) represents the DRB1*04:03:01 allele of the HLA-DRB1 gene, as represented in the alternate locus group ALT_REF_LOCI_7 of the reference genome."[34]
- NP_002115.2 major histocompatibility complex, class II, DR beta 1 precursor precursor: "Transcript Variant: This variant (1) represents the DRB1*15:01:01 allele of the HLA-DRB1 gene, as represented in the assembled chromosome 6 in the primary assembly of the reference genome and the CHM1_1.1 genome."[34]
Gene ID: 3125 is HLA-DRB3 major histocompatibility complex, class II, DR beta 3, on 6p21.3: "HLA-DRB3 belongs to the HLA class II beta chain paralogues. This class II molecule is a heterodimer consisting of an alpha (DRA) and a beta (DRB) chain, both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells. The beta chain is approximately 26-28 kDa and its gene contains 6 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, exon 4 encodes the transmembrane domain and exon 5 encodes the cytoplasmic tail. Within the DR molecule the beta chain contains all the polymorphisms specifying the peptide binding specificities. Typing for these polymorphisms is routinely done for bone marrow and kidney transplantation. There are multiple pseudogenes of this gene."[35]
- NP_072049.2 major histocompatibility complex, class II, DR beta 3 precursor.[35]
Gene ID: 3126 is HLA-DRB4 major histocompatibility complex, class II, DR beta 4, on 6p21.3: "HLA-DRB4 belongs to the HLA class II beta chain paralogues. This class II molecule is a heterodimer consisting of an alpha (DRA) and a beta (DRB) chain, both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells. The beta chain is approximately 26-28 kDa and its gene contains 6 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, exon 4 encodes the transmembrane domain and exon 5 encodes the cytoplasmic tail. Within the DR molecule the beta chain contains all the polymorphisms specifying the peptide binding specificities. Typing for these polymorphisms is routinely done for bone marrow and kidney transplantation. There are multiple pseudogenes of this gene."[36]
- NP_068818.4 major histocompatibility complex, class II, DR beta 4 precursor.[36]
Gene ID: 3127 is HLA-DRB5 major histocompatibility complex, class II, DR beta 5, on 6p21.32: "HLA-DRB5 belongs to the HLA class II beta chain paralogues. This class II molecule is a heterodimer consisting of an alpha (DRA) and a beta (DRB) chain, both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells. The beta chain is approximately 26-28 kDa and its gene contains 6 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, exon 4 encodes the transmembrane domain and exon 5 encodes the cytoplasmic tail. Within the DR molecule the beta chain contains all the polymorphisms specifying the peptide binding specificities. Typing for these polymorphisms is routinely done for bone marrow and kidney transplantation. There are multiple pseudogenes of this gene."[37]
- NP_002116.2 major histocompatibility complex, class II, DR beta 5 precursor.[37]
Gene ID: 4261 is CIITA class II major histocompatibility complex transactivator on 16p13.13: "This gene encodes a protein with an acidic transcriptional activation domain, 4 LRRs (leucine-rich repeats) and a GTP binding domain. The protein is located in the nucleus and acts as a positive regulator of class II major histocompatibility complex gene transcription, and is referred to as the "master control factor" for the expression of these genes. The protein also binds GTP and uses GTP binding to facilitate its own transport into the nucleus. Once in the nucleus it does not bind DNA but rather uses an intrinsic acetyltransferase (AT) activity to act in a coactivator-like fashion. Mutations in this gene have been associated with bare lymphocyte syndrome type II (also known as hereditary MHC class II deficiency or HLA class II-deficient combined immunodeficiency), increased susceptibility to rheumatoid arthritis, multiple sclerosis, and possibly myocardial infarction. Several transcript variants encoding different isoforms have been found for this gene."[38]
- NP_000237.2 MHC class II transactivator isoform 2: "Transcript Variant: This variant (2) uses an alternate in-frame splice junction at the 5' end of an exon compared to variant 1. The resulting isoform (2) is 1 aa shorter compared to isoform 1."[38]
- NP_001273331.1 MHC class II transactivator isoform 1: "Transcript Variant: This variant (1) represents the longest transcript and encodes the longest isoform (1)."[38]
- NP_001273332.1 MHC class II transactivator isoform 3: "Transcript Variant: This variant (3) uses two alternate splice junctions and lacks two alternate coding exons compared to variant 1. The resulting isoform (3) has the same N- and C-termini but is shorter compared to isoform 1."[38]
- NR_104444.2 RNA Sequence: "Transcript Variant: This variant (4) uses an alternate splice junction and lacks an alternate exon compared to variant 1. This variant is represented as non-coding because the use of the 5'-most expected translational start codon, as used in variant 1, renders the transcript a candidate for nonsense-mediated mRNA decay (NMD)."[38]
Gene ID: 4904 is YBX1 Y-box binding protein 1 aka CCAAT-binding transcription factor I subunit A, DNA-binding protein B, Y-box transcription factor, enhancer factor I subunit A, major histocompatibility complex, class II, Y box-binding protein I, nuclease-sensitive element-binding protein 1 on 1p34.2: "This gene encodes a highly conserved cold shock domain protein that has broad nucleic acid binding properties. The encoded protein functions as both a DNA and RNA binding protein and has been implicated in numerous cellular processes including regulation of transcription and translation, pre-mRNA splicing, DNA reparation and mRNA packaging. This protein is also a component of messenger ribonucleoprotein (mRNP) complexes and may have a role in microRNA processing. This protein can be secreted through non-classical pathways and functions as an extracellular mitogen. Aberrant expression of the gene is associated with cancer proliferation in numerous tissues. This gene may be a prognostic marker for poor outcome and drug resistance in certain cancers. Alternate splicing results in multiple transcript variants. Pseudogenes of this gene are found on multiple chromosomes."[39]
- NP_004550.2 Y-box-binding protein 1: "Transcript Variant: This variant (1) represents the longer transcript and encodes the functional protein."[39]
- NR_132737.2 RNA Sequence: "Transcript Variant: This variant (2) contains an alternate 5' exon and lacks an internal exon, compared to variant 1. This variant is represented as non-coding because the predicted protein does not meet RefSeq quality criteria."[39]
Class III
MHC class III region encodes for other immune components, such as complement components (e.g., C2, C4, factor B) and some that encode cytokines (e.g., TNF-α) and also HSPs. They are mainly known from their genes because their gene cluster is present between those of class I and class II.[40] The gene cluster was discovered in between class I and class II genes on the short (p) arm of human chromosome 6. It was later found that it contains many genes for different signalling molecules such as tumour necrosis factors (TNFs) and heat shock proteins. More than 60 MHC class III genes are described, which is about 28% of the total MHC genes (224).[41]
MHC class III genes are located on chromosome 6 (6p21.3) in humans. It covers 700 kb and contains 61 genes. The gene cluster is the most gene-dense region of the human genome. They are basically similar with those of other animals. The functions of many genes are yet unknown.[42] Many retroelements such as human endogenous retrovirus (HERV) and Alu elements are located in the cluster.[43] The region containing genes G11/C4/Z/CYP21/X/Y, varying in size from 142 to 214 kb, is known as the most complex gene cluster in the human genome.[44]
MHC class III genes are similar in humans, mouse, frog (Xenopus tropicalis), and gray short-tailed opossum, but not all genes are common. For example, human NCR3, MIC and MCCD1 are absent in mouse. Human NCR3 and LST1 are absent in opossum.[45] However, birds (chicken and quail) have only a single gene, which codes for a complement component gene (C4).[46] In fishes, the genes are distributed in different chromosomes.[47]
Gene ID: 177 is AGER advanced glycosylation end-product specific receptor on 6p21.32: "The advanced glycosylation end product (AGE) receptor encoded by this gene is a member of the immunoglobulin superfamily of cell surface receptors. It is a multiligand receptor, and besides AGE, interacts with other molecules implicated in homeostasis, development, and inflammation, and certain diseases, such as diabetes and Alzheimer's disease. Many alternatively spliced transcript variants encoding different isoforms, as well as non-protein-coding variants, have been described for this gene (PMID:18089847)."[48]
- NP_001127.1 advanced glycosylation end product-specific receptor isoform 1 precursor: "Transcript Variant: This variant (1, also known as RAGE) represents the predominant transcript, and encodes isoform 1."[48]
- NP_001193858.1 advanced glycosylation end product-specific receptor isoform 2 precursor: "Transcript Variant: This variant (2, also known as RAGE_v5) uses an alternate in-frame donor splice site at an internal coding exon compared to variant 1. This results in a longer isoform (2) containing an additional protein segment compared to isoform 1."[48]
- NP_001193861.1 advanced glycosylation end product-specific receptor isoform 3 precursor: "Transcript Variant: This variant (3, also known as RAGE_v4) uses alternate in-frame acceptor and donor splice sites at two internal coding exons compared to variant 1. This results in a shorter isoform (2) missing two internal protein segments compared to isoform 1."[48]
- NP_001193863.1 advanced glycosylation end product-specific receptor isoform 4 precursor: "Transcript Variant: This variant (4, also known as RAGE_v6) lacks the penultimate coding exon, and uses alternate donor splice sites at two internal coding exons compared to variant 1. This results in a frame-shift and a shorter isoform (4) with a distinct C-terminus compared to isoform 1."[48]
- NP_001193865.1 advanced glycosylation end product-specific receptor isoform 5 precursor: "Transcript Variant: This variant (5, also known as RAGE_v9) lacks the penultimate coding exon, and uses alternate acceptor and donor splice sites at two internal coding exons compared to variant 1. This results in a frame-shift and a shorter isoform (5) with a distinct C-terminus compared to isoform 1."[48]
- NP_001193869.1 advanced glycosylation end product-specific receptor isoform 6 precursor: "Transcript Variant: This variant (6, also known as RAGE_v1) lacks the penultimate coding exon, and uses an alternate donor splice site at another coding exon compared to variant 1. This results in a frame-shift and a shorter isoform (6, also known as esRAGE and soluble RAGE) with a distinct C-terminus compared to isoform 1. This isoform lacks the transmembrane and intracellular domains, is secreted (PMID:18089847), and thought to function as a decoy receptor that inhibits RAGE signaling, and thus prevent the pathological progression of some pathologic conditions, such as Alzheimer's disease (PMID:18431028). Variants 6 and 9 encode the same isoform."[48]
- NP_001193883.1 advanced glycosylation end product-specific receptor isoform 8 precursor: "Transcript Variant: This variant (8, also known as RAGE_v8) lacks two internal coding exons, and uses an alternate donor splice site at another coding exon compared to variant 1. This results in a frame-shift, and a shorter isoform (8) with a distinct C-terminus compared to isoform 1."[48]
- NP_001193895.1 advanced glycosylation end product-specific receptor isoform 6 precursor: "Transcript Variant: This variant (9, also known as RAGE_v10) lacks the penultimate coding exon, and uses alternate splice sites at other exons at the 3' end compared to variant 1. This results in a frame-shift and a shorter isoform (6, also known as esRAGE and soluble RAGE) with a distinct C-terminus compared to isoform 1. Variants 6 and 9 encode the same isoform."[48]
- NP_751947.1 advanced glycosylation end product-specific receptor isoform 7 precursor: "Transcript Variant: This variant (7, also known as RAGE_v16) uses alternate splice sites at several internal coding exons compared to variant 1. This results in a frame-shift and a shorter isoform (7, also known as hRAGEsec) with a distinct C-terminus compared to isoform 1."[48]
Gene ID: 629 is CFB complement factor B on 6p21.33: "This gene encodes complement factor B, a component of the alternative pathway of complement activation. Factor B circulates in the blood as a single chain polypeptide. Upon activation of the alternative pathway, it is cleaved by complement factor D yielding the noncatalytic chain Ba and the catalytic subunit Bb. The active subunit Bb is a serine protease which associates with C3b to form the alternative pathway C3 convertase. Bb is involved in the proliferation of preactivated B lymphocytes, while Ba inhibits their proliferation. This gene localizes to the major histocompatibility complex (MHC) class III region on chromosome 6. This cluster includes several genes involved in regulation of the immune reaction. Polymorphisms in this gene are associated with a reduced risk of age-related macular degeneration. The polyadenylation site of this gene is 421 bp from the 5' end of the gene for complement component 2."[49]
- NP_001701.2 complement factor B preproprotein.[49]
Gene ID: 717 is C2 complement C2 on 6p21.33: "Component C2 is a serum glycoprotein that functions as part of the classical pathway of the complement system. Activated C1 cleaves C2 into C2a and C2b. The serine proteinase C2a then combines with complement factor 4b to create the C3 or C5 convertase. Deficiency of C2 has been reported to associated with certain autoimmune diseases and SNPs in this gene have been associated with altered susceptibility to age-related macular degeneration. This gene localizes within the class III region of the MHC on the short arm of chromosome 6. Alternative splicing results in multiple transcript variants encoding distinct isoforms. Additional transcript variants have been described in publications but their full-length sequence has not been determined."[50]
- NP_000054.2 complement C2 isoform 1 preproprotein: "Transcript Variant: This variant (1) encodes the longest isoform (1)."[50]
- NP_001139375.1 complement C2 isoform 2 precursor: "Transcript Variant: This variant (2) lacks two in-frame exons in the 5' coding region, compared to variant 1, that results in an isoform (2) with a shorter N-terminus that lacks one of two SUSHI repeat domains, compared to isoform 1."[50]
- NP_001171534.1 complement C2 isoform 3: "Transcript Variant: This variant (3) has an alternate 5' sequence and lacks an in-frame internal segment, as compared to variant 1. The resulting isoform (3) is shorter; it has a distinct N-terminus and lacks an internal segment, as compared to isoform 1."[50]
- NP_001269386.1 complement C2 isoform 4: "Transcript Variant: This variant (4) represents use of an alternate promoter and has multiple differences in the coding region compared to variant 1. The resulting protein (isoform 4) has a distinct N-terminus and is shorter than isoform 1."[50]
- NP_001269387.1 complement C2 isoform 5: "Transcript Variant: This variant (5) differs in the 5' UTR, lacks a portion of the 5' coding region, and initiates translation at an alternate start codon, compared to variant 1. The encoded isoform (5) has a shorter and distinct N-terminus compared to isoform 1."[50]
- NP_001269388.1 complement C2 isoform 6 precursor: "Transcript Variant: This variant (6) uses an alternate 3' exon structure, and thus differs in the 3' coding region and 3' UTR compared to variant 1. It encodes isoform 6 which is shorter and has a distinct C-terminus, compared to isoform 1."[50]
Gene ID: 720 is C4A complement C4A (Rodgers blood group) aka MHC class III region complement on 6p21.33: "This gene encodes the acidic form of complement factor 4, part of the classical activation pathway. The protein is expressed as a single chain precursor which is proteolytically cleaved into a trimer of alpha, beta, and gamma chains prior to secretion. The trimer provides a surface for interaction between the antigen-antibody complex and other complement components. The alpha chain is cleaved to release C4 anaphylatoxin, an antimicrobial peptide and a mediator of local inflammation. Deficiency of this protein is associated with systemic lupus erythematosus and type I diabetes mellitus. This gene localizes to the major histocompatibility complex (MHC) class III region on chromosome 6. Varying haplotypes of this gene cluster exist, such that individuals may have 1, 2, or 3 copies of this gene. Two transcript variants encoding different isoforms have been found for this gene."[51]
- NP_001239133.1 complement C4-A isoform 2 preproprotein: "Transcript Variant: This variant (2) lacks an alternate in-frame segment compared to variant 1. The resulting isoform (2) has the same N- and C-termini but is shorter compared to isoform 1."[51]
- NP_009224.2 complement C4-A isoform 1 preproprotein: "Transcript Variant: This variant (1) represents the longer transcript and encodes the longer isoform (1)."[51]
Gene ID: 721 is C4B complement C4B (Chido blood group) on 6p21.33: "This gene encodes the basic form of complement factor 4, part of the classical activation pathway. The protein is expressed as a single chain precursor which is proteolytically cleaved into a trimer of alpha, beta, and gamma chains prior to secretion. The trimer provides a surface for interaction between the antigen-antibody complex and other complement components. The alpha chain may be cleaved to release C4 anaphylatoxin, a mediator of local inflammation. Deficiency of this protein is associated with systemic lupus erythematosus. This gene localizes to the major histocompatibility complex (MHC) class III region on chromosome 6. Varying haplotypes of this gene cluster exist, such that individuals may have 1, 2, or 3 copies of this gene. In addition, this gene exists as a long form and a short form due to the presence or absence of a 6.4 kb endogenous HERV-K retrovirus in intron 9."[52]
- NP_001002029.3 complement C4-B preproprotein.[52]
Gene ID: 1432 is MAPK14 mitogen-activated protein kinase 14 on 6p21.31: "The protein encoded by this gene is a member of the MAP kinase family. MAP kinases act as an integration point for multiple biochemical signals, and are involved in a wide variety of cellular processes such as proliferation, differentiation, transcription regulation and development. This kinase is activated by various environmental stresses and proinflammatory cytokines. The activation requires its phosphorylation by MAP kinase kinases (MKKs), or its autophosphorylation triggered by the interaction of MAP3K7IP1/TAB1 protein with this kinase. The substrates of this kinase include transcription regulator ATF2, MEF2C, and MAX, cell cycle regulator CDC25B, and tumor suppressor p53, which suggest the roles of this kinase in stress related transcription and cell cycle regulation, as well as in genotoxic stress response. Four alternatively spliced transcript variants of this gene encoding distinct isoforms have been reported."[53]
- NP_001306.1 mitogen-activated protein kinase 14 isoform 1: "Transcript Variant: This variant (1) encodes the longest isoform (1) [...] STKc_p38alpha; Catalytic domain of the Serine/Threonine Kinase, p38alpha Mitogen-Activated Protein Kinase (also called MAPK14)."[53]
- NP_620581.1 mitogen-activated protein kinase 14 isoform 2: "Transcript Variant: This variant (2) contains a different segment within the coding region when compared to variant 1. The translation frame remains the same, and the resulting isoform 2 has an internal segment different from that of isoform 1."[53]
- NP_620582.1 mitogen-activated protein kinase 14 isoform 3: "Transcript Variant: This variant (3) contains a different internal segment within the coding region, and a different 3' coding region as well as a different 3' UTR, when compared to variant 1. It thus encodes an isoform that has a different internal segment, and a distinct C-terminus, as compared to isoform 1."[53]
- NP_620583.1 mitogen-activated protein kinase 14 isoform 4: "Transcript Variant: This variant (4) contains a different internal segment when compared to variant 1. It thus encodes an isoform that has a different and shorter internal segment, as compared to isoform 1."[53]
Gene ID: 1589 is CYP21A2 cytochrome P450 family 21 subfamily A member 2 on 6p21.33: "This gene encodes a member of the cytochrome P450 superfamily of enzymes. The cytochrome P450 proteins are monooxygenases which catalyze many reactions involved in drug metabolism and synthesis of cholesterol, steroids and other lipids. This protein localizes to the endoplasmic reticulum and hydroxylates steroids at the 21 position. Its activity is required for the synthesis of steroid hormones including cortisol and aldosterone. Mutations in this gene cause congenital adrenal hyperplasia. A related pseudogene is located near this gene; gene conversion events involving the functional gene and the pseudogene are thought to account for many cases of steroid 21-hydroxylase deficiency. Two transcript variants encoding different isoforms have been found for this gene."[54]
- NP_000491.4 steroid 21-hydroxylase isoform a: "Transcript Variant: This variant (1) encodes the longer isoform (a)."[54]
- NP_001122062.3 steroid 21-hydroxylase isoform b: "Transcript Variant: This variant (2) lacks an alternate in-frame exon compared to variant 1. The resulting isoform (b) has the same N- and C-termini but is shorter compared to isoform a."[54]
- NP_001355072.1 steroid 21-hydroxylase isoform c: "Transcript Variant: This variant (3), as well as variant 4, encodes isoform c."[54]
- NP_001355073.1 steroid 21-hydroxylase isoform c: "Transcript Variant: This variant (4), as well as variant 3, encodes isoform c."[54]
Gene ID: 4855 is NOTCH4 notch receptor 4 on 6p21.32: "This gene encodes a member of the NOTCH family of proteins. Members of this Type I transmembrane protein family share structural characteristics including an extracellular domain consisting of multiple epidermal growth factor-like (EGF) repeats, and an intracellular domain consisting of multiple different domain types. Notch signaling is an evolutionarily conserved intercellular signaling pathway that regulates interactions between physically adjacent cells through binding of Notch family receptors to their cognate ligands. The encoded preproprotein is proteolytically processed in the trans-Golgi network to generate two polypeptide chains that heterodimerize to form the mature cell-surface receptor. This receptor may play a role in vascular, renal and hepatic development. Mutations in this gene may be associated with schizophrenia. Alternative splicing results in multiple transcript variants, at least one of which encodes an isoform that is proteolytically processed."[55]
- NP_004548.3 neurogenic locus notch homolog protein 4 preproprotein: "Transcript Variant: This variant (1) represents the longest transcript and encodes the protein."[55]
Gene ID: 7148 is TNXB tenascin XB on 6p21.33-p21.32: "This gene encodes a member of the tenascin family of extracellular matrix glycoproteins. The tenascins have anti-adhesive effects, as opposed to fibronectin which is adhesive. This protein is thought to function in matrix maturation during wound healing, and its deficiency has been associated with the connective tissue disorder Ehlers-Danlos syndrome. This gene localizes to the major histocompatibility complex (MHC) class III region on chromosome 6. It is one of four genes in this cluster which have been duplicated. The duplicated copy of this gene is incomplete and is a pseudogene which is transcribed but does not encode a protein. The structure of this gene is unusual in that it overlaps the CREBL1 and CYP21A2 genes at its 5' and 3' ends, respectively. Multiple transcript variants encoding different isoforms have been found for this gene."[56]
- NP_001352205.1 tenascin-X isoform 3 precursor: "Transcript Variant: This variant (3) represents the longest transcript and encodes the longest isoform (3). It should be noted that the exon combination of this variant lacks full-length transcript support in human; it is predicted based on a combination of partial human and homologous transcript alignments."[56]
- NP_061978.6 tenascin-X isoform 1 precursor: "Transcript Variant: This variant (XB) uses an alternate in-frame splice junction compared to variant 3. The resulting isoform (1) has the same N- and C-termini but is shorter compared to isoform 3. It should be noted that the exon combination of this variant lacks full-length transcript support in human; it is predicted based on a combination of partial human and homologous transcript alignments."[56]
- NP_115859.2 tenascin-X isoform 2: "Transcript Variant: This variant (XB-S) is transcribed from a cryptic internal promoter sequence and is substantially shorter than variant 3 at the 5' end. It encodes isoform 2, which is identical to the C-terminus of the full-length protein, isoform 3."[56]
Gene ID: 7936 is NELFE negative elongation factor complex member E aka major histocompatibility complex gene RD on 6p21.33: "The protein encoded by this gene is part of a complex termed negative elongation factor (NELF) which represses RNA polymerase II transcript elongation. This protein bears similarity to nuclear RNA-binding proteins; however, it has not been demonstrated that this protein binds RNA. The protein contains a tract of alternating basic and acidic residues, largely arginine (R) and aspartic acid (D). The gene localizes to the major histocompatibility complex (MHC) class III region on chromosome 6."[57]
- NP_002895.3 negative elongation factor E.[57]
Gene ID: 8859 is STK19 serine/threonine kinase 19 aka MHC class III HLA-RP1 on 6p21.33: "This gene encodes a serine/threonine kinase which localizes predominantly to the nucleus. Its specific function is unknown; it is possible that phosphorylation of this protein is involved in transcriptional regulation. This gene localizes to the major histocompatibility complex (MHC) class III region on chromosome 6 and expresses two transcript variants."[58]
- NP_004188.1 serine/threonine-protein kinase 19 isoform 1: "Transcript Variant: This variant (1) uses an alternate splice site in the coding region, compared to variant 2. It encodes isoform 1 which is shorter compared to isoform 2. Although isoforms 1 and 2 differ in the kinase domain, it appears that there is no difference in kinase activity between isoforms 1 and 2."[58]
- NP_115830.1 serine/threonine-protein kinase 19 isoform 2: "Transcript Variant: This variant (2) represents the longest transcript and encodes the longest isoform (2). Although isoforms 1 and 2 differ in the kinase domain, it appears that there is no difference in kinase activity between isoforms 1 and 2."[58]
Gene ID: 259197 is NCR3 natural cytotoxicity triggering receptor 3 on 6p21.33: "The protein encoded by this gene is a natural cytotoxicity receptor (NCR) that may aid NK cells in the lysis of tumor cells. The encoded protein interacts with CD3-zeta (CD247), a T-cell receptor. A single nucleotide polymorphism in the 5' untranslated region of this gene has been associated with mild malaria suceptibility. Three transcript variants encoding different isoforms have been found for this gene."[59]
- NP_001138938.1 natural cytotoxicity triggering receptor 3 isoform b: "Transcript Variant: This variant (2) uses an alternate splice site in the 3' coding region compared to variant 1, that results in a frameshift. It encodes isoform b, which has a shorter and distinct C-terminus compared to isoform a."[59]
- NP_001138939.1 natural cytotoxicity triggering receptor 3 isoform c: "Transcript Variant: This variant (3) uses an alternate splice site in the 3' coding region compared to variant 1, that results in a frameshift. It encodes isoform c, which has a shorter and distinct C-terminus compared to isoform a."[59]
- NP_667341.1 natural cytotoxicity triggering receptor 3 isoform a precursor: "Transcript Variant: This variant (1) encodes the longest isoform (a). [...] Ig; Immunoglobulin domain".[59]
Class IV
Several "genes have been described that are encoded in the telomeric end of the Class III region and that appear to be involved in both global and specific inflammatory responses. Due to this commonality of function this gene-rich region was dubbed Class IV, and includes the TNF family, AIF1, and HSP70."[40]
"A cluster of genes for three related cytokines/cytokine receptors, tumor necrosis factor (TNF, formerly known as TNF-alpha or cachectin), lymphotoxin alpha (LTA), and lymphotoxin beta (LTB), lies in the Class IV region shortly before the most centromeric Class I related genes. TNF has been very extensively studied(5) and plays an important role in inflammation, bacterial(6) and viral infection,(7) tumor cachexia and the immune response. It is produced by a variety of cells including prominently monocytes, macrophages, and some T cell subsets."[40]
"LTB (also called TNF C) is a membrane bound molecule that forms a heterotrimer with LTA.(12) This LTA-LTB complex can then induce activation of NF kappa B in certain cell lines by binding with the LTB receptor, a member of the TNF receptor family.(13) (14) NF kappa B is a pleiotropic transcription factor capable of activating the expression of a great variety of genes critical for the Immunoin flammatory response.(14)"[40]
The B144/LST1 protein [...] is expressed in T cell, monocytic, and macrophage cell lines, and is also substantially expressed in both murine and human dendritic cells in culture."[40]
"The 1C7 gene [...] is located immediately adjacent to the B144 gene. RNA for B144 and 1C7 are transcribed in convergent directions such that there is a slight overlap between the 3' ends of the two mRNAs. [Human] 1C7 also shows multiple splice forms with 9 forms of the human mRNA reported so far.(21) The major forms encode proteins containing a leader sequence, a probable trans-membrane segment, an external sequence including an immunoglobulin-like domain, and at least three alternative forms of the putative intracellular segment of the protein. One alternative splice modifies the structure of the immunoglobulin-like domain, changing it from a sequence more closely resembling those of the V regions of Ig molecules to one that is more similar to IgC2 regions. Of the three alternative putative intracellular domains, one encodes multiple proline repeats suggestive of SH3 binding domains."[40]
"The existence of the G1 gene was initially noted as a part of a screen of MHC cosmids for embedded genes. The G1 and AIF1 transcripts appear to be derived by alternative splicing from partially overlapping genomic templates. A third human interferon gamma-responsive transcript, IRT-1, has been noted that shares some internal sequences with both G1 and AIF1, but on the basis of the predicted open reading frame it shares only limited amino acid sequences with G1."[40]
Gene ID: 199 is AIF1 allograft inflammatory factor 1 on 6p21.33: "This gene encodes a protein that binds actin and calcium. This gene is induced by cytokines and interferon and may promote macrophage activation and growth of vascular smooth muscle cells and T-lymphocytes. Polymorphisms in this gene may be associated with systemic sclerosis. Alternative splicing results in multiple transcript variants, but the full-length and coding nature of some of these variants is not certain."[60]
- NP_001305899.1 allograft inflammatory factor 1 isoform 1: "Transcript Variant: This variant (4) uses an alternate splice site in the 5' region and initiates translation at a downstream start codon compared to variant 3. The encoded isoform (1) has a shorter N-terminus than isoform 3. Variants 1 and 4 encode the same isoform (1)."[60]
- NP_001614.3 allograft inflammatory factor 1 isoform 3: "Transcript Variant: This variant (3) encodes the longest isoform (3)."[60]
- NP_116573.1 allograft inflammatory factor 1 isoform 1: "Transcript Variant: This variant (1, also known as G1) differs in the 5' UTR, lacks a portion of the 5' coding region, and initiates translation at a downstream start codon compared to variant 3. The encoded isoform (1) has a shorter N-terminus than isoform 3. Variants 1 and 4 encode the same isoform (1)."[60]
"AIF-1 (allograft inflammatory factor-1) is a Ca2+ binding protein predominantly expressed by activated monocytes, originally identified in rat cardiac allografts with chronic rejection.(22) The human cDNA homologue is 86% identical to the rat (90% identical to the amino acid sequence) and was identified by reverse transcriptase-PCR of endomyocardial biopsy specimens from human heart transplants and in macrophage cell lines.(23)"[40]
Gene ID: 3303 is HSPA1A heat shock protein family A (Hsp70) member 1A on 6p21.33: "This intronless gene encodes a 70kDa heat shock protein which is a member of the heat shock protein 70 family. In conjuction with other heat shock proteins, this protein stabilizes existing proteins against aggregation and mediates the folding of newly translated proteins in the cytosol and in organelles. It is also involved in the ubiquitin-proteasome pathway through interaction with the AU-rich element RNA-binding protein 1. The gene is located in the major histocompatibility complex class III region, in a cluster with two closely related genes which encode similar proteins."[61]
Gene ID: 3304 is HSPA1B heat shock protein family A (Hsp70) member 1B on 6p21.33: "This intronless gene encodes a 70kDa heat shock protein which is a member of the heat shock protein 70 family. In conjuction with other heat shock proteins, this protein stabilizes existing proteins against aggregation and mediates the folding of newly translated proteins in the cytosol and in organelles. It is also involved in the ubiquitin-proteasome pathway through interaction with the AU-rich element RNA-binding protein 1. The gene is located in the major histocompatibility complex class III region, in a cluster with two closely related genes which encode similar proteins."[62]
Gene ID: 3305 is HSPA1L heat shock protein family A (Hsp70) member 1 like on 6p21.33: "This gene encodes a 70kDa heat shock protein. In conjunction with other heat shock proteins, this protein stabilizes existing proteins against aggregation and mediates the folding of newly translated proteins in the cytosol and in organelles. The gene is located in the major histocompatibility complex class III region, in a cluster with two closely related genes which also encode isoforms of the 70kDa heat shock protein."[63]
Gene ID: 6892 is TAPBP TAP binding protein on 6p21.32: "This gene encodes a transmembrane glycoprotein which mediates interaction between newly assembled major histocompatibility complex (MHC) class I molecules and the transporter associated with antigen processing (TAP), which is required for the transport of antigenic peptides across the endoplasmic reticulum membrane. This interaction is essential for optimal peptide loading on the MHC class I molecule. Up to four complexes of MHC class I and this protein may be bound to a single TAP molecule. This protein contains a C-terminal double-lysine motif (KKKAE) known to maintain membrane proteins in the endoplasmic reticulum. This gene lies within the major histocompatibility complex on chromosome 6. Alternative splicing results in three transcript variants encoding different isoforms."[64]
- NP_003181.3 tapasin isoform 1 precursor: "Transcript Variant: This variant (1) represents the longest transcript and encodes isoform 1. [...] Ig; Immunoglobulin domain"[64]
- NP_757345.2 tapasin isoform 2 precursor: "Transcript Variant: This variant (2) differs in the 3' coding region and 3' UTR, compared to variant 1. The encoded isoform (2) has a distinct C-terminus and is longer than isoform 1."[64]
- NP_757346.2 tapasin isoform 3 precursor: "Transcript Variant: This variant (3) lacks an alternate in-frame exon in the central coding region, compared to variant 1, resulting in an isoform (3) that is shorter than isoform 1."[64]
Class V
Class VI
The region within the MHC class III gene cluster that contains genes for TNFs is also known as MHC class VI or the inflammatory region.[45]
Gene ID: 7124 is TNF tumor necrosis factor on 6p21.33: "This gene encodes a multifunctional proinflammatory cytokine that belongs to the tumor necrosis factor (TNF) superfamily. This cytokine is mainly secreted by macrophages. It can bind to, and thus functions through its receptors TNFRSF1A/TNFR1 and TNFRSF1B/TNFBR. This cytokine is involved in the regulation of a wide spectrum of biological processes including cell proliferation, differentiation, apoptosis, lipid metabolism, and coagulation. This cytokine has been implicated in a variety of diseases, including autoimmune diseases, insulin resistance, and cancer. Knockout studies in mice also suggested the neuroprotective function of this cytokine."[65]
ATP-binding cassette (ABC) transporters
Gene ID: 23 is ABCF1 ATP binding cassette subfamily F member 1, on 6p21.33: "The protein encoded by this gene is a member of the superfamily of ATP-binding cassette (ABC) transporters. ABC proteins transport various molecules across extra- and intra-cellular membranes. ABC genes are divided into seven distinct subfamilies (ABC1, MDR/TAP, MRP, ALD, OABP, GCN20, White). This protein is a member of the GCN20 subfamily. Unlike other members of the superfamily, this protein lacks the transmembrane domains which are characteristic of most ABC transporters. This protein may be regulated by tumor necrosis factor-alpha and play a role in enhancement of protein synthesis and the inflammation process."[66]
- NP_001020262.1 ATP-binding cassette sub-family F member 1 isoform a: "Transcript Variant: This variant (1) represents the longer transcript and encodes the longer isoform (a)."[66]
- NP_001081.1 ATP-binding cassette sub-family F member 1 isoform b: "Transcript Variant: This variant (2) lacks an alternate in-frame exon, compared to variant 1. The resulting protein (isoform b) is shorter than isoform a."[66]
Gene ID: 6890 is TAP1 transporter 1, ATP binding cassette subfamily B member aka transporter, ATP-binding cassette, major histocompatibility complex, 1 on 6p21.32: "The membrane-associated protein encoded by this gene is a member of the superfamily of ATP-binding cassette (ABC) transporters. ABC proteins transport various molecules across extra- and intra-cellular membranes. ABC genes are divided into seven distinct subfamilies (ABC1, MDR/TAP, MRP, ALD, OABP, GCN20, White). This protein is a member of the MDR/TAP subfamily. Members of the MDR/TAP subfamily are involved in multidrug resistance. The protein encoded by this gene is involved in the pumping of degraded cytosolic peptides across the endoplasmic reticulum into the membrane-bound compartment where class I molecules assemble. Mutations in this gene may be associated with ankylosing spondylitis, insulin-dependent diabetes mellitus, and celiac disease. Two transcript variants encoding different isoforms have been found for this gene."[67]
- NP_000584.3 antigen peptide transporter 1 isoform 1: "Transcript Variant: This variant (1) represents the longer transcript and encodes the longer isoform (1)."[67]
- NP_001278951.1 antigen peptide transporter 1 isoform 2: "Transcript Variant: This variant (2) differs in the 5' UTR and coding sequence compared to variant 1. The resulting isoform (2) is shorter at the N-terminus compared to isoform 1."[67]
Gene ID: 6891 is TAP2 transporter 2, ATP binding cassette subfamily B member on 6p21.32: "The membrane-associated protein encoded by this gene is a member of the superfamily of ATP-binding cassette (ABC) transporters. ABC proteins transport various molecules across extra- and intra-cellular membranes. ABC genes are divided into seven distinct subfamilies (ABC1, MDR/TAP, MRP, ALD, OABP, GCN20, White). This protein is a member of the MDR/TAP subfamily. Members of the MDR/TAP subfamily are involved in multidrug resistance. This gene is located 7 kb telomeric to gene family member ABCB2. The protein encoded by this gene is involved in antigen presentation. This protein forms a heterodimer with ABCB2 in order to transport peptides from the cytoplasm to the endoplasmic reticulum. Mutations in this gene may be associated with ankylosing spondylitis, insulin-dependent diabetes mellitus, and celiac disease. Alternative splicing of this gene produces products which differ in peptide selectivity and level of restoration of surface expression of MHC class I molecules."[68]
- NP_000535.3 antigen peptide transporter 2 isoform 1: "Transcript Variant: This variant (1, B allele) represents the longer transcript and encodes the longest isoform (1). An allele (variant 1, A allele) exists in which a single nt change creates an internal stop codon, leading to a protein that is 17 aa shorter at the C-terminus."[68]
- NP_001276972.1 antigen peptide transporter 2 isoform 3: "Transcript Variant: This variant (1, A allele) differs at 3 nt positions compared to variant 1, B allele. The resulting isoform (3) is shorter at the C-terminus compared to isoform 1."[68]
- NP_061313.2 antigen peptide transporter 2 isoform 2: "Transcript Variant: This variant (2) differs in the 5' UTR and coding region compared to variant 1. The resulting isoform (2) is shorter and has a distinct C-terminus compared to isoform 1."[68]
NF-kappa-B inhibitor family
Gene ID: 4792 is NFKBIA NFKB inhibitor alpha aka major histocompatibility complex enhancer-binding protein [mitotic arrest deficient 3] MAD3 on 14q13.2: "This gene encodes a member of the NF-kappa-B inhibitor family, which contain multiple ankrin repeat domains. The encoded protein interacts with REL dimers to inhibit NF-kappa-B/REL complexes which are involved in inflammatory responses. The encoded protein moves between the cytoplasm and the nucleus via a nuclear localization signal and CRM1-mediated nuclear export. Mutations in this gene have been found in ectodermal dysplasia anhidrotic with T-cell immunodeficiency autosomal dominant disease."[69]
ZAS family
Gene ID: 3096 is HIVEP1 HIVEP zinc finger 1 aka major histocompatibility complex binding protein 1 on 6p24.1: "This gene encodes a transcription factor belonging to the ZAS family, members of which are large proteins that contain a ZAS domain - a modular protein structure consisting of a pair of C2H2 zinc fingers with an acidic-rich region and a serine/threonine-rich sequence. These proteins bind specifically to the DNA sequence motif, GGGACTTTCC, found in the enhancer elements of several viral promoters, including human immunodeficiency virus (HIV), and to related sequences found in the enhancer elements of a number of cellular promoters. This protein binds to this sequence motif, suggesting a role in the transcriptional regulation of both viral and cellular genes."[70]
- NP_002105.3 zinc finger protein 40.[70]
Hypotheses
- Downstream core promoters may work as transcription factors even as their complements or inverses.
- In addition to the DNA binding sequences listed above, the transcription factors that can open up and attach through the local epigenome need to be known and specified.
See also
References
- ↑ 1.0 1.1 1.2 Noriaki Ishioka, Nobuhiro Takahashi, and Frank W. Putnam (April 1986). "Amino acid sequence of human plasma 𝛂1B-glycoprotein: Homology to the immunoglobulin supergene family" (PDF). Proceedings of the National Academy of Sciences USA. 83 (8): 2363–7. doi:10.1073/pnas.83.8.2363. PMID 3458201. Retrieved 9 March 2020.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 RefSeq (May 2010). "CEACAM1 CEA cell adhesion molecule 1 [ Homo sapiens (human) ]". U.S. National Library of Medicine, 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information. Retrieved 27 March 2020.
- ↑ 3.0 3.1 RefSeq (13 March 2020). "AZGP1 alpha-2-glycoprotein 1, zinc-binding [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 5 April 2020.
- ↑ 4.0 4.1 RefSeq (August 2014). "B2M beta-2-microglobulin [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 3 April 2020.
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 5.12 RefSeq (June 2018). "CANX calnexin [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 1 April 2020.
- ↑ 6.0 6.1 6.2 RefSeq (March 2016). "CD1A CD1a molecule [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 5 April 2020.
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 RefSeq (January 2016). "CD1D CD1d molecule [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 4 April 2020.
- ↑ 8.0 8.1 8.2 RefSeq (April 2009). "FCGRT Fc fragment of IgG receptor and transporter [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2 April 2020.
- ↑ 9.00 9.01 9.02 9.03 9.04 9.05 9.06 9.07 9.08 9.09 9.10 RefSeq (July 2008). "HFE homeostatic iron regulator [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2 April 2020.
- ↑ 10.0 10.1 10.2 RefSeq (July 2008). "HLA-A major histocompatibility complex, class I, A [ Homo sapiens (human) ]". U.S. National Library of Medicine, 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information. Retrieved 27 March 2020.
- ↑ 11.0 11.1 RefSeq (July 2008). "HLA-B major histocompatibility complex, class I, B [ Homo sapiens (human) ]". U.S. National Library of Medicine, 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information. Retrieved 27 March 2020.
- ↑ 12.0 12.1 12.2 RefSeq (July 2008). "HLA-C major histocompatibility complex, class I, C [ Homo sapiens (human) ]". U.S. National Library of Medicine, 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information. Retrieved 27 March 2020.
- ↑ 13.0 13.1 RefSeq (July 2008). "HLA-E major histocompatibility complex, class I, E [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 28 March 2020.
- ↑ 14.0 14.1 14.2 14.3 RefSeq (July 2008). "HLA-F major histocompatibility complex, class I, F [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 1 April 2020.
- ↑ 15.0 15.1 15.2 RefSeq (July 2008). "HLA-G major histocompatibility complex, class I, G [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 28 March 2020.
- ↑ 16.0 16.1 16.2 16.3 16.4 16.5 RefSeq (July 2015). "MR1 major histocompatibility complex, class I-related [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2 April 2020.
- ↑ 17.0 17.1 RefSeq (July 2008). "KIR2DS1 killer cell immunoglobulin like receptor, two Ig domains and short cytoplasmic tail 1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 6 April 2020.
- ↑ 18.0 18.1 18.2 18.3 RefSeq (January 2014). "MICB MHC class I polypeptide-related sequence B [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 5 April 2020.
- ↑ 19.0 19.1 19.2 19.3 19.4 19.5 RefSeq (January 2014). "MICA MHC class I polypeptide-related sequence A [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 28 March 2020.
- ↑ 20.0 20.1 20.2 20.3 20.4 20.5 20.6 RefSeq (August 2011). "CD74 CD74 molecule [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 1 April 2020.
- ↑ RefSeq (13 March 2020). "GTF2H4 general transcription factor IIH subunit 4 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 6 April 2020.
- ↑ 22.0 22.1 RefSeq (July 2008). "HLA-DMA major histocompatibility complex, class II, DM alpha [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2 April 2020.
- ↑ 23.0 23.1 RefSeq (July 2008). "HLA-DMB major histocompatibility complex, class II, DM beta [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2 April 2020.
- ↑ 24.0 24.1 RefSeq (July 2008). "HLA-DOA major histocompatibility complex, class II, DO alpha [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2 April 2020.
- ↑ 25.0 25.1 RefSeq (July 2008). "HLA-DOB major histocompatibility complex, class II, DO beta [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2 April 2020.
- ↑ 26.0 26.1 26.2 26.3 RefSeq (July 2008). "HLA-DPA1 major histocompatibility complex, class II, DP alpha 1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 1 April 2020.
- ↑ 27.0 27.1 RefSeq (July 2008). "HLA-DPB1 major histocompatibility complex, class II, DP beta 1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 28 March 2020.
- ↑ 28.0 28.1 RefSeq (July 2008). "HLA-DQA1 major histocompatibility complex, class II, DQ alpha 1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 28 March 2020.
- ↑ 29.0 29.1 RefSeq (June 2010). "HLA-DQA2 major histocompatibility complex, class II, DQ alpha 2 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2 April 2020.
- ↑ 30.0 30.1 30.2 30.3 RefSeq (September 2011). "HLA-DQB1 major histocompatibility complex, class II, DQ beta 1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 28 March 2020.
- ↑ 31.0 31.1 31.2 RefSeq (October 2010). "HLA-DQB2 major histocompatibility complex, class II, DQ beta 2 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2 April 2020.
- ↑ RefSeq (24 March 2019). "HLA-DQB3 major histocompatibility complex, class II, DQ beta 3 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 5 April 2020.
- ↑ 33.0 33.1 RefSeq (July 2008). "HLA-DRA major histocompatibility complex, class II, DR alpha [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 30 March 2020.
- ↑ 34.0 34.1 34.2 34.3 34.4 RefSeq (February 2020). "HLA-DRB1 major histocompatibility complex, class II, DR beta 1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 28 March 2020.
- ↑ 35.0 35.1 RefSeq (February 2020). "HLA-DRB3 major histocompatibility complex, class II, DR beta 3 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 30 March 2020.
- ↑ 36.0 36.1 RefSeq (February 2020). "HLA-DRB4 major histocompatibility complex, class II, DR beta 4 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 28 March 2020.
- ↑ 37.0 37.1 RefSeq (February 2020). "HLA-DRB5 major histocompatibility complex, class II, DR beta 5 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 30 March 2020.
- ↑ 38.0 38.1 38.2 38.3 38.4 RefSeq (November 2013). "CIITA class II major histocompatibility complex transactivator [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 30 March 2020.
- ↑ 39.0 39.1 39.2 RefSeq (September 2015). "YBX1 Y-box binding protein 1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 19 November 2018.
- ↑ 40.0 40.1 40.2 40.3 40.4 40.5 40.6 40.7 Gruen, JR; Weissman, SM (2001). "Human MHC class III and IV genes and disease associations". Frontiers in Bioscience. 6 (3): D960–172. doi:10.2741/A658. PMID 11487469.
- ↑ The MHC sequencing consortium (1999). "Complete sequence and gene map of a human major histocompatibility complex". Nature. 401 (6756): 921–923. Bibcode:1999Natur.401..921T. doi:10.1038/44853. PMID 10553908.
- ↑ Xie, T; Rowen, L; Aguado, B; Ahearn, ME; Madan, A; Qin, S; Campbell, RD; Hood, L (2003). "Analysis of the gene-dense major histocompatibility complex class III region and its comparison to mouse". Genome Research. 13 (12): 2621–36. doi:10.1101/gr.1736803. PMC 403804. PMID 14656967.
- ↑ Dawkins, R; Leelayuwat, C; Gaudieri, S; Tay, G; Hui, J; Cattley, S; Martinez, P; Kulski, J (1999). "Genomics of the major histocompatibility complex: haplotypes, duplication, retroviruses and disease". Immunological Reviews. 167: 275–304. doi:10.1111/j.1600-065X.1999.tb01399.x. PMID 10319268.
- ↑ Milner, CM; Campbell, RD (2001). "Genetic organization of the human MHC class III region". Frontiers in Bioscience. 6 (3): D914–926. doi:10.2741/A653. PMID 11487476.
- ↑ 45.0 45.1 Deakin, Janine E; Papenfuss, Anthony T; Belov, Katherine; Cross, Joseph GR; Coggill, Penny; Palmer, Sophie; Sims, Sarah; Speed, Terence P; Beck, Stephan; Graves, Jennifer (2006). "Evolution and comparative analysis of the MHC Class III inflammatory region". BMC Genomics. 7 (1): 281. doi:10.1186/1471-2164-7-281. PMC 1654159. PMID 17081307.
- ↑ Shiina, T; Shimizu, S; Hosomichi, K; Kohara, S; Watanabe, S; Hanzawa, K; Beck, S; Kulski, JK; Inoko, H (2004). "Comparative genomic analysis of two avian (quail and chicken) MHC regions". Journal of Immunology. 172 (11): 6751–63. doi:10.4049/jimmunol.172.11.6751. PMID 15153492.
- ↑ Sambrook, JG; Figueroa, F; Beck, S (2005). "A genome-wide survey of Major Histocompatibility Complex (MHC) genes and their paralogues in zebrafish". BMC Genomics. 6: 152. doi:10.1186/1471-2164-6-152. PMC 1309616. PMID 16271140.
- ↑ 48.0 48.1 48.2 48.3 48.4 48.5 48.6 48.7 48.8 48.9 RefSeq (May 2011). "AGER advanced glycosylation end-product specific receptor [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 7 April 2020.
- ↑ 49.0 49.1 RefSeq (July 2008). "CFB complement factor B [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 6 April 2020.
- ↑ 50.0 50.1 50.2 50.3 50.4 50.5 50.6 RefSeq (March 2009). "C2 complement C2 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 6 April 2020.
- ↑ 51.0 51.1 51.2 RefSeq (November 2014). "C4A complement C4A (Rodgers blood group) [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 6 April 2020.
- ↑ 52.0 52.1 RefSeq (July 2008). "C4B complement C4B (Chido blood group) [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 5 April 2020.
- ↑ 53.0 53.1 53.2 53.3 53.4 RefSeq (July 2008). "MAPK14 mitogen-activated protein kinase 14 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 6 April 2020.
- ↑ 54.0 54.1 54.2 54.3 54.4 RefSeq (July 2008). "CYP21A2 cytochrome P450 family 21 subfamily A member 2 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 6 April 2020.
- ↑ 55.0 55.1 RefSeq (January 2016). "NOTCH4 notch receptor 4 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 6 April 2020.
- ↑ 56.0 56.1 56.2 56.3 RefSeq (July 2008). "TNXB tenascin XB [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 6 April 2020.
- ↑ 57.0 57.1 RefSeq (July 2008). "NELFE negative elongation factor complex member E [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2 April 2020.
- ↑ 58.0 58.1 58.2 RefSeq (July 2008). "STK19 serine/threonine kinase 19 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 6 April 2020.
- ↑ 59.0 59.1 59.2 59.3 RefSeq (May 2010). "NCR3 natural cytotoxicity triggering receptor 3 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 6 April 2020.
- ↑ 60.0 60.1 60.2 60.3 RefSeq (January 2016). "AIF1 allograft inflammatory factor 1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 7 April 2020.
- ↑ RefSeq (July 2008). "HSPA1A heat shock protein family A (Hsp70) member 1A [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 4 April 2020.
- ↑ RefSeq (July 2008). "HSPA1B heat shock protein family A (Hsp70) member 1B [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 5 April 2020.
- ↑ RefSeq (July 2008). "HSPA1L heat shock protein family A (Hsp70) member 1 like [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 6 April 2020.
- ↑ 64.0 64.1 64.2 64.3 RefSeq (July 2008). "TAPBP TAP binding protein [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 6 April 2020.
- ↑ RefSeq (July 2008). "TNF tumor necrosis factor [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 6 April 2020.
- ↑ 66.0 66.1 66.2 RefSeq (July 2008). "ABCF1 ATP binding cassette subfamily F member 1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 7 April 2020.
- ↑ 67.0 67.1 67.2 RefSeq (May 2014). "TAP1 transporter 1, ATP binding cassette subfamily B member [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 1 April 2020.
- ↑ 68.0 68.1 68.2 68.3 RefSeq (February 2014). "TAP2 transporter 2, ATP binding cassette subfamily B member [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 6 April 2020.
- ↑ RefSeq (August 2011). "NFKBIA NFKB inhibitor alpha [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 28 March 2020.
- ↑ 70.0 70.1 RefSeq (October 2011). "HIVEP1 HIVEP zinc finger 1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 3 April 2020.