Major histocompatibility complex class II gene family
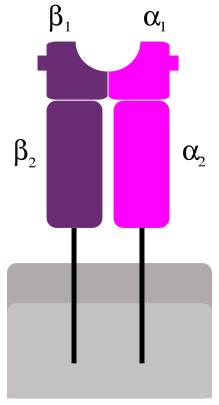

MHC Class II molecules are found only on a few specialized cell types, including macrophages, dendritic cells and B cells, all of which are professional antigen-presenting cells (APCs).
The peptides presented by class II molecules are derived from extracellular proteins (not cytosolic as in class I); hence, the MHC class II-dependent pathway of antigen presentation is called the endocytic or exogenous pathway.
Loading of class II molecules must still occur inside the cell; extracellular proteins are endocytosed, digested in lysosomes, and bound by the class II MHC molecule prior to the molecule's migration to the plasma membrane.
Gene ID: 972 is CD74 CD74 molecule, aka HLA class II histocompatibility antigen gamma chain, on 5q33.1: "The protein encoded by this gene associates with class II major histocompatibility complex (MHC) and is an important chaperone that regulates antigen presentation for immune response. It also serves as cell surface receptor for the cytokine macrophage migration inhibitory factor (MIF) which, when bound to the encoded protein, initiates survival pathways and cell proliferation. This protein also interacts with amyloid precursor protein (APP) and suppresses the production of amyloid beta (Abeta). Multiple alternatively spliced transcript variants encoding different isoforms have been identified."[1]
- NP_001020329.1 HLA class II histocompatibility antigen gamma chain isoform c: "Transcript Variant: This variant (3) lacks three consecutive exons in the 3' coding region, which results in a frame-shift, compared to variant 1. The resulting isoform (c) has a shorter and distinct C-terminus, compared to isoform a."[1]
- NP_001020330.1 HLA class II histocompatibility antigen gamma chain isoform a: "Transcript Variant: This variant (1) encodes the longest isoform (a)."[1]
- NP_001351012.1 HLA class II histocompatibility antigen gamma chain isoform d.[1]
- NP_001351013.1 HLA class II histocompatibility antigen gamma chain isoform e.[1]
- NP_004346.1 HLA class II histocompatibility antigen gamma chain isoform b: "Transcript Variant: This variant (2) lacks an in-frame exon in the 3' coding region, compared to variant 1. The resulting isoform (b) lacks an internal segment in the C-terminal region, compared to isoform a."[1]
- NR_157074.2 RNA Sequence.[1]
Gene ID: 1302 is COL11A2 collagen type XI alpha 2 chain on 6p21.32: "This gene encodes one of the two alpha chains of type XI collagen, a minor fibrillar collagen. It is located on chromosome 6 very close to but separate from the gene for retinoid X receptor beta. Type XI collagen is a heterotrimer but the third alpha chain is a post-translationally modified alpha 1 type II chain. Proteolytic processing of this type XI chain produces PARP, a proline/arginine-rich protein that is an amino terminal domain. Mutations in this gene are associated with type III Stickler syndrome, otospondylomegaepiphyseal dysplasia (OSMED syndrome), Weissenbacher-Zweymuller syndrome, autosomal dominant non-syndromic sensorineural type 13 deafness (DFNA13), and autosomal recessive non-syndromic sensorineural type 53 deafness (DFNB53). Alternative splicing results in multiple transcript variants. A related pseudogene is located nearby on chromosome 6."[2]
- NP_001157243.1 collagen alpha-2(XI) chain isoform 4 precursor: "Transcript Variant: This variant (4) lacks several exons at the 3' end and has a distinct 3' coding region and 3' UTR, compared to variant 1. The encoded isoform (4) has a distinct C-terminus and is considerably shorter than isoform 1."[2]
- NP_542410.2 collagen alpha-2(XI) chain isoform 3 preproprotein: "Transcript Variant: This variant (3) lacks three alternate in-frame exons, compared to variant 1, resulting in a shorter protein (isoform 3), compared to isoform 1. There are no publicly available human transcripts representing the full-length exon combination of this variant, but the region of variation is supported by experimental data in PMIDs 8663204 and 8838804, by homology data in mouse, and the full-length sequence is represented in accession U32169.1:AAC50215.1."[2]
- NP_542411.2 collagen alpha-2(XI) chain isoform 1 preproprotein: "Transcript Variant: This variant (1) represents the longest transcript and encodes the longest isoform (1). There are no publicly available human transcripts representing the full-length exon combination of this variant, but the region of variation is supported by experimental data in PMIDs 8663204 and 8838804, by homology data in mouse, and the full-length sequence is represented in accession U32169.1:AAC50214.1."[2]
- NP_542412.2 collagen alpha-2(XI) chain isoform 2 preproprotein: "Transcript Variant: This variant (2) lacks two alternate in-frame exons, compared to variant 1, resulting in a shorter protein (isoform 2), compared to isoform 1. There are no publicly available human transcripts representing the full-length exon combination of this variant, but the region of variation is supported by experimental data in PMIDs 8663204 and 8838804, by homology data in mouse, and the full-length sequence is represented in accession U32169.1:AAC50213.1."[2]
Gene ID: 1388 is ATF6B activating transcription factor 6 beta on 6p21.32: "The protein encoded by this gene is a transcription factor in the unfolded protein response (UPR) pathway during ER stress. Either as a homodimer or as a heterodimer with ATF6-alpha, the encoded protein binds to the ER stress response element, interacting with nuclear transcription factor Y to activate UPR target genes. The protein is normally found in the membrane of the endoplasmic reticulum; however, under ER stress, the N-terminal cytoplasmic domain is cleaved from the rest of the protein and translocates to the nucleus. Two transcript variants encoding different isoforms have been found for this gene."[3]
- NP_001129625.1 cyclic AMP-dependent transcription factor ATF-6 beta isoform b: "Transcript Variant: This variant (2) uses an alternate in-frame splice site at the 3' end of exon 1 compared to variant 1. The resulting isoform (b) has the same N- and C-termini but is 3 aa shorter compared to isoform a."[3]
- NP_004372.3 cyclic AMP-dependent transcription factor ATF-6 beta isoform a: "Transcript Variant: This variant (1) represents the longer transcript and encodes the longer isoform (a)."[3]
Gene ID: 1616 is DAXX death domain associated protein on 6p21.32: "This gene encodes a multifunctional protein that resides in multiple locations in the nucleus and in the cytoplasm. It interacts with a wide variety of proteins, such as apoptosis antigen Fas, centromere protein C, and transcription factor erythroblastosis virus E26 oncogene homolog 1. In the nucleus, the encoded protein functions as a potent transcription repressor that binds to sumoylated transcription factors. Its repression can be relieved by the sequestration of this protein into promyelocytic leukemia nuclear bodies or nucleoli. This protein also associates with centromeres in G2 phase. In the cytoplasm, the encoded protein may function to regulate apoptosis. The subcellular localization and function of this protein are modulated by post-translational modifications, including sumoylation, phosphorylation and polyubiquitination. Alternative splicing results in multiple transcript variants."[4]
- NP_001135441.1 death domain-associated protein 6 isoform a: "Transcript Variant: This variant (1) represents the longest transcript but encodes the shorter isoform (a). Variants 1 and 2 encode the same isoform (a)."[4]
- NP_001135442.1 death domain-associated protein 6 isoform b: "Transcript Variant: This variant (3) lacks an in-frame segment in a 5' coding exon and uses an upstream start codon, compared to variant 1. The resulting protein (isoform b) has a distinct and longer N-terminus, compared to isoform a."[4]
- NP_001241646.1 death domain-associated protein 6 isoform c: "Transcript Variant: This variant (4) differs in the 5' UTR and lacks an exon in the coding region, compared to variant 1. These differences cause translation initiation at a downstream AUG and result in an isoform (c) with a shorter N-terminus, compared to isoform a."[4]
- NP_001341.1 death domain-associated protein 6 isoform a: "Transcript Variant: This variant (2) uses an alternate splice site in the 5' UTR compared to variant 1. Variants 1 and 2 encode the same isoform (a)."[4]
Gene ID: 2968 is GTF2H4 general transcription factor IIH subunit 4 on 6p21.33.[5]
Gene ID: 3108 is HLA-DMA major histocompatibility complex, class II, DM alpha, on 6p21.32: "HLA-DMA belongs to the HLA class II alpha chain paralogues. This class II molecule is a heterodimer consisting of an alpha (DMA) and a beta chain (DMB), both anchored in the membrane. It is located in intracellular vesicles. DM plays a central role in the peptide loading of MHC class II molecules by helping to release the CLIP molecule from the peptide binding site. Class II molecules are expressed in antigen presenting cells (APC: B lymphocytes, dendritic cells, macrophages). The alpha chain is approximately 33-35 kDa and its gene contains 5 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, exon 4 encodes the transmembrane domain and the cytoplasmic tail."[6]
- NP_006111.2 HLA class II histocompatibility antigen, DM alpha chain precursor.[6]
Gene ID: 3109 is HLA-DMB major histocompatibility complex, class II, DM beta, on 6p21.32: "HLA-DMB belongs to the HLA class II beta chain paralogues. This class II molecule is a heterodimer consisting of an alpha (DMA) and a beta (DMB) chain, both anchored in the membrane. It is located in intracellular vesicles. DM plays a central role in the peptide loading of MHC class II molecules by helping to release the CLIP (class II-associated invariant chain peptide) molecule from the peptide binding site. Class II molecules are expressed in antigen presenting cells (APC: B lymphocytes, dendritic cells, macrophages). The beta chain is approximately 26-28 kDa and its gene contains 6 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, exon 4 encodes the transmembrane domain and exon 5 encodes the cytoplasmic tail."[7]
- NP_002109.2 HLA class II histocompatibility antigen, DM beta chain precursor.[7]
Gene ID: 3111 is HLA-DOA major histocompatibility complex, class II, DO alpha, on 6p21.32: "HLA-DOA belongs to the HLA class II alpha chain paralogues. HLA-DOA forms a heterodimer with HLA-DOB. The heterodimer, HLA-DO, is found in lysosomes in B cells and regulates HLA-DM-mediated peptide loading on MHC class II molecules. In comparison with classical HLA class II molecules, this gene exhibits very little sequence variation, especially at the protein level."[8]
- NP_002110.1 HLA class II histocompatibility antigen, DO alpha chain precursor.[8]
Gene ID: 3112 is HLA-DOB major histocompatibility complex, class II, DO beta, on 6p21.32: "HLA-DOB belongs to the HLA class II beta chain paralogues. This class II molecule is a heterodimer consisting of an alpha (DOA) and a beta chain (DOB), both anchored in the membrane. It is located in intracellular vesicles. DO suppresses peptide loading of MHC class II molecules by inhibiting HLA-DM. Class II molecules are expressed in antigen presenting cells (APC: B lymphocytes, dendritic cells, macrophages). The beta chain is approximately 26-28 kDa and its gene contains 6 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, exon 4 encodes the transmembrane domain and exon 5 encodes the cytoplasmic tail."[9]
- NP_002111.1 HLA class II histocompatibility antigen, DO beta chain precursor.[9]
Gene ID: 3113 is HLA-DPA1 major histocompatibility complex, class II, DP alpha 1 on 6p21.32: "HLA-DPA1 belongs to the HLA class II alpha chain paralogues. This class II molecule is a heterodimer consisting of an alpha (DPA) and a beta (DPB) chain, both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells (APC: B lymphocytes, dendritic cells, macrophages). The alpha chain is approximately 33-35 kDa and its gene contains 5 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, exon 4 encodes the transmembrane domain and the cytoplasmic tail. Within the DP molecule both the alpha chain and the beta chain contain the polymorphisms specifying the peptide binding specificities, resulting in up to 4 different molecules."[10]
- NP_001229453.1 HLA class II histocompatibility antigen, DP alpha 1 chain precursor: "Transcript Variant: This variant (2) differs in the 5' UTR compared to variant 1. Variants 1, 2 and 3 encode the same protein."[10]
- NP_001229454.1 HLA class II histocompatibility antigen, DP alpha 1 chain precursor: "Transcript Variant: This variant (3) differs in the 5' UTR compared to variant 1. Variants 1, 2 and 3 encode the same protein."[10]
- NP_291032.2 HLA class II histocompatibility antigen, DP alpha 1 chain precursor: "Transcript Variant: This variant (1) represents the shortest transcript. Variants 1, 2 and 3 encode the same protein."[10]
Gene ID: 3115 is HLA-DPB1 major histocompatibility complex, class II, DP beta 1, on 6p21.32: "HLA-DPB belongs to the HLA class II beta chain paralogues. This class II molecule is a heterodimer consisting of an alpha (DPA) and a beta chain (DPB), both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells (APC: B lymphocytes, dendritic cells, macrophages). The beta chain is approximately 26-28 kDa and its gene contains 6 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, exon 4 encodes the transmembrane domain and exon 5 encodes the cytoplasmic tail. Within the DP molecule both the alpha chain and the beta chain contain the polymorphisms specifying the peptide binding specificities, resulting in up to 4 different molecules."[11]
- NP_002112.3 HLA class II histocompatibility antigen, DP beta 1 chain precursor.[11]
Gene ID: 3117 is HLA-DQA1 major histocompatibility complex, class II, DQ alpha 1, on 6p21.32: "HLA-DQA1 belongs to the HLA class II alpha chain paralogues. The class II molecule is a heterodimer consisting of an alpha (DQA) and a beta chain (DQB), both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells (APC: B Lymphocytes, dendritic cells, macrophages). The alpha chain is approximately 33-35 kDa. It is encoded by 5 exons; exon 1 encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, and exon 4 encodes the transmembrane domain and the cytoplasmic tail. Within the DQ molecule both the alpha chain and the beta chain contain the polymorphisms specifying the peptide binding specificities, resulting in up to four different molecules. Typing for these polymorphisms is routinely done for bone marrow transplantation."[12]
- NP_002113.2 HLA class II histocompatibility antigen, DQ alpha 1 chain precursor.[12]
Gene ID: 3118 is HLA-DQA2 major histocompatibility complex, class II, DQ alpha 2, on 6p21.32: "This gene belongs to the HLA class II alpha chain family. The encoded protein forms a heterodimer with a class II beta chain. It is located in intracellular vesicles and plays a central role in the peptide loading of MHC class II molecules by helping to release the CLIP molecule from the peptide binding site. Class II molecules are expressed in antigen presenting cells (B lymphocytes, dendritic cells, macrophages) and are used to present antigenic peptides on the cell surface to be recognized by CD4 T-cells."[13]
- NP_064440.1 HLA class II histocompatibility antigen, DQ alpha 2 chain precursor.[13]
Gene ID: 3119 is HLA-DQB1 major histocompatibility complex, class II, DQ beta 1, on 6p21.32: "HLA-DQB1 belongs to the HLA class II beta chain paralogs. This class II molecule is a heterodimer consisting of an alpha (DQA) and a beta chain (DQB), both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells (APC: B lymphocytes, dendritic cells, macrophages). The beta chain is approximately 26-28 kDa and it contains six exons. Exon 1 encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, exon 4 encodes the transmembrane domain and exon 5 encodes the cytoplasmic tail. Within the DQ molecule both the alpha chain and the beta chain contain the polymorphisms specifying the peptide binding specificities, resulting in up to four different molecules. Typing for these polymorphisms is routinely done for bone marrow transplantation. Alternative splicing results in multiple transcript variants."[14]
- NP_001230890.1 HLA class II histocompatibility antigen, DQ beta 1 chain isoform 2 precursor: "Transcript Variant: This variant (2) includes an alternate in-frame exon in the coding region, compared to variant 1. It encodes isoform 2 which is longer than isoform 1. This transcript represents the DQB1*06:02:01:01 allele of the HLA-DQB1 gene, as represented in the assembled chromosome 6 in the primary assembly of the reference genome."[14]
- NP_001230891.1 HLA class II histocompatibility antigen, DQ beta 1 chain isoform 1 precursor: "Transcript Variant: This variant (3) has the same exon combination as variant 1 but represents the DQB1*02:01:01:01 allele of the HLA-DQB1 gene, as represented in the alternate locus group ALT_REF_LOCI_2 of the reference genome. It encodes isoform 1."[14]
- NP_002114.3 HLA class II histocompatibility antigen, DQ beta 1 chain isoform 1 precursor: "Transcript Variant: This variant (1) is the predominant transcript and encodes isoform 1. This transcript represents the DQB1*06:02:01:01 allele of the HLA-DQB1 gene, as represented in the assembled chromosome 6 in the primary assembly of the reference genome."[14]
Gene ID: 3120 is HLA-DQB2 major histocompatibility complex, class II, DQ beta 2, on 6p21.32: "HLA-DQB2 belongs to the family of HLA class II beta chain paralogs. Class II molecules are heterodimers consisting of an alpha (DQA) and a beta chain (DQB), both anchored in the membrane. They play a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells (APC: B lymphocytes, dendritic cells, macrophages). Polymorphisms in the alpha and beta chains specify the peptide binding specificity, and typing for these polymorphisms is routinely done for bone marrow transplantation. However this gene, HLA-DQB2, is not routinely typed, as it is not thought to have an effect on transplantation. There is conflicting evidence in the literature and public sequence databases for the protein-coding capacity of HLA-DQB2. Because there is evidence of transcription and an intact ORF, HLA-DQB2 is represented in Entrez Gene and in RefSeq as a protein-coding locus."[15]
- NP_001185787.1 HLA class II histocompatibility antigen, DQ beta 2 chain isoform 2 precursor: "Transcript Variant: This variant (2) lacks an in-frame exon in the 3' coding region, compared to variant 1. It encodes a shorter isoform (2), compared to isoform 1."[15]
- NP_001287719.1 HLA class II histocompatibility antigen, DQ beta 2 chain isoform 1 precursor: "Transcript Variant: This variant (1) represents the longer transcript and encodes the longer isoform (1)."[15]
Gene ID: 3121 is HLA-DQB3 major histocompatibility complex, class II, DQ beta 3, on 6p21.3: "not in current annotation release"[16]
Gene ID: 3122 is HLA-DRA major histocompatibility complex, class II, DR alpha, on 6p21.32: "HLA-DRA is one of the HLA class II alpha chain paralogues. This class II molecule is a heterodimer consisting of an alpha and a beta chain, both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells (APC: B lymphocytes, dendritic cells, macrophages). The alpha chain is approximately 33-35 kDa and its gene contains 5 exons. Exon 1 encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, and exon 4 encodes the transmembrane domain and the cytoplasmic tail. DRA does not have polymorphisms in the peptide binding part and acts as the sole alpha chain for DRB1, DRB3, DRB4 and DRB5."[17]
- NP_061984.2 HLA class II histocompatibility antigen, DR alpha chain precursor.[17]
Gene ID: 3123 is HLA-DRB1 major histocompatibility complex, class II, DR beta 1, on 6p21.32: "HLA-DRB1 belongs to the HLA class II beta chain paralogs. The class II molecule is a heterodimer consisting of an alpha (DRA) and a beta chain (DRB), both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells. The beta chain is approximately 26-28 kDa. It is encoded by 6 exons. Exon one encodes the leader peptide; exons 2 and 3 encode the two extracellular domains; exon 4 encodes the transmembrane domain; and exon 5 encodes the cytoplasmic tail. Within the DR molecule the beta chain contains all the polymorphisms specifying the peptide binding specificities. Hundreds of DRB1 alleles have been described and some alleles have increased frequencies associated with certain diseases. There are multiple pseudogenes of this gene."[18]
- NP_001230894.1 major histocompatibility complex, class II, DR beta 1 precursor precursor: "Transcript Variant: This variant (2) represents the DRB1*03:01:01 allele of the HLA-DRB1 gene, as represented in the alternate locus groups ALT_REF_LOCI_2 and ALT_REF_LOCI_6 of the reference genome."[18]
- NP_001346122.1 major histocompatibility complex, class II, DR beta 1 precursor: "Transcript Variant: This variant (3) represents the DRB1*07:01:01 allele of the HLA-DRB1 gene, as represented in the alternate locus groups ALT_REF_LOCI_3 and ALT_REF_LOCI_4 of the reference genome."[18]
- NP_001346123.1 major histocompatibility complex, class II, DR beta 1 precursor: "Transcript Variant: This variant (4) represents the DRB1*04:03:01 allele of the HLA-DRB1 gene, as represented in the alternate locus group ALT_REF_LOCI_7 of the reference genome."[18]
- NP_002115.2 major histocompatibility complex, class II, DR beta 1 precursor precursor: "Transcript Variant: This variant (1) represents the DRB1*15:01:01 allele of the HLA-DRB1 gene, as represented in the assembled chromosome 6 in the primary assembly of the reference genome and the CHM1_1.1 genome."[18]
Gene ID: 3125 is HLA-DRB3 major histocompatibility complex, class II, DR beta 3, on 6p21.3: "HLA-DRB3 belongs to the HLA class II beta chain paralogues. This class II molecule is a heterodimer consisting of an alpha (DRA) and a beta (DRB) chain, both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells. The beta chain is approximately 26-28 kDa and its gene contains 6 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, exon 4 encodes the transmembrane domain and exon 5 encodes the cytoplasmic tail. Within the DR molecule the beta chain contains all the polymorphisms specifying the peptide binding specificities. Typing for these polymorphisms is routinely done for bone marrow and kidney transplantation. There are multiple pseudogenes of this gene."[19]
- NP_072049.2 major histocompatibility complex, class II, DR beta 3 precursor.[19]
Gene ID: 3126 is HLA-DRB4 major histocompatibility complex, class II, DR beta 4, on 6p21.3: "HLA-DRB4 belongs to the HLA class II beta chain paralogues. This class II molecule is a heterodimer consisting of an alpha (DRA) and a beta (DRB) chain, both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells. The beta chain is approximately 26-28 kDa and its gene contains 6 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, exon 4 encodes the transmembrane domain and exon 5 encodes the cytoplasmic tail. Within the DR molecule the beta chain contains all the polymorphisms specifying the peptide binding specificities. Typing for these polymorphisms is routinely done for bone marrow and kidney transplantation. There are multiple pseudogenes of this gene."[20]
- NP_068818.4 major histocompatibility complex, class II, DR beta 4 precursor.[20]
Gene ID: 3127 is HLA-DRB5 major histocompatibility complex, class II, DR beta 5, on 6p21.32: "HLA-DRB5 belongs to the HLA class II beta chain paralogues. This class II molecule is a heterodimer consisting of an alpha (DRA) and a beta (DRB) chain, both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells. The beta chain is approximately 26-28 kDa and its gene contains 6 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the two extracellular domains, exon 4 encodes the transmembrane domain and exon 5 encodes the cytoplasmic tail. Within the DR molecule the beta chain contains all the polymorphisms specifying the peptide binding specificities. Typing for these polymorphisms is routinely done for bone marrow and kidney transplantation. There are multiple pseudogenes of this gene."[21]
- NP_002116.2 major histocompatibility complex, class II, DR beta 5 precursor.[21]
Gene ID: 3833 is KIFC1 kinesin family member C1 on 6p21.32.[22]
Gene ID: 4261 is CIITA class II major histocompatibility complex transactivator on 16p13.13: "This gene encodes a protein with an acidic transcriptional activation domain, 4 LRRs (leucine-rich repeats) and a GTP binding domain. The protein is located in the nucleus and acts as a positive regulator of class II major histocompatibility complex gene transcription, and is referred to as the "master control factor" for the expression of these genes. The protein also binds GTP and uses GTP binding to facilitate its own transport into the nucleus. Once in the nucleus it does not bind DNA but rather uses an intrinsic acetyltransferase (AT) activity to act in a coactivator-like fashion. Mutations in this gene have been associated with bare lymphocyte syndrome type II (also known as hereditary MHC class II deficiency or HLA class II-deficient combined immunodeficiency), increased susceptibility to rheumatoid arthritis, multiple sclerosis, and possibly myocardial infarction. Several transcript variants encoding different isoforms have been found for this gene."[23]
- NP_000237.2 MHC class II transactivator isoform 2: "Transcript Variant: This variant (2) uses an alternate in-frame splice junction at the 5' end of an exon compared to variant 1. The resulting isoform (2) is 1 aa shorter compared to isoform 1."[23]
- NP_001273331.1 MHC class II transactivator isoform 1: "Transcript Variant: This variant (1) represents the longest transcript and encodes the longest isoform (1)."[23]
- NP_001273332.1 MHC class II transactivator isoform 3: "Transcript Variant: This variant (3) uses two alternate splice junctions and lacks two alternate coding exons compared to variant 1. The resulting isoform (3) has the same N- and C-termini but is shorter compared to isoform 1."[23]
- NR_104444.2 RNA Sequence: "Transcript Variant: This variant (4) uses an alternate splice junction and lacks an alternate exon compared to variant 1. This variant is represented as non-coding because the use of the 5'-most expected translational start codon, as used in variant 1, renders the transcript a candidate for nonsense-mediated mRNA decay (NMD)."[23]
Gene ID: 4904 is YBX1 Y-box binding protein 1 aka CCAAT-binding transcription factor I subunit A, DNA-binding protein B, Y-box transcription factor, enhancer factor I subunit A, major histocompatibility complex, class II, Y box-binding protein I, nuclease-sensitive element-binding protein 1 on 1p34.2: "This gene encodes a highly conserved cold shock domain protein that has broad nucleic acid binding properties. The encoded protein functions as both a DNA and RNA binding protein and has been implicated in numerous cellular processes including regulation of transcription and translation, pre-mRNA splicing, DNA reparation and mRNA packaging. This protein is also a component of messenger ribonucleoprotein (mRNP) complexes and may have a role in microRNA processing. This protein can be secreted through non-classical pathways and functions as an extracellular mitogen. Aberrant expression of the gene is associated with cancer proliferation in numerous tissues. This gene may be a prognostic marker for poor outcome and drug resistance in certain cancers. Alternate splicing results in multiple transcript variants. Pseudogenes of this gene are found on multiple chromosomes."[24]
- NP_004550.2 Y-box-binding protein 1: "Transcript Variant: This variant (1) represents the longer transcript and encodes the functional protein."[24]
- NR_132737.2 RNA Sequence: "Transcript Variant: This variant (2) contains an alternate 5' exon and lacks an internal exon, compared to variant 1. This variant is represented as non-coding because the predicted protein does not meet RefSeq quality criteria."[24]
Gene ID: 5089 is PBX2 PBX homeobox 2 on 6p21.32: "This gene encodes a ubiquitously expressed member of the TALE/PBX homeobox family. It was identified by its similarity to a homeobox gene which is involved in t(1;19) translocation in acute pre-B-cell leukemias. This protein is a transcriptional activator which binds to the TLX1 promoter. The gene is located within the major histocompatibility complex (MHC) on chromosome 6."[25]
Gene ID: 5252 is PHF1 PHD finger protein 1 on 6p21.32: "This gene encodes a Polycomb group protein. The protein is a component of a histone H3 lysine-27 (H3K27)-specific methyltransferase complex, and functions in transcriptional repression of homeotic genes. The protein is also recruited to double-strand breaks, and reduced protein levels results in X-ray sensitivity and increased homologous recombination. Multiple transcript variants encoding different isoforms have been found for this gene."[26]
- NP_002627.2 PHD finger protein 1 isoform a: "Transcript Variant: This variant (1), uses an alternate splice site and lacks an alternate exon in the 3' coding region resulting in a frameshift, compared to variant 2. The resulting isoform (a) has a shorter and distinct C-terminus, compared to isoform b."[26]
- NP_077084.2 PHD finger protein 1 isoform b: "Transcript Variant: This variant (2), also called variant PHF2, represents the longest transcript and encodes the longer isoform (b)."[26]
Gene ID: 5696 is PSMB8 proteasome 20S subunit beta 8 on 6p21.32: "The proteasome is a multicatalytic proteinase complex with a highly ordered ring-shaped 20S core structure. The core structure is composed of 4 rings of 28 non-identical subunits; 2 rings are composed of 7 alpha subunits and 2 rings are composed of 7 beta subunits. Proteasomes are distributed throughout eukaryotic cells at a high concentration and cleave peptides in an ATP/ubiquitin-dependent process in a non-lysosomal pathway. An essential function of a modified proteasome, the immunoproteasome, is the processing of class I MHC peptides. This gene encodes a member of the proteasome B-type family, also known as the T1B family, that is a 20S core beta subunit. This gene is located in the class II region of the MHC (major histocompatibility complex). Expression of this gene is induced by gamma interferon and this gene product replaces catalytic subunit 3 (proteasome beta 5 subunit) in the immunoproteasome. Proteolytic processing is required to generate a mature subunit. Two alternative transcripts encoding two isoforms have been identified; both isoforms are processed to yield the same mature subunit."[27]
- NP_004150.1 proteasome subunit beta type-8 isoform E1: "Transcript Variant: This variant (1) represents the longer transcript but encodes the shorter isoform (E1)."[27]
- NP_683720.2 proteasome subunit beta type-8 isoform E2 precursor: "Transcript Variant: This variant (2) differs in the 5' UTR and contains an alternate in-frame exon in the 5' coding region, compared to variant 1. Isoform E2 has a distinct N-terminus, compared to isoform E1."[27]
Gene ID: 5698 is PSMB9 proteasome 20S subunit beta 9 on 6p21.32: "The proteasome is a multicatalytic proteinase complex with a highly ordered ring-shaped 20S core structure. The core structure is composed of 4 rings of 28 non-identical subunits; 2 rings are composed of 7 alpha subunits and 2 rings are composed of 7 beta subunits. Proteasomes are distributed throughout eukaryotic cells at a high concentration and cleave peptides in an ATP/ubiquitin-dependent process in a non-lysosomal pathway. An essential function of a modified proteasome, the immunoproteasome, is the processing of class I MHC peptides. This gene encodes a member of the proteasome B-type family, also known as the T1B family, that is a 20S core beta subunit. This gene is located in the class II region of the MHC (major histocompatibility complex). Expression of this gene is induced by gamma interferon and this gene product replaces catalytic subunit 1 (proteasome beta 6 subunit) in the immunoproteasome. Proteolytic processing is required to generate a mature subunit."[28]
Gene ID: 5863 is RGL2 ral guanine nucleotide dissociation stimulator like 2 on 6p21.32.[29]
- NP_001230667.1 ral guanine nucleotide dissociation stimulator-like 2 isoform 2: "Transcript Variant: This variant (2) lacks an internal exon and uses a downstream, in-frame start codon, compared to variant 1. The encoded isoform (2) has a shorter N-terminus, compared to isoform 1."[29]
- NP_004752.1 ral guanine nucleotide dissociation stimulator-like 2 isoform 1: "Transcript Variant: This variant (1) represents the longer transcript and encodes the longer isoform (1)."[29]
Gene ID: 6015 is RING1 ring finger protein 1 on 6p21.32: "This gene belongs to the RING finger family, members of which encode proteins characterized by a RING domain, a zinc-binding motif related to the zinc finger domain. The gene product can bind DNA and can act as a transcriptional repressor. It is associated with the multimeric polycomb group protein complex. The gene product interacts with the polycomb group proteins BMI1, EDR1, and CBX4, and colocalizes with these proteins in large nuclear domains. It interacts with the CBX4 protein via its glycine-rich C-terminal domain. The gene maps to the HLA class II region, where it is contiguous with the RING finger genes FABGL and HKE4."[30]
Gene ID: 6046 is BRD2 bromodomain containing 2 on 6p21.32: "This gene encodes a transcriptional regulator that belongs to the BET (bromodomains and extra terminal domain) family of proteins. This protein associates with transcription complexes and with acetylated chromatin during mitosis, and it selectively binds to the acetylated lysine-12 residue of histone H4 via its two bromodomains. The gene maps to the major histocompatability complex (MHC) class II region on chromosome 6p21.3, but sequence comparison suggests that the protein is not involved in the immune response. This gene has been implicated in juvenile myoclonic epilepsy, a common form of epilepsy that becomes apparent in adolescence. Multiple alternatively spliced variants have been described for this gene."[31]
- NP_001106653.1 bromodomain-containing protein 2 isoform 1: "Transcript Variant: This variant (2) has an alternate 5' UTR compared to variant 1. Both variants 1 and 2 encode the same isoform (1)."[31]
- NP_001186384.1 bromodomain-containing protein 2 isoform 2: "Transcript Variant: This variant (3) has an additional in-frame exon in the CDS compared to variant 1. The resulting isoform (2) is longer than isoform 1."[31]
- NP_001186385.1 bromodomain-containing protein 2 isoform 3: "Transcript Variant: This variant (4) has an alternate 5' exon, resulting in a downstream AUG start codon compared to variant 1. The resulting isoform (3) is shorter at the N-terminus, as compared to isoform 1."[31]
- NP_001278915.1 bromodomain-containing protein 2 isoform 4: "Transcript Variant: This variant (5) has an alternate 5' exon and an additional internal exon compared to variant 1. The resulting isoform (4) is shorter at the N-terminus compared to isoform 1."[31]
- NP_005095.1 bromodomain-containing protein 2 isoform 1: "Transcript Variant: This variant (1) and variant 2 encode the same predominant isoform (1)."[31]
Gene ID: 6048 is RNF5 ring finger protein 5 on 6p21.32: "The protein encoded by this gene contains a RING finger, which is a motif known to be involved in protein-protein interactions. This protein is a membrane-bound ubiquitin ligase. It can regulate cell motility by targeting paxillin ubiquitination and altering the distribution and localization of paxillin in cytoplasm and cell focal adhesions."[32]
Gene ID: 6222 is RPS18 ribosomal protein S18 on 6p21.32: "Ribosomes, the organelles that catalyze protein synthesis, consist of a small 40S subunit and a large 60S subunit. Together these subunits are composed of 4 RNA species and approximately 80 structurally distinct proteins. This gene encodes a ribosomal protein that is a component of the 40S subunit. The protein belongs to the S13P family of ribosomal proteins. It is located in the cytoplasm. The gene product of the E. coli ortholog (ribosomal protein S13) is involved in the binding of fMet-tRNA, and thus, in the initiation of translation. This gene is an ortholog of mouse Ke3. As is typical for genes encoding ribosomal proteins, there are multiple processed pseudogenes of this gene dispersed through the genome."[33]
Gene ID: 6257 is RXRB retinoid X receptor beta on 6p21.32: "This gene encodes a member of the retinoid X receptor (RXR) family of nuclear receptors which are involved in mediating the effects of retinoic acid (RA). The encoded protein forms homodimers with the retinoic acid, thyroid hormone, and vitamin D receptors, increasing both DNA binding and transcriptional function on their respective response elements. This gene lies within the major histocompatibility complex (MHC) class II region on chromosome 6. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene."[34]
- NP_001257330.1 retinoic acid receptor RXR-beta isoform 1: "Transcript Variant: This variant (1) represents the longer transcript and encodes the longer isoform (1)."[34]
- NP_001278918.1 retinoic acid receptor RXR-beta isoform 3: "Transcript Variant: This variant (3) differs in the 5' UTR and coding sequence compared to variant 1. The resulting isoform (3) has a shorter and distinct N-terminus compared to isoform 1."[34]
- NP_068811.1 retinoic acid receptor RXR-beta isoform 2: "Transcript Variant: This variant (2) uses an alternate splice site in the coding region, but maintains the reading frame, compared to variant 1. The encoded isoform (2) is shorter than isoform 1."[34]
Gene ID: 6293 is VPS52 VPS52 subunit of GARP complex on 6p21.32: "This gene encodes a protein that is similar to the yeast suppressor of actin mutations 2 gene. The yeast protein forms a subunit of the tetrameric Golgi-associated retrograde protein complex that is involved in vesicle trafficking from from both early and late endosomes, back to the trans-Golgi network. This gene is located on chromosome 6 in a head-to-head orientation with the gene encoding ribosomal protein S18. Alternative splicing results in multiple transcript variants."[35]
- NP_001276103.1 vacuolar protein sorting-associated protein 52 homolog isoform 2: "Transcript Variant: This variant (2) differs in the 5' UTR, initiates translation at an alternate start codon and lacks an alternate exon in the 5' coding region, compared to variant 1. It encodes isoform 2, which is shorter and has a distinct N-terminus, compared to isoform 1."[35]
- NP_001276104.1 vacuolar protein sorting-associated protein 52 homolog isoform 3: "Transcript Variant: This variant (3) differs in the 5' UTR, lacks a portion of the 5' coding region and initiates translation at a downstream start codon, compared to variant 1. It encodes isoform 3, which is shorter at the N-terminus, compared to isoform 1."[35]
- NP_001276105.1 vacuolar protein sorting-associated protein 52 homolog isoform 4: "Transcript Variant: This variant (4) differs in the 5' UTR and the 5' coding region and initiates translation at a downstream start codon, compared to variant 1. It encodes isoform 4, which is shorter than isoform 1."[35]
- NP_072047.4 vacuolar protein sorting-associated protein 52 homolog isoform 1: "Transcript Variant: This variant (1) encodes the longest isoform (1)."[35]
Gene ID: 7922 is SLC39A7 solute carrier family 39 member 7 on 6p21.32: "The protein encoded by this gene transports zinc from the Golgi and endoplasmic reticulum to the cytoplasm. This transport may be important for activation of tyrosine kinases, some of which could be involved in cancer progression. Therefore, modulation of the encoded protein could be useful as a therapeutic agent against cancer. Alternative splicing results in multiple transcript variants."[36]
- NP_001070984.1 zinc transporter SLC39A7 isoform 1 precursor: "Transcript Variant: This variant (2) differs in the 5' UTR, compared to variant 1. Variants 1 and 2 encode the same protein (isoform 1)."[36]
- NP_001275706.1 zinc transporter SLC39A7 isoform 2: "Transcript Variant: This variant (3) differs in the 5' UTR, lacks a portion of the 5' coding region, and initiates translation at a downstream start codon, compared to variant 1. It encodes isoform 2, which contains a shorter N-terminus, compared to isoform 1."[36]
- NP_008910.2 zinc transporter SLC39A7 isoform 1 precursor: "Transcript Variant: This variant (1) represents the longest transcript and encodes the longer isoform (1). Variants 1 and 2 encode the same protein."[36]
Gene ID: 7923 is HSD17B8 hydroxysteroid 17-beta dehydrogenase 8 on 6p21.32: "In mice, the Ke6 protein is a 17-beta-hydroxysteroid dehydrogenase that can regulate the concentration of biologically active estrogens and androgens. It is preferentially an oxidative enzyme and inactivates estradiol, testosterone, and dihydrotestosterone. However, the enzyme has some reductive activity and can synthesize estradiol from estrone. The protein encoded by this gene is similar to Ke6 and is a member of the short-chain dehydrogenase superfamily. An alternatively spliced transcript of this gene has been detected, but the full-length nature of this variant has not been determined."[37]
Gene ID: 8705 is B3GALT4 beta-1,3-galactosyltransferase 4 on 6p21.32: "his gene is a member of the beta-1,3-galactosyltransferase (beta3GalT) gene family. This family encodes type II membrane-bound glycoproteins with diverse enzymatic functions using different donor substrates (UDP-galactose and UDP-N-acetylglucosamine) and different acceptor sugars (N-acetylglucosamine, galactose, N-acetylgalactosamine). The beta3GalT genes are distantly related to the Drosophila Brainiac gene and have the protein coding sequence contained in a single exon. The beta3GalT proteins also contain conserved sequences not found in the beta4GalT or alpha3GalT proteins. The carbohydrate chains synthesized by these enzymes are designated as type 1, whereas beta4GalT enzymes synthesize type 2 carbohydrate chains. The ratio of type 1:type 2 chains changes during embryogenesis. By sequence similarity, the beta3GalT genes fall into at least two groups: beta3GalT4 and 4 other beta3GalT genes (beta3GalT1-3, beta3GalT5). This gene is oriented telomere to centromere in close proximity to the ribosomal protein S18 gene. The functionality of the encoded protein is limited to ganglioseries glycolipid biosynthesis."[38]
Gene ID: 8831 is SYNGAP1 synaptic Ras GTPase activating protein 1 on 6p21.32: "This gene encodes a Ras GTPase activating protein that is a member of the N-methyl-D-aspartate receptor complex. The N-terminal domain of the protein contains a Ras-GAP domain, a pleckstrin homology domain, and a C2 domain that may be involved in binding of calcium and phospholipids. The C-terminal domain consists of a ten histidine repeat region, serine and tyrosine phosphorylation sites, and a T/SXV motif required for postsynaptic scaffold protein interaction. The encoded protein negatively regulates Ras, Rap and alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor trafficking to the postsynaptic membrane to regulate synaptic plasticity and neuronal homeostasis. Allelic variants of this gene are associated with intellectual disability and autism spectrum disorder. Alternative splicing results in multiple transcript variants."[39]
- NP_001123538.1 ras/Rap GTPase-activating protein SynGAP isoform 2.[39]
- NP_006763.2 ras/Rap GTPase-activating protein SynGAP isoform 1.[39]
Gene ID: 9277 is WDR46 WD repeat domain 46 on 6p21.32.[40]
- NP_001157739.1 WD repeat-containing protein 46 isoform 2: "Transcript Variant: This variant (2) uses an alternate in-frame splice site in the 5' coding region, compared to variant 1. This results in a shorter protein (isoform 2), compared to isoform 1."[40]
- NP_005443.3 WD repeat-containing protein 46 isoform 1: "Transcript Variant: This variant (1) represents the longer transcript and encodes the longer isoform (1)."[40]
Gene ID: 9278 is ZBTB22 zinc finger and BTB domain containing 22 on 6p21.32.[41]
- NP_001138810.1 zinc finger and BTB domain-containing protein 22: "Transcript Variant: This variant differs in the 5' UTR compared to variant 1. Both variants 1 and 2 encode the same protein."[41]
- NP_005444.4 zinc finger and BTB domain-containing protein 22: "Transcript Variant: This variant (1) represents the longer transcript. Both variants 1 and 2 encode the same protein."[41]
Gene ID: 9374 is PPT2 palmitoyl-protein thioesterase 2 on 6p21.32: "This gene encodes a member of the palmitoyl-protein thioesterase family. The encoded glycosylated lysosomal protein has palmitoyl-CoA hydrolase activity in vitro, but does not hydrolyze palmitate from cysteine residues in proteins. Alternative splicing results in multiple transcript variants. Read-through transcription also exists between this gene and the downstream EGFL8 (EGF-like-domain, multiple 8) gene."[42]
- NP_001191032.1 lysosomal thioesterase PPT2 isoform a precursor: "Transcript Variant: This variant (3) differs in the 5' UTR compared to variant 1. Both variants 1 and 3 encode the same isoform (a)."[42]
- NP_005146.4 lysosomal thioesterase PPT2 isoform a precursor: "Transcript Variant: This variant (1) represents the longest transcript and encodes the shorter isoform (a). Both variants 1 and 3 encode the same isoform."[42]
- NP_619731.2 lysosomal thioesterase PPT2 isoform b precursor: "Transcript Variant: This variant (2) differs in the 5' UTR and 5' coding region, and uses an alternate start codon, compared to variant 1. The resulting isoform (b) is longer at the N-terminus, compared to isoform a."[42]
Gene ID: 10471 is PFDN6 prefoldin subunit 6 aka HLA class II region expressed gene KE2 on 6p21.32: "PFDN6 is a subunit of the heteromeric prefoldin complex that chaperones nascent actin (see MIM 102560) and alpha- and beta-tubulin (see MIM 602529 and MIM 191130, respectively) chains pending their transfer to the cytosolic chaperonin containing TCP1 (MIM 186980) (CCT) complex (Hansen et al., 1999 [PubMed 10209023])."[43]
- NP_001172110.1 prefoldin subunit 6: "Transcript Variant: This variant (1) represents the longest transcript. All variants encode the same protein."[43]
- NP_001252524.1 prefoldin subunit 6: "Transcript Variant: This variant (3) differs in the 5' UTR, compared to variant 1. All variants encode the same protein."[43]
- NP_001252525.1 prefoldin subunit 6: "Transcript Variant: This variant (4) differs in the 5' UTR, compared to variant 1. All variants encode the same protein."[43]
- NP_055075.1 prefoldin subunit 6: "Transcript Variant: This variant (2) differs in the 5' UTR, compared to variant 1. All variants encode the same protein."[43]
See also
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 RefSeq (August 2011). "CD74 CD74 molecule [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 1 April 2020.
- ↑ 2.0 2.1 2.2 2.3 2.4 RefSeq (July 2009). "COL11A2 collagen type XI alpha 2 chain [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 7 April 2020.
- ↑ 3.0 3.1 3.2 RefSeq (October 2008). "ATF6B activating transcription factor 6 beta [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 8 April 2020.
- ↑ 4.0 4.1 4.2 4.3 4.4 RefSeq (November 2008). "DAXX death domain associated protein [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 8 April 2020.
- ↑ RefSeq (13 March 2020). "GTF2H4 general transcription factor IIH subunit 4 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 6 April 2020.
- ↑ 6.0 6.1 RefSeq (July 2008). "HLA-DMA major histocompatibility complex, class II, DM alpha [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2 April 2020.
- ↑ 7.0 7.1 RefSeq (July 2008). "HLA-DMB major histocompatibility complex, class II, DM beta [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2 April 2020.
- ↑ 8.0 8.1 RefSeq (July 2008). "HLA-DOA major histocompatibility complex, class II, DO alpha [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2 April 2020.
- ↑ 9.0 9.1 RefSeq (July 2008). "HLA-DOB major histocompatibility complex, class II, DO beta [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2 April 2020.
- ↑ 10.0 10.1 10.2 10.3 RefSeq (July 2008). "HLA-DPA1 major histocompatibility complex, class II, DP alpha 1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 1 April 2020.
- ↑ 11.0 11.1 RefSeq (July 2008). "HLA-DPB1 major histocompatibility complex, class II, DP beta 1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 28 March 2020.
- ↑ 12.0 12.1 RefSeq (July 2008). "HLA-DQA1 major histocompatibility complex, class II, DQ alpha 1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 28 March 2020.
- ↑ 13.0 13.1 RefSeq (June 2010). "HLA-DQA2 major histocompatibility complex, class II, DQ alpha 2 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2 April 2020.
- ↑ 14.0 14.1 14.2 14.3 RefSeq (September 2011). "HLA-DQB1 major histocompatibility complex, class II, DQ beta 1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 28 March 2020.
- ↑ 15.0 15.1 15.2 RefSeq (October 2010). "HLA-DQB2 major histocompatibility complex, class II, DQ beta 2 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2 April 2020.
- ↑ RefSeq (24 March 2019). "HLA-DQB3 major histocompatibility complex, class II, DQ beta 3 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 5 April 2020.
- ↑ 17.0 17.1 RefSeq (July 2008). "HLA-DRA major histocompatibility complex, class II, DR alpha [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 30 March 2020.
- ↑ 18.0 18.1 18.2 18.3 18.4 RefSeq (February 2020). "HLA-DRB1 major histocompatibility complex, class II, DR beta 1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 28 March 2020.
- ↑ 19.0 19.1 RefSeq (February 2020). "HLA-DRB3 major histocompatibility complex, class II, DR beta 3 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 30 March 2020.
- ↑ 20.0 20.1 RefSeq (February 2020). "HLA-DRB4 major histocompatibility complex, class II, DR beta 4 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 28 March 2020.
- ↑ 21.0 21.1 RefSeq (February 2020). "HLA-DRB5 major histocompatibility complex, class II, DR beta 5 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 30 March 2020.
- ↑ RefSeq (5 April 2020). "KIFC1 kinesin family member C1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 9 April 2020.
- ↑ 23.0 23.1 23.2 23.3 23.4 RefSeq (November 2013). "CIITA class II major histocompatibility complex transactivator [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 30 March 2020.
- ↑ 24.0 24.1 24.2 RefSeq (September 2015). "YBX1 Y-box binding protein 1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 19 November 2018.
- ↑ RefSeq (July 2008). "PBX2 PBX homeobox 2 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 9 April 2020.
- ↑ 26.0 26.1 26.2 RefSeq (May 2009). "PHF1 PHD finger protein 1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 9 April 2020.
- ↑ 27.0 27.1 27.2 RefSeq (July 2008). "PSMB8 proteasome 20S subunit beta 8 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 9 April 2020.
- ↑ RefSeq (March 2010). "PSMB9 proteasome 20S subunit beta 9 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 9 April 2020.
- ↑ 29.0 29.1 29.2 RefSeq (20 March 2020). "RGL2 ral guanine nucleotide dissociation stimulator like 2 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 9 April 2020.
- ↑ RefSeq (July 2008). "RING1 ring finger protein 1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 10 April 2020.
- ↑ 31.0 31.1 31.2 31.3 31.4 31.5 RefSeq (December 2010). "BRD2 bromodomain containing 2 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 10 April 2020.
- ↑ RefSeq (July 2008). "RNF5 ring finger protein 5 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 10 April 2020.
- ↑ RefSeq (July 2008). "RPS18 ribosomal protein S18 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 10 April 2020.
- ↑ 34.0 34.1 34.2 34.3 RefSeq (July 2008). "RXRB retinoid X receptor beta [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 10 April 2020.
- ↑ 35.0 35.1 35.2 35.3 35.4 RefSeq (January 2014). "VPS52 VPS52 subunit of GARP complex [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 10 April 2020.
- ↑ 36.0 36.1 36.2 36.3 RefSeq (January 2014). "SLC39A7 solute carrier family 39 member 7 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 11 April 2020.
- ↑ RefSeq (July 2008). "HSD17B8 hydroxysteroid 17-beta dehydrogenase 8 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 11 April 2020.
- ↑ RefSeq (July 2008). "B3GALT4 beta-1,3-galactosyltransferase 4 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 11 April 2020.
- ↑ 39.0 39.1 39.2 RefSeq (November 2016). "SYNGAP1 synaptic Ras GTPase activating protein 1 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 11 April 2020.
- ↑ 40.0 40.1 40.2 RefSeq (5 April 2020). "WDR46 WD repeat domain 46 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 11 April 2020.
- ↑ 41.0 41.1 41.2 RefSeq (13 March 2020). "ZBTB22 zinc finger and BTB domain containing 22 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 11 April 2020.
- ↑ 42.0 42.1 42.2 42.3 RefSeq (February 2011). "PPT2 palmitoyl-protein thioesterase 2 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 11 April 2020.
- ↑ 43.0 43.1 43.2 43.3 43.4 RefSeq (July 2010). "PFDN6 prefoldin subunit 6 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 11 April 2020.