A1BG gene transcriptions
Editor-In-Chief: Henry A. Hoff
A1BG is the official symbol for GeneID: 1, "A1BG alpha-1-B glycoprotein [ Homo sapiens ]", in the National Center for Biotechnology Information (NCBI) gene database for gene-specific information. To access information about this gene, use the "Home - Gene - NCBI" external link and enter "1[uid]", without the quotes, or the number one after the "/". The official name of this gene is "alpha-1-B glycoprotein". The protein encoded by this gene is a plasma glycoprotein of unknown function. The protein shows sequence similarity to the variable regions of some immunoglobulin supergene family member proteins.
Transcription of eukaryotic genes is believed to be reasonably well understood. Not all the facts are known but phenomenology is probably complete. Each possibility to cause a simple computer program to transcribe the gene for A1BG is to be tested.
Most undergraduate university courses describe transcription, so many of the terms used here to analyze the transcription should be reasonably understood. Some usage of state-of-the-art refereed journal articles is included.
Although there are computer programs being used to predict the effects of gene expression, up-regulation or down-regulation, most of these are quite advanced and are not readily available for students or researchers outside specific universities or companies performing drug research and testing.
Just about any student with access to a computer in various forms is able to write small programs in a variety of computer languages to test for themselves whether or not a specific algorithm works to transcribe this gene.
The transcription start site for A1BG has probably been discovered through the use of reverse transcriptase. The publication demonstrating this has been found. Further, the product of the gene has been detected, but exactly how the product has its messenger RNA initially transcripted is unspecified and perhaps unknown.
Notations
Notation: let the symbol / replace or.
For example, (A or C or G) becomes (A/C/G).
Notation: let the following table of International Union of Pure and Applied Chemistry (IUPAC) stand for the nucleotides indicated.
| IUPAC nucleotide code | Base |
|---|---|
| A | Adenine |
| B | C/G/T |
| C | Cytosine |
| D | A/G/T |
| G | Guanine |
| H | A/C/T |
| K | G/T |
| M | A/C |
| N | any base (A/C/G/T) |
| R | A/G |
| S | G/C |
| T | Thymine |
| V | A/C/G |
| W | A/T |
| Y | C/T |
Control groups
For the transcription of A1BG, a control group would likely have a DNA segment that contains at least one transcription start site, e.g., G/A/T-G/C+1-G-T/C-G-G-G/A-A-G/C. This segment directs the RNA polymerase II holoenzyme complex to the exact TSS.
In terms of a promoter, this transcription by definition should occur in either the core promoter or the proximal promoter. If not fully understood, the TSS may occur in either a focused promoter or a dispersed promoter.
A transcriptional characteristic of a promoter element may be to be located between -8 and +2 relative to at least one TSS.
Each promoter element may be found in at least one gene or isoform.
Genomes

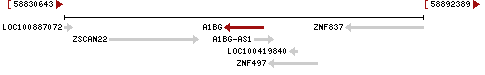
Shown on the top diagram is the genomic context of A1BG as of 2010. The gene is overlapped by a non-coding RNA (NCRNA00181), and has nearby genes ZSCAN22 at the 5' end and ZNF497 (GeneID: 162968) at the 3' end. The nucleotides between genes ZSCAN22 (GeneID: 342945) and the 5'-end of A1BG, should contain the A1BG promoter. NCRNA00181, which overlaps A1BG, has numbered nucleotides 58863336 to 58866548.
In the lower diagram NCRNA00181 has undergone a name change to A1BG antisense RNA 1 (A1BG-AS1), GeneID: 503538.
Orthologs of A1BG occur in
- Acinonyx jubatus (cheetah),
- Ailuropoda melanoleuca (giant panda),
- Aotus nancymaae (Ma's night monkey),
- Balaenoptera acutorostrata scammoni,
- Bison bison bison,
- Bos taurus (cattle),
- Bubalus bubalis (water buffalo),
- Callithrix jacchus (white-tufted-ear marmoset),
- Camelus bactrianus (Bactrian camel),
- Camelus dromedarius (Arabian camel),
- Camelus ferus (Wild Bactrian camel),
- Canis lupus familiaris (dog),
- Capra hircus (goat),
- Cavia porcellus (domestic guinea pig),
- Ceratotherium simum simum (southern white rhinoceros),
- Cercocebus atys (sooty mangabey),
- Chinchilla lanigera (long-tailed chinchilla),
- Chlorocebus sabaeus (green monkey),
- Chrysochloris asiatica (Cape golden mole),
- Colobus angolensis palliatus,
- Dasypus novemcinctus (nine-banded armadillo),
- Echinops telfairi (small Madagascar hedgehog),
- Eptesicus fuscus (big brown bat),
- Equus asinus (ass),
- Equus caballus (horse),
- Equus przewalskii (Przewalski's horse),
- Felis catus (domestic cat),
- Fukomys damarensis (Damara mole-rat),
- Galeopterus variegatus (Sunda flying lemur),
- Gorilla gorilla (western gorilla),
- Heterocephalus glaber (naked mole-rat),
- Homo sapiens (human),
- Jaculus jaculus (lesser Egyptian jerboa),
- Leptonychotes weddellii (Weddell seal),
- Loxodonta africana (African savanna elephant),
- Macaca fascicularis (crab-eating macaque),
- Macaca mulatta (Rhesus monkey),
- Macaca nemestrina (pig-tailed macaque),
- Mandrillus leucophaeus (drill),
- Marmota marmota marmota (Alpine marmot),
- Mesocricetus auratus (golden hamster),
- Microcebus murinus (gray mouse lemur),
- Microtus ochrogaster (prairie vole),
- Miniopterus natalensis,
- Mus musculus (house mouse),
- Mustela putorius furo (domestic ferret),
- Myotis brandtii (Brandt's bat),
- Myotis davidii,
- Myotis lucifugus (little brown bat),
- Nannospalax galili (Upper Galilee mountains blind mole rat),
- Nomascus leucogenys (northern white-cheeked gibbon),
- Ochotona princeps (American pika),
- Octodon degus (degu),
- Odobenus rosmarus divergens (Pacific walrus),
- Orcinus orca (killer whale),
- Orycteropus afer afer,
- Oryctolagus cuniculus (rabbit),
- Otolemur garnettii (small-eared galago),
- Ovis aries (sheep),
- Pan paniscus (pygmy chimpanzee),
- Pan troglodytes (chimpanzee),
- Panthera tigris altaica (Amur tiger),
- Papio anubis (olive baboon),
- Peromyscus maniculatus bairdii (prairie deer mouse),
- Physeter catodon (sperm whale),
- Pongo abelii (Sumatran orangutan),
- Propithecus coquereli (Coquerel's sifaka),
- Pteropus alecto (black flying fox),
- Pteropus vampyrus (large flying fox),
- Rhinopithecus roxellana (golden snub-nosed monkey),
- Rousettus aegyptiacus (Egyptian rousette),
- Saimiri boliviensis (Bolivian squirrel monkey),
- Sarcophilus harrisii (Tasmanian devil),
- Sorex araneus (European shrew),
- Sus scrofa (pig),
- Trichechus manatus latirostris (Florida manatee),
- Tupaia chinensis (Chinese tree shrew),
- Ursus maritimus (polar bear),
- Vicugna pacos (alpaca).
Genes
Def. a unit of heredity; a segment of DNA or RNA that is transmitted from one generation to the next, and that carries genetic information such as the sequence of amino acids for a protein is called a gene.
A1BG seems to be the central, intermediary link for differentially expressed proteins (DEPs) in the plasma of autoimmune hepatitis (AIH) and normal mice that can mimic the pathological process of AIH in the human body.[1]
Gene expressions
{{fairuse}}Although it is harder to regulate the transcription of genes with multiple transcription start sites, "variations in the expression of a constitutive gene would be minimized by the use of multiple start sites."[2]
Earlier "studies led to the design of a super core promoter (SCP) that contains a TATA, Inr, MTE, and DPE in a single promoter (Juven-Gershon et al., 2006b). The SCP is the strongest core promoter observed in vitro and in cultured cells and yields high levels of transcription in conjunction with transcriptional enhancers. These findings indicate that gene expression levels can be modulated via the core promoter."[2]
Gene expressions for A1BG is a suite of genes, and their isoforms, that appear to be biochemically involved in the appearance of A1BG products.
A concept search of the National Institutes of Health NCBI gene database (http://www.ncbi.nlm.nih.gov/gene/) may provide possible genes (and associated isoforms and variants) participating in or causing altered expressions of A1BG.
Cancers up-regulated A1BG
Pancreatic ductal adenocarcinoma
Pancreatic juice is "an exceptionally rich source of proteins which are released from pancreatic cells in the physiological state".[3] Matrix metalloproteinase-9 (MMP-9), A1BG, and oncogene DJ-1 occur in the pancreatic cancer juice.[3] "MMP-9, DJ-1 and A1BG [are] positively expressed in 82.4%, 72.5% and 86.3% of pancreatic cancer tissues, significantly higher than that in normal pancreas tissues."[3]
Silicosis or dust-exposed workers without silicosis
"Compared with [healthy individuals] HI, 42 proteins were more abundant and 8 were less abundant in [dust-exposed workers without silicosis] DEW, and these were also differentially accumulated in [silicosis patients] SP. Closer inspection revealed that serine protease granzyme A, alpha-1-B-glycoprotein (A1BG) and the T4 surface glycoprotein precursor (TSGP) were among the up-regulated proteins in DEW and SP. Significant changes in serine proteases, glycoproteins and proto-oncogenes may be associated with the response to cytotoxicity and infectious pathogens by activation of T cells, positive regulation of extracellular matrix structural constituents and immune response, and fibroblast proliferation."[4]
Endometrial cancer, cervical cancer and cervical squamous cell carcinoma
"A1BG was reportedly involved in a number of diseases such as endometrial cancer, cervical cancer and cervical squamous cell carcinoma (30), and was also elevated in hepatic fibrosis patients, which allowed such patients to be easily distinguished from HI (31)."[4]
Lung fibrosis
"Up-regulation of both A1BG and TSGP in DEW indicated that these proteins may be involved in the early regulation of fibrosis in the lung and may be potentially useful for diagnosis factors."[4]
Cervical intraepithelial neoplasia
"The present study investigated five sera samples from females who were Human Papilloma Virus (HPV) 16+ and who had been histopathologically diagnosed with CIN III, as well as five sera samples from healthy control females who were HPV-negative. Protein separation was performed using two-dimensional (2D) gel electrophoresis and the proteins were stained with Colloidal Coommassie Blue. Quantitative analysis was performed using ImageMaster 2D Platinum 6.0 software. Peptide sequence identification was performed using a nano-LC ESIMS/MS system. The proteins with the highest Mascot score were validated using western blot analysis in an additional 55 sera samples from the control and CIN III groups. The eight highest score spots that were found to be overexpressed in the CIN III sera group were identified as α-1-B glycoprotein (A1BG), complement component 3 (C3), a pro-apolipoprotein, two apolipoproteins and three haptoglobins. Only A1BG and C3 were validated using western blot analysis, and the bands were compared between the two groups using densitometry analysis. The relative density of the bands of A1BG and C3 was found to be greater in all of the serum samples from the females with CIN III, compared with those of the individuals in the control group."[5]
Colon carcinoma
Liver metastasis-associated A1BG in human colon carcinoma is up-regulated.[6]
Cancers down-regulated A1BG
A1BG is generally down-regulated in small-cell lung cancer (SCLC) samples.[7]
"Downregulation of serum A1BG and afamin in non-131I-avid lung metastases of [papillary thyroid carcinoma] PTC was validated in an independent set of serum samples (serum samples from 25 PTC patients with 131I-avid lung metastases and 25 PTC patients with non-131I-avid lung metastases) by western blotting to evaluate their potential as serum biomarkers."[8]
Female expressions
"Expression of the a1bg gene in rat liver is specifically induced by the female pattern of [growth hormone] GH secretion."[9]
"Expression of reporter constructs showed that the 160 bp proximal part of the a1bg promoter contained elements directing sex-specific expression."[9]
"a1bg gene sequences [are] more represented in the [suppressive subtractive hybridization] SSH assay than CYP2C12 sequences, which indicates a strong female expression of a1bg. In a subsequent study, we confirmed a female-specific and GH-dependent expression of the a1bg gene (Gardmo et al. 2001). The gene encodes a plasma protein, α1B-glycoprotein (A1BG), for which a function has remained elusive for a long time. However, A1BG was recently shown to bind cystein-rich secretory protein-3 (CRISP-3) with high affinity, and was suggested to protect against potentially toxic effects of CRISP-3 in the circulation (Udby et al. 2004)."[9]
"Genome walking (Rat GenomeWalker Kit, Clontech) was used to clone 2.3 kb of the 5′ flanking region of the rat a1bg gene into a T/A vector (AdvanTAge PCR Cloning Kit, Clontech). The following gene-specific primers, GSP1-ACCAGTGTGAGGTTTGCCCAGGGT-TCCA and GSP2-GAGGCTCTGCCCGTAGTTCAGGTT-CACT were used."[9]
"A hydrodynamics-based procedure for in vivo transfection of mice (Liu et al. 1999, Zhang et al. 1999) was adapted for rats. Isofluran-anaesthetized rats were injected in the tail vein with 100 μg plasmid in saline, 50 μg a1bg reporter construct and 50 μg transfection control vector pRL-TK (Promega; the vector contains the herpes simplex thymidine kinase promotor infront of the Renilla luciferase gene). The injected saline volume corresponded to approximately 8–9% of the rats’ body weight and the injection was completed in < 15 s. After 20-h transfection, the animals were decapitated and the liver excised. Samples were prepared by homogenizing 400 μg liver in 2 ml passive lysis buffer (Promega) using a glass teflon homogenizer. The homogenates were centrifuged for 5 min at 12 000 g and 4 °C, and the supernatants were stored at −70 °C."[9]
"The template for the footprint was obtained by PCR amplification of the rat a1bg sequence from −199 to +29 using the 5′ primer GATCTCAGCTCTGGTGCATGCT and the 3′ primer used for subcloning of the a1bg promoter following genome walking. The 5′ oligonucleotide was end-labelled with γ-32P ATP (Amersham) using T4 polynucleotide kinase and purified with QiaQuick nucleotide Purification Kit (Qiagen) prior to the PCR. The 5′ primer was furnished with an extra G-nucleotide at the 5′ end in order to improve the end-labelling. For the binding reactions 2, 5 and 10 μg nuclear protein extract from female rats were mixed with 1 μg poly(dI-dC), 25 μg BSA and 150 000 c.p.m. of labelled template in a buffer containing 20 mM Tris pH 8, 10 mM MgCl2, 40 mM KCl and 0.2 mM EDTA [ethylenediaminetetraacetic acid]. The binding reactions were incubated on ice for 45 min followed by DNase I treatment (no. 104132, Roche Diagnostics) for 2 min at room temperature. The concentrations of DNase I were 0.6 and 0.3 ng/μl for samples with and without nuclear extracts respectively. To terminate the digestion, 1 vol. of buffer (0.06 M EDTA, 0.4 M ammonium acetate and 12.5 μg/ml ssDNA) was added after which the reactions were phenol/chloroform extracted and precipitated. The pellets were dissolved in gel-loading buffer (80%(v/v) deionized formamide, 1 mM EDTA pH 8 and 0.1%(w/v) bromophenol blue), heated at 95 °C before separation on a 6% sequencing gel alongside with sequencing reactions. The gels were dried on Whatmann paper and exposed to X-ray film. The sequencing reactions were made with Sequenase version 2.0 DNA polymerase and T7 Sequencing mixes, both from USB Corp (Cleveland, OH, USA), together with the end-labelled primer used for the PCR amplification of the footprint template."[9]
The "a1bg promoter and rapid tail vein injection for transfection of hepatocytes in vivo can be used to investigate molecular mechanisms whereby GH induces female-specific expression of a gene."[9]
Both "the 2.3 kb and the 160 bp proximal parts of the a1bg promoter direct sex-specific expression of the reporter gene, and that a negative regulatory element resides in the −1 kb to −160 bp region."[9]
"Computer analysis of the 2.3 kb a1bg promoter fragment revealed two putative Stat5 sites and one [hepatic nuclear factor 6] HNF6/HNF3 binding site at −2077/−2069, −69/−61 and −137/−128 respectively [...]."[9]
The "GH-dependent sexually dimorphic expression conveyed by the 2.3 kb a1bg promoter is enhanced by the HNF6/HNF3 site and, if anything, reduced by the proximal Stat5 site in that the impact of the 3′Stat5 mutation was more pronounced in males."[9]
The "binding of Stat5 and HNF6 to the respective site by electromobility shift analysis (EMSA) [was verified] using female-derived liver nuclear extracts. [...] Stat5 bound to the a1bg proximal [signal transducer and activator of transcriptions] Stat5 site, 3′Stat5 and the mutated oligonucleotide was unable to compete for the binding. Similarly, HNF6 bound to the a1bg HNF6 oligonucleotide, but in this case, the mutated oligonucleotide was able to compete for binding when added in large excess [...]. However, [...] the HNF6 binding capacity of the mutated oligonucleotide was clearly reduced. A 20 molar excess of the mutated oligonucleotide had only a marginal effect on the binding of HNF6 (Fig. 3C⇓, lane 6), whereas a 20 molar excess of unlabelled probe [...] completely abolished binding. Supershift analysis with an HNF6 antibody revealed a complex with a slightly lower mobility than the HNF6 complex [...]. By extending the electrophoresis run and including nuclear extract from hypophysectomized rats, devoid of GH and thereby lacking HNF6 (Lahuna et al. 1997), the two different complexes were clearly visualized. The complex with the lower mobility is most probably due to the binding of HNF3, in analogy with what was shown by Lahuna et al. for the CYP2C12 HNF6 binding site; HNF3 can bind to the site in the absence of HNF6 (Lahuna et al. 1997). To summarize the EMSA results, Stat5 and HNF6 could bind to their respective site in the a1bg promoter in vitro, and the mutations introduced in respective site abolished binding of the corresponding factor."[9]
The "expression of a −116/−89 deletion construct in which also the HNF6 site was mutated, (−116/−89)delmutHNF6-Luc, [...] the generated luciferase activities were reduced in both sexes [...]. This is in contrast to that mutation/deletion of the sites separately only affected the expression in female livers."[9]
The "−116/−89 region contains a site(s) of importance for the GH-dependent and female-specific expression of the a1bg gene, and that the impact of this region together with the HNF6 site is more complex than mere enhancement of the expression in females."[9]
The "Stat5 site conveys expression of a1bg to higher extent in male than in female livers, thereby reducing the sex difference. [...] On the other hand, HNF6 is expressed at higher levels in female than in male rat liver (Lahuna et al. 1997). Indeed, following mutation of the HNF6-binding element, mutHNF6-Luc, the sex-differentiated expression was attenuated due to reduced expression in females. Thus, for a1bg, the sex-related difference in amount of HNF6 is likely to contribute to the sex-differentiated and female characteristic expression."[9]
Chromatin "condensation [may have] a role in sex-differentiated expression of a1bg."[9]
The "−116/−89 region together with the HNF6 site are regions of importance for the female characteristic expression of the a1bg gene."[9]
Nuclear "proteins binding to the a1bg −116/−89 region [are] members of the [nuclear factor 1] NF1 and the [octamer transcription factor] Oct families of transcription factors. NF1 genes are expressed in most adult tissues (Osada et al. 1999). It is not known how NF1 modulates transcriptional activity, and both activation and repression of transcription have been reported (Gronostajski 2000). Cofactors such as CBP/p300 and HDAC have been shown to interact with NF1 proteins suggesting modulation of chromatin structure (Chaudhry et al. 1999). NF1 factors have also been shown to interact directly with the basal transcription machinery as well as with other transcription factors, including Stat5 (Kim & Roeder 1994, Mukhopadhyay et al. 2001) and synergistic effects with HNF4 have been reported (Ulvila et al. 2004). In addition to the HNF6, Stat5 and NF1/Oct sites, the a1bg promoter harbours an imperfect HNF4 site at −51/−39 with two mismatches compared with the HNF4 consensus site. HNF4 is clearly important for the expression of CYP2C12 (Sasaki et al. 1999), however, the −51/−39 region in a1bg was not protected in the footprinting analysis and was therefore not analysed further. Like NF1, Oct proteins have been reported to be involved in activation as well as repression of gene expression (Phillips & Luisi 2000). Stat5 has been shown to form a stable complex with Oct-1 (Magne et al. 2003). Moreover, NF1 and Oct-1 have been shown to, reciprocally, facilitate each other’s binding (O’Connor & Bernard 1995, Belikov et al. 2004)."[9]
In the diagram on the right is liver "expression of a1bg-luciferase constructs. (A) Stat5 and HNF6 consensus sequences and corresponding sites in the 2.3 kb a1bg promoter alongside with the used mutations. (B) Female (black bars) and male (open bars) rats [results]."[9]
Gene IDs
- GeneID: 1 A1BG alpha-1-B glycoprotein.
- GeneID: 310 ANXA7 annexin A7.
- GeneID: 368 ABCC6 ATP binding cassette subfamily C member 6.
- GeneID: 1026 CDKN1A cyclin-dependent kinase inhibitor 1A.
- GeneID: 2886 GRB7 growth factor receptor bound protein 7.
- GeneID: 5980 REV3L REV3 like, DNA directed polymerase zeta catalytic subunit.
- GeneID: 6472 SHMT2 serine hydroxymethyltransferase 2.
- GeneID: 6606 SMN1 survival of motor neuron 1, telomeric.
- GeneID: 6622 SNCA synuclein alpha.
- GeneID: 7083 TK1 thymidine kinase 1.
- GeneID: 10321 CRISP3 cysteine rich secretory protein 3.
- GeneID: 10549 PRDX4 peroxiredoxin 4.
- GeneID: 80854 SETD7 SET domain containing lysine methyltransferase 7.
- GeneID: 284161 GDPD1 glycerophosphodiester phosphodiesterase domain containing 1.
Gene clusters
GeneID: 348 APOE apolipoprotein E description also contains this: "This gene maps to chromosome 19 in a cluster with the related apolipoprotein C1 and C2 genes."
Although genes on chromosome 19 may not be expressed when A1BG is expressed, they may be close enough or part of the cluster that is activated. Each of these would then be checked against the NASA database and the open literature searchable with Google Scholar or other search engines.
GeneID: 718, C3, complement component 3, 19p13.3-p13.2.
GeneID: 2524, SE, fucosyltransferase 2, 19q13.3.
GeneID: 4059, LU, basal cell adhesion molecule (Lutheran blood group), 19q13.2.
GeneID: 1, A1BG, alpha-1-B glycoprotein, 19q13.4.
"The most likely order would appear to be C3-SE-LU-A1BG."[10]
Gene regulations
Each gene, or its isoforms, is likely to have upregulation and downregulation transcription factors. As each gene is investigated, these enhancers and inhibitors are noted as discovered.
Gene similarities
There are genes on other chromosomes that are similar to each gene being considered.
A "metalloproteinase inhibitor designated oprin (opossum [Didelphis virginiana] proteinase inhibitor) [...] is a single-chain glycoprotein (26% carbohydrate) with an estimated Mr = 52,000, pI = 3.5, and E(1%/1 cm) = 11. Oprin inhibited snake venom metalloproteinases [...] Incubation of Crotalus atrox alpha-proteinase (EC 3.4.24.1) with oprin, and analysis of the reaction products by chromatography on Mono Q HR 5/5 and by electrophoresis under nondenaturing conditions, indicated formation of an inactive enzyme/inhibitor complex. The complex dissociated during SDS/polyacrylamide gel electrophoresis. An opossum liver cDNA library was immunoscreened, and clones containing cDNA encoding for part of the open reading frame for oprin were isolated. The cDNA inserts contained nucleotide sequences corresponding to two internal amino acid sequences of oprin which had been separately determined by protein sequence analysis. Protein database screening using a 211 amino acid sequence deduced from one of the cDNA inserts showed no significant homology to known proteinase inhibitors. There was, however, a 36% identity with human alpha 1B-glycoprotein, a plasma protein of unknown function related to the immunoglobulin supergene family. In addition, the amino-terminal sequence of oprin showed 46% identity with human alpha 1B-glycoprotein in a 26 amino acid residue overlap."[11]
"DM64 is an acidic protein showing 15% glycosylation and with a molecular mass of 63 659 Da when analysed by MALDI-TOF MS. It was cloned and the amino acid sequence was found to be homologous to DM43, a metalloproteinase inhibitor from D. marsupialis serum, and to human alpha1B-glycoprotein, indicating the presence of five immunoglobulin-like domains."[12]
"DM64 has the same high similarity (78%) with DM43 and oprin. In addition, 50% similarity was found with human α1B-glycoprotein, a plasma protein of unknown function and a member of the immunoglobulin supergene family [41] [...]. Each domain of these proteins possesses two cysteine residues at conserved positions [...]. DM64 also presented four putative N-glycosylation sites [...], three of them aligning to the same DM43 sites [...]. A gap of four amino acids beginning after residue 242 of DM64 is also present in human α1B-glycoprotein. Such gap was not found on the third domain of DM43."[12]
Human DNA
"[H]uman DNA has millions of on-off switches and complex networks that control the genes' activities. ... [A]t least 80% of the human genome is active, which opposed the previously held idea that most of the DNA are useless."[13]
"DNA contains genes, which hold the instructions for [life. But, these] take up only about 2 percent of the genome ... The human genome is made up of about 3 billion “letters” along strands that make up the familiar double helix structure of DNA. Particular sequences of these letters form genes, which tell cells how to make proteins. People have about 20,000 genes, but the vast majority of DNA lies outside of genes. ... [A]t least three-quarters of the genome is involved in making RNA [...] it appears to help regulate gene activity."[14]
There are "more than 4 million sites where proteins bind to DNA to regulate genetic function, sort of like a switch."[14]
Over 50% of human DNA consists of non-coding repetitive sequences.[15]
Some DNA sequences may encode functional non-coding RNA molecules, which are involved in the regulation of gene expression.[16]
About 2700 formerly active genes are now pseudogenes. Additional DNA is used in introns and for centromeres and telomeres.
Some introns themselves encode specific proteins or can be further processed after splicing to generate noncoding RNA molecules.[17]
Single-nucleotide polymorphisms
"Hypertension is the most common chronic disease in the United States, affecting approximately one third of the adult population, and is a major risk factor for acute myocardial infarction (MI), stroke, heart failure, and renal failure.1 Numerous antihypertensive drugs are considered appropriate first-line therapy to lower blood pressure (BP), including diuretics, β-blockers (βB), calcium channel blockers (CCB), angiotensin-converting enzyme inhibitors, and angiotensin receptor blockers. These drugs are ultimately prescribed to prevent the long-term cardiovascular complications of hypertension.2 However, there is great interpatient variability in antihypertensive drug response, with only ≈50% of patients achieving an adequate BP response to any 1 drug,3 and limited data are available to guide treatment selection. Why patients respond differently to the same drug and why some patients experience adverse cardiovascular outcomes, despite BP control, while others do not, remains poorly understood. Pharmacogenomics aims to identify genetic markers that are associated with drug response, outcomes, and adverse events."[18]
A nonsynonymous (ns) single-nucleotide polymorphism (SNP) "rs893184, located in A1BG (α-1-B glycoprotein; white interaction P=0.0248, Hispanic interaction P=0.0310, and combined interaction P=0.0036; Table 2; Figure S1B). rs893184 causes a histidine (His) to arginine (Arg) substitution at amino acid position 52 in A1BG. His carriers had a lower risk for the [primary outcome] PO in the [calcium channel blockers] CCB strategy in whites and Hispanics, whereas Arg/Arg patients had a higher risk for the PO in the CCB strategy."[18]
"The α-1-B glycoprotein precursor (A1BG) is also located at 19q13.4 (6.85 Mb away from SIGLEC12) and encodes a plasma glycoprotein with unknown function. The A1BG protein is a member of the immunoglobulin superfamily and has been shown to form a complex with cysteine-rich secretory protein 3, a protein present in neutrophilic granulocytes that is thought to play a role in innate immunity.15"[18]
Alleles
Def. one of a number of alternative forms of the same gene occupying a given position on a chromosome is called an allele.
"The distribution of plasma α1B-glycoprotein (A1BG) was determined by a two-dimensional electrophoresis (agarose-polyacrylamide gel) followed by protein staining in a group of 1099 individuals from 11 populations of the Indian subcontinent. The sample comprised 454 from several tribes of Arunachal Pradesh; 76 Bengali Hindus and 88 Bengali Muslims; 179 Tamil Hindus from Singapore and 107 from India; 81 Tamil Muslims, 48 Sinhalese from Sri Lanka and 66 North Indians. Three common A1BG phenotypes (1–1, 1–2 and 2–2) were observed in this study. One each of a new allele (A1BG*7) in heterozygous form (1–7) was detected respectively among Tamil Hindus of India and Singapore. The phenotypic distribution of A1BG alleles was at Hardy-Weinberg equilibrium in all the populations. The frequency of A1BG*2 was in general lower in the Mongoloid tribes of Arunachal Pradesh (0·043–0·104) and North Indians (0·068) compared to that in other Indian populations (0·130–0·171) and Sinhalese (0·208)."[19]
Epigenomes
Inside each eukaryote nucleus is genetic material (DNA) surrounded by protective and regulatory proteins. These protective and regulatory proteins and the dynamic changes to them that occur during the course of a eukaryote's existence are the epigenome.
There are "nearly 50,000 acetylated sites [punctate sites of modified histones] in the human genome that correlate with active transcription start sites and CpG islands and tend to cluster within gene-rich loci."[20]
Any of the epigenome sites may be influenced during or before transcription to modify gene expressions.
Alternate sites
Alternate transcription start sites may be needed. These include those on the coding strand, from the opposite direction (other end of the gene) on the same strand, additional start sites of isoforms, other start sites for the same product, ncRNA or miscRNA start sites within the gene, and pseudogenes.
To induce the transcription mechanism to relocate to an alternate site, a modification to a transcription factor may be required.
Strand transfers
These two genes produce versions of integrase which appear to facilitate strand transfer from the coding strand to the template strand using a retrotransposon:
- GeneID: 54826 GIN1 gypsy retrotransposon integrase 1 and
- GeneID: 57523 NYNRN NYN domain and retroviral integrase containing.
"The human testis Rad51 protein, a structural homolog of E. coli RecA, binds single- and double-stranded DNA and exhibits DNA-dependent ATPase activity. [...] hRad51 promotes homologous pairing and strand exchange reactions in vitro. Joint molecule formation was dependent upon ATP hydrolysis and DNA homology and was stimulated by the single-strand DNA-binding protein RP-A. In these reactions, the 5′ terminus of the complementary strand of the linear duplex was efficiently transferred to the ssDNA. However, under standard conditions, extensive strand exchange was not observed. These results establish hRad51 as a functional homolog of RecA, but indicate that the human protein and its bacterial counterpart differ in their ability to promote extensive strand transfer. It is proposed that hRad51 mediates homology recognition and initiates strand exchange, but that extensive heteroduplex formation in higher organisms requires the actions of additional proteins."[21]
Mutational "asymmetry has acted over long periods of time to produce a compositional asymmetry, an excess of G+T over A+C on the coding strand, in most genes [of mammals]."[22]
Double strand transcriptions
However, in plants, most siRNAs are generated by RNA-dependent RNA polymerase.[23]
Single strand RNA transcripts: ssRNA. RNA interference requires that two base pair-complementary strands of RNA to come together to form double stranded RNA.[24]
Gene transcriptions
The detection of the gene product presumes that transcription occurs and may suffice as proof of concept.
Def. "the factors, including RNA polymerase II itself, that are minimally essential for transcription in vitro from an isolated core promoter"[25] are called the basal transcription machinery.
RNA polymerase II holoenzyme complex finds and uses the transcription start site. Once the DNA double helix and its associated epigenome have been melted so that the template strand is available for binding, a transcription factor binds to a specific nucleotide sequence to biochemically influence gene transcription.
| Transcription factor glossary |
|---|
| • coactivator – a protein that works with transcription factors to increase the rate of gene transcription |
| • corepressor – a protein that works with transcription factors to decrease the rate of gene transcription |
| • downregulation, repression, or suppression – decrease the rate of gene transcription |
| • factor – a substance, such as a protein, that contributes to the cause of a specific biochemical reaction or bodily process |
| • general transcription factor – a transcription factor that activates gene transcription |
| • gene transcription - copying of DNA into messenger RNA by RNA polymerase |
| • transcriptional regulation – modulating the rate of gene transcription |
| • upregulation, activation, or promotion – increase the rate of gene transcription |
| edit |
Immunoglobulin domains
The immunoglobulin domain is a type of protein domain that consists of a 2-layer sandwich of between 7 and 9 antiparallel β-strands arranged in two β-sheets with a Greek key topology.[26][27]
Angiotensin-converting enzyme
The angiotensin I converting enzyme (ACE) is involved in catalyzing the conversion of angiotensin I into a physiologically active peptide angiotensin II and has a testicular form variant (2).
ACE plays a key role in the renin-angiotensin system.
"CRISP proteins have been shown to be involved in various functions related to sperm–oocyte fusion, innate host defense function and ion channel blockage."[28]
"Multiple members of the CRISP family have been identified in the mammalian male genital tract (CRISP1, CRISP2 and CRISP3)."[28]
"[T]here is evidence that prostate cancer patients with higher levels of CRISP3 have a smaller probability of recurrence-free outcomes [13]."[29]
A1BG-CRISP3
The human cysteine-rich secretory protein (CRISP3) "is present in exocrine secretions and in secretory granules of neutrophilic granulocytes and is believed to play a role in innate immunity."[30] CRISP3 has a relatively high content in human plasma.[30]
"The A1BG-CRISP-3 complex is noncovalent with a 1:1 stoichiometry and is held together by strong electrostatic forces."[30] "Similar [complex formation] between toxins from snake venom and A1BG-like plasma proteins ... inhibits the toxic effect of snake venom metalloproteinases or myotoxins and protects the animal from envenomation."[30]
Opossums [such as shown at top of the article] have a remarkably robust immune system, and show partial or total immunity to the venom of rattlesnakes, [Agkistrodon piscivorus] cottonmouths, and other [Crotalinae] pit vipers.[31][32]
"Crisp3 [is] mainly [expressed] in the salivary glands, pancreas, and prostate."[33] "CRISP3 is highly expressed in the human cauda epididymidis and ampulla of vas deferens (Udby et al. 2005)."[33]
Genomic product
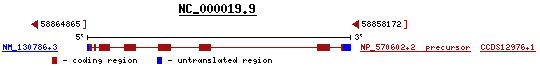
As indicated in the diagram at left, there are eight exons (red rectangles) and seven introns (red lines) between the 5' untranslated region (UTR, 5'-UTR) and the 3'-UTR.
When A1BG is transcribed by the RNA polymerase II holoenzyme, the pre-mRNA (messenger RNA, mRNA) consists of eight exons and seven introns which are spliced to yield the final mRNA. A1BG is a minus strand transcription. The included nucleotides as numbered in the human genome go from 3'-58858172 to 58864864-5' inclusive and are so transcribed. When the mRNA template 5'-58858172 to 58864864-3' is used to create the protein, the translation proceeds 3'-58864864 to 58858172-5' rather than 5'-58858172 to 58864864-3'.
Strands
Eukaryotes have a double helix of DNA surrounded by an epigenome.
A single strand of DNA [has a positive sense (+)] if an RNA version of the same sequence is translated or translatable into protein. Its complementary strand is called antisense (or negative (-) sense).
The two complementary strands of double-stranded DNA (dsDNA) are usually differentiated as the "sense" strand and the "antisense" strand. The DNA antisense strand serves as the source for the protein code, because, with bases complementary to the DNA sense strand, it is used as a template for the mRNA.
The only real biological information that is important for labeling strands is the location of the 5' phosphate group and the 3' hydroxyl group because these ends determine the direction of transcription and translation.
From a molecular point of view, a transcription complex may not have an obvious way to choose the "antisense" strand from the "sense" strand. It also may not have an obvious way to chose the direction of transcription once a strand has been chosen as the "antisense" strand.
To choose which strand, then which direction, may require chemical cues. A1BG has two sections of nucleotides between itself and neighboring genes:
- between ZSCAN22 and A1BG and
- between ZNF497 and A1BG.
The NCBI gene database conveniently labels the genomic context with increasing nucleotide numbers in the direction of transcription 3'-5' on the template strand. When viewing the genomic regions, transcripts, and products under tools, click on "Sequence Text View". The database informs you which strand you are looking at (negative strand) or by "Flip Strands" the (positive strand).
Eukaryotic transcription of the A1BG "protein-coding gene is preceded by ...
- decondensation of the locus,
- nucleosome remodeling,
- histone modifications,
- binding of transcriptional activators and coactivators to enhancers and promoters, and
- recruitment of the basal transcription machinery to the core promoter."[25]
Preinitiation complexes
"A stable preinitiation complex can form in vitro on TATA-dependent core promoters by association of the basal factors in the following order: TFIID/TFIIA, TFIIB, RNA polymerase II/TFIIF, TFIIE, and then TFIIH."[25]
Promoters
The A1BG promoter is a region of the DNA adjacent to the gene itself that facilitates gene transcription. Within the overall promoter are response elements which provide a secure initial binding site for the RNA polyermase II holoenzyme.
Positions in the promoter are designated relative to the transcription start site (N+1, TSS), with positions upstream having negative numbers counting back from -1 away from the TSS, for example, -100 is 100 nucleotides (nts) before 3'-58858172 (specifically pre-3' at 58858072).
Focused promoters
"In focused transcription, there is either a single major transcription start site or several start sites within a narrow region of several nucleotides. Focused transcription is the predominant mode of transcription in simpler organisms."[2]
"Focused transcription initiation occurs in all organisms, and appears to be the predominant or exclusive mode of transcription in simpler organisms."[2]
"In vertebrates, focused transcription tends to be associated with regulated promoters".[2]
"The analysis of focused core promoters has led to the discovery of sequence motifs such as the TATA box, BREu (upstream TFIIBrecognition element), Inr (initiator), MTE (motif ten element), DPE (downstream promoter element), DCE (downstream core element), and XCPE1 (Xcore promoter element 1) [...]."[2]
Dispersed promoters
"In dispersed transcription, there are several weak transcription start sites over a broad region of about 50 to 100 nucleotides. Dispersed transcription is the most common mode of transcription in vertebrates. For instance, dispersed transcription is observed in about two-thirds of human genes."[2]
In vertebrates, "dispersed transcription is typically observed in constitutive promoters in CpG islands."[2]
Long-non-coding RNAs
Long-non-coding RNAs (lncRNAs) can act either as a tumor suppressor or oncogenes in hepatocellular carcinoma (HCC) via regulating tumor‐related genes or miRNA expression in tumorigenesis.[34]
The lncRNA A1BG antisense RNA 1 (A1BG‐AS1) acts as a competing endogenous RNAs (ceRNAs), inhibiting HCC proliferation and invasion via sponging miR‐216a‐5p.[34]
Expression of lncRNA A1BG-AS1 is up-regulated in breast cancer tissues compared to their normal counterpart, whereas depletion of A1BG-AS1 suppressed the proliferation, migration, and invasion of breast cancer cells.[35]
Sex-differential "chromatin interactions involving sex-biased gene promoters, enhancers, and lncRNAs were associated with sex-biased binding of cohesin and/or CTCF."[36]
"3D genome organization impacts sex-biased gene expression in a non-reproductive tissue through both direct and indirect effects of cohesin and CTCF looping on distal enhancer interactions with sex-differentially expressed genes."[36]
"Distal enhancer viewpoint near [mouse] A1bg and female-biased [long, non-coding RNAs] lncRNAs [...] includes 12 female-biased and nuclear-enriched mono-exonic lncRNAs, which fall into three clusters. The lncRNAs in each cluster are all transcribed from the same strand, [...] and [positive] direction of transcription of the most upstream lncRNA in each cluster."[36]
"Robust interactions were observed in female but not male livers between the viewpoint enhancer and three genomic regions [...]: a strong female-biased enhancer [...], the promoter of A1bg [...], and a region downstream of A1bg that contains a cluster of four female-specific lncRNAs [...], where [...] the strongest interactions [occurred]. The lncRNAs in this cluster are more highly expressed [...] and are more consistently female-biased across various RNA-seq datasets than the other two lncRNA clusters [...]."[36]
"The precise relationship between the female-biased expression of these lncRNAs and the female-bias in 3D interactions with the distal enhancer is not known. The interaction may be regulatory in nature (e.g., an enhancer–promoter interaction, as with any gene) or it could be facilitated by one or more of the 12 nuclear-enriched, female-biased lncRNAs, as was described for the lncRNAs Xist [65], Firre [66], and Haunt [67]. Alternatively, the female-specific interactions shown may be primarily those of regulatory enhancers driving expression of several female-specific genes—including A1bg and multiple lncRNAs. The female-biased CTCF binding seen at both interacting regions [...] lends mechanistic support for the latter proposal, with CTCF mediating enhancer–promoter and enhancer–enhancer interactions. As CTCF is known to interact with lncRNAs in a functional manner, and with high affinity [68], these two mechanisms are not mutually exclusive; one or more of these highly female-specific lncRNAs [...] could function in a cis-acting manner to selectively guide CTCF binding and interactions unique to female liver."[36]
Mouse a1bg
The mouse A1bg gene is not conserved in human and it is not expressed in mouse mammary gland.[37]
Female-biased "proximal enhancer–promoter interactions in [a] gene region associated with female-biased cohesin binding, [and] female-biased interactions between the A1bg promoter, a far distal (> 100 kb) enhancer, and distal female-biased CTCF binding sites [were revealed]."[36]
"A1bg exemplifies direct regulation, where female-biased CTCF binding can explain the observed female-bias in looping interactions."[36]
"In [mouse liver], the highly female-biased gene A1bg is nearby several strongly female-biased, nuclear-enriched mono-exonic lncRNAs, several of which are transcribed from genomic loci that show female-specific interactions with the distal female-specific enhancer viewpoint [...]."[36]
While "most sex-biased binding sites for CTCF and cohesin were found to be distal from sex-biased genes, a subset likely contributes to sex-biased looping between regulatory elements in cis, as exemplified by the female-biased DNA looping interactions observed for A1bg."[36]
Distal promoters
A distal promoter is a portion of the promoter for a particular gene. This distal sequence upstream of the gene is a region of DNA that may contain additional regulatory elements, often with a weaker influence than the proximal promoter.
"[T]he distal promoter ... can range several thousands of nucleotides upstream of the TSS and contains additional regulatory elements called enhancers and silencers."[38]
"Several thousand sex-differential distal enhancers have been identified in mouse liver".[36] Some "1847 mouse liver genomic regions [show] significant sex differential occupancy by cohesin and [CCCTC-binding factor] CTCF, two key 3D nuclear organizing factors. These sex-differential binding sites were primarily distal to sex-biased genes but rarely generated sex-differential TAD (topologically associating domain) or intra-TAD loop anchors, and were sometimes found in TADs without sex-biased genes."[36]
"Cohesin depletion reduced the expression of male-biased genes with distal, but not proximal, sex-biased enhancers by >10-fold, implicating cohesin in long-range enhancer interactions regulating sex-biased genes."[36]
Sex "differences in distal sex-biased enhancer–promoter interactions are common. Intra-TAD loops with sex-independent cohesin-and-CTCF anchors conferred sex specificity to chromatin interactions indirectly, by insulating sex-biased enhancer–promoter contacts and by bringing sex-biased genes into closer proximity to sex-biased enhancers."[36]
AGC boxes
An AGC box has the consensus sequence AGCCGCC in the direction of transcription. It may also occur as TCGGCGG in the direction of transcription, or inverted which has been reported: CCGCCGA and GGCGGCT. Ideally, each of these four should be tested on each of the four possible transcription directions.
A1BG has four possible transcription directions:
- on the negative strand from ZSCAN22 to A1BG,
- on the positive strand from ZSCAN22 to A1BG,
- on the negative strand from ZNF497 to A1BG, and
- on the positive strand from ZNF497 to A1BG.
For each transcription promoter that interacts directly with RNA polymerase II holoenzyme, the four possible consensus sequences need to be tested on the four possible transcription directions, even though some genes may only be transcribed from the negative strand in the direction on the transcribed strand.
For the Basic programs (starting with SuccessablesAGC.bas) written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), including extending the number of nts from 958 to 4445, the programs are, are looking for, and found:
- negative strand in the negative direction is SuccessablesAGC--.bas, looking for AGCCGCC, 0,
- negative strand in the positive direction is SuccessablesAGC-+.bas, looking for AGCCGCC, 0,
- positive strand in the negative direction is SuccessablesAGC+-.bas, looking for AGCCGCC, 0,
- positive strand in the positive direction is SuccessablesAGC++.bas, looking for AGCCGCC, 0,
- complement, negative strand, negative direction is SuccessablesAGCc--.bas, looking for TCGGCGG, 0,
- complement, negative strand, positive direction is SuccessablesAGCc-+.bas, looking for TCGGCGG, 0,
- complement, positive strand, negative direction is SuccessablesAGCc+-.bas, looking for TCGGCGG, 0,
- complement, positive strand, negative direction is SuccessablesAGCc++.bas, looking for TCGGCGG, 0,
- inverse complement, negative strand, negative direction is SuccessablesAGCci--.bas, looking for GGCGGCT, 0,
- inverse complement, negative strand, positive direction is SuccessablesAGCci-+.bas, looking for GGCGGCT, 0,
- inverse complement, positive strand, negative direction is SuccessablesAGCci+-.bas, looking for GGCGGCT, 1, GGCGGCT, 1754,
- inverse complement, positive strand, positive direction is SuccessablesAGCci++.bas, looking for GGCGGCT, 0,
- inverse, negative strand, negative direction, is SuccessablesAGCi--.bas, looking for CCGCCGA, 1, CCGCCGA, 1754,
- inverse, negative strand, positive direction, is SuccessablesAGCi-+.bas, looking for CCGCCGA, 0,
- inverse, positive strand, negative direction, is SuccessablesAGCi+-.bas, looking for CCGCCGA, 0,
- inverse, positive strand, positive direction, is SuccessablesAGCi++.bas, looking for CCGCCGA, 0.
Enhancer boxes
An E-box (Enhancer Box) is a DNA sequence which usually lies upstream of a gene in a promoter region. It is a transcription factor binding site where the specific sequence of DNA, CANNTG, is recognized by proteins that can bind to it to help initiate its transcription. Once transcription factors bind to promoters, they allow for association of other enzymes which will copy the DNA into mRNA. The consensus sequence for the E-box element is CANNTG, with a palindromic canonical sequence of CACGTG.[39] Transcription factors containing the basic helix-loop-helix protein structural motif typically bind to E-boxes or related variant sequences and enhance transcription of the downstream gene.
For the Basic programs (starting with SuccessablesE.bas) written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), including extending the number of nts from 958 to 4445, the programs are looking for, and found:
- negative strand in the negative direction is SuccessablesE--.bas, looking for C-A-(A/C/G/T)-(A/C/G/T)-T-G, 4, CACATG at 324, CACATG at 797, CACATG at 2213, and CACATG at 2342,
- negative strand in the positive direction is SuccessablesE-+.bas, looking for C-A-(A/C/G/T)-(A/C/G/T)-T-G, 10, CACATG at 141, CACATG at 268, CACATG at 303, CACATG at 388, CACATG at 461, CACATG at 517, CACATG at 714, CACATG at 782, CACATG at 925, CACATG at 931,
- positive strand in the negative direction is SuccessablesE+-.bas, looking for C-A-(A/C/G/T)-(A/C/G/T)-T-G, 15, CACATG at 123, CACATG at 200, CACATG at 952, CACATG at 1206, CACATG at 1849, CACATG at 1952, CACATG at 2151, CACATG at 2276, CACATG at 2322, CACATG at 2533, CACATG at 2613, CACATG at 2667, CACATG at 2751, CACATG at 2783, CACATG at 4106, CACATG at 4116, CACATG at 4247,
- positive strand in the positive direction is SuccessablesE++.bas, looking for C-A-(A/C/G/T)-(A/C/G/T)-T-G, 0,
- complement, negative strand, negative direction is SuccessablesEc--.bas, looking for G-T-(A/C/G/T)-(A/C/G/T)-A-C, 17, GTGTAC at 123, GTGTAC at 200, GTGTAC at 952, GTGTAC at 1206, GTGTAC at 1849, GTGTAC at 1952, GTGTAC at 2151, GTGTAC at 2276, GTGTAC at 2322, GTGTAC at 2533, GTGTAC at 2613, GTGTAC at 2667, GTGTAC at 2751, GTGTAC at 2783, GTGTAC at 4106, GTGTAC at 4116, GTGTAC at 4247,
- complement, negative strand, positive direction is SuccessablesEc-+.bas, looking for G-T-(A/C/G/T)-(A/C/G/T)-A-C, 0,
- complement, positive strand, negative direction is SuccessablesEc+-.bas, looking for G-T-(A/C/G/T)-(A/C/G/T)-A-C, 4, GTGTAC at 324, GTGTAC at 797, GTGTAC at 2213, GTGTAC at 2342,
- complement, positive strand, positive direction is SuccessablesEc++.bas, looking for G-T-(A/C/G/T)-(A/C/G/T)-A-C, 10, GTGTAC, at 141, GTGTAC, at 268, GTGTAC, at 303, GTGTAC, at 388, GTGTAC, at 461, GTGTAC, at 517, GTGTAC, at 714, GTGTAC, at 782, GTGTAC, at 925, GTGTAC, at 931,
- inverse complement, negative strand, negative direction is SuccessablesEci--.bas, looking for C-A-(A/C/G/T)-(A/C/G/T)-T-G, 4, CACATG at 324, CACATG at 797, CACATG at 2213, and CACATG at 2342,
- inverse complement, negative strand, positive direction is SuccessablesEci-+.bas, looking for C-A-(A/C/G/T)-(A/C/G/T)-T-G, 10, CACATG at 141, CACATG at 268, CACATG at 303, CACATG at 388, CACATG at 461, CACATG at 517, CACATG at 714, CACATG at 782, CACATG at 925, CACATG at 931,
- inverse complement, positive strand, negative direction is SuccessablesEci+-.bas, looking for C-A-(A/C/G/T)-(A/C/G/T)-T-G, 15, CACATG at 123, CACATG at 200, CACATG at 952, CACATG at 1206, CACATG at 1849, CACATG at 1952, CACATG at 2151, CACATG at 2276, CACATG at 2322, CACATG at 2533, CACATG at 2613, CACATG at 2667, CACATG at 2751, CACATG at 2783, CACATG at 4106, CACATG at 4116, CACATG at 4247,
- inverse complement, positive strand, positive direction is SuccessablesEci++.bas, looking for C-A-(A/C/G/T)-(A/C/G/T)-T-G, 0,
- inverse, negative strand, negative direction, is SuccessablesEi--.bas, looking for G-T-(A/C/G/T)-(A/C/G/T)-A-C, 17, GTGTAC at 123, GTGTAC at 200, GTGTAC at 952, GTGTAC at 1206, GTGTAC at 1849, GTGTAC at 1952, GTGTAC at 2151, GTGTAC at 2276, GTGTAC at 2322, GTGTAC at 2533, GTGTAC at 2613, GTGTAC at 2667, GTGTAC at 2751, GTGTAC at 2783, GTGTAC at 4106, GTGTAC at 4116, GTGTAC at 4247,
- inverse, negative strand, positive direction, is SuccessablesEi--.bas, looking for G-T-(A/C/G/T)-(A/C/G/T)-A-C, 0,
- inverse, positive strand, negative direction, is SuccessablesEi+-.bas, looking for G-T-(A/C/G/T)-(A/C/G/T)-A-C, 4, GTGTAC at 324, GTGTAC at 797, GTGTAC at 2213, GTGTAC at 2342,
- inverse, positive strand, positive direction, is SuccessablesEi++.bas, looking for G-T-(A/C/G/T)-(A/C/G/T)-A-C, 10, GTGTAC, at 141, GTGTAC, at 268, GTGTAC, at 303, GTGTAC, at 388, GTGTAC, at 461, GTGTAC, at 517, GTGTAC, at 714, GTGTAC, at 782, GTGTAC, at 925, GTGTAC, at 931.
Proximal promoters
A 'proximal promoter' is a proximal sequence upstream of the gene, specifically the transcription start site (TSS) of the gene, that tends to contain primary regulatory elements. It is approximately 250 nucleotides (nts) upstream (signified by a negative sign before the number of nucleotides, eg. -250 nts) of the TSS and has specific transcription factor binding sites.
"[T]he proximal promoter [is] a region containing several regulatory elements, which ranges up to a few hundred nucleotides upstream of the TSS"[38].
CArG boxes
"CArG box [CC(A/T)6GG] DNA [consensus] sequences present within the promoters of SMC genes play a pivotal role in controlling their transcription".[40]
"Serum response factor (SRF) controls [smooth muscle cell] SMC gene transcription via binding to CArG box DNA sequences found within genes that exhibit SMC-restricted expression."[40]
"SMC genes examined in this study display SMC-specific histone modifications at the 5′-CArG boxes."[40]
"The SRF-CArG association is required for transcriptional activation of SMC genes [...] the SMC genes examined in this study display SMC-specific histone modifications at the 5′-CArG boxes. [...] enrichment of H4 and H3 acetylation [...] were relatively low from positions –2,800 to –1,600 in the 5′ region. However, at position –1,600 to –1,200, there was a sharp rise in these modifications, which was increased even further at +400 in the coding region. We observed similar patterns for H3K4dMe and H3 Lys79 di-methylation [...]. SRF, TFIID, and RNA polymerase II displayed enrichments that were consistent with the positions of the CArG boxes, TATA box, and coding region, respectively".[40]
The CArG boxes occur between -400 and -200 nts, between the E boxes and the TCE element.[40]
For the Basic programs (starting with SuccessablesCArG.bas) written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), including extending the number of nts from 958 to 4445, the programs are, are looking for, and found:
- negative strand in the negative direction (from ZSCAN22 to A1BG) is SuccessablesCArG--.bas, looking for 3'-CC(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)GG-5', 0,
- negative strand in the positive direction (from ZNF497 to A1BG) is SuccessablesCArG-+.bas, looking for 3'-CC(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)GG-5', 0,
- positive strand in the negative direction is SuccessablesCArG+-.bas, looking for 3'-CC(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)GG-5', 0,
- positive strand in the positive direction is SuccessablesCArG++.bas, looking for 3'-CC(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)GG-5', 0,
- complement, negative strand, negative direction is SuccessablesCArGc--.bas, looking for 3'-GG(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)CC-5', 0,
- complement, negative strand, positive direction is SuccessablesCArGc-+.bas, looking for 3'-GG(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)CC-5', 0,
- complement, positive strand, negative direction is SuccessablesCArGc+-.bas, looking for 3'-GG(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)CC-5', 0,
- complement, positive strand, positive direction is SuccessablesCArGc++.bas, looking for 3'-GG(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)CC-5', 0,
- inverse complement, negative strand, negative direction is SuccessablesCArGci--.bas, looking for 3'-CC(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)GG-5', 0,
- inverse complement, negative strand, positive direction is SuccessablesCArGci-+.bas, looking for 3'-CC(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)GG-5', 0,
- inverse complement, positive strand, negative direction is SuccessablesCArGci+-.bas, looking for 3'-CC(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)GG-5', 0,
- inverse complement, positive strand, positive direction is SuccessablesCArGci++.bas, looking for 3'-CC(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)GG-5', 0,
- inverse, negative strand, negative direction, is SuccessablesCArGi--.bas, looking for 3'-GG(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)CC-5', 0,
- inverse, negative strand, positive direction, is SuccessablesCArGi-+.bas, looking for 3'-GG(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)CC-5', 0,
- inverse, positive strand, negative direction, is SuccessablesCArGi+-.bas, looking for 3'-GG(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)CC-5', 0,
- inverse, positive strand, positive direction, is SuccessablesCArGi++.bas, looking for 3'-GG(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)CC-5', 0.
Testing the more general 3'-C(C/A/T)(A/T)6(A/G)G-5':
- negative strand in the negative direction (from ZSCAN22 to A1BG) is SuccessablesCArG--.bas, looking for 3'-C(C/A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/G)G-5', 0,
- negative strand in the positive direction (from ZNF497 to A1BG) is SuccessablesCArG-+.bas, looking for 3'-C(C/A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/G)G-5', 0,
- positive strand in the negative direction is SuccessablesCArG+-.bas, looking for 3'-C(C/A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/G)G-5', 2, 3'-CAAAAAAAAG-5', 1399, 3'-CATTAAAAGG-5', 3441,
- positive strand in the positive direction is SuccessablesCArG++.bas, looking for 3'-C(C/A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/G)G-5', 0,
- complement, negative strand, negative direction is SuccessablesCArGc--.bas, looking for 3'-G(A/G/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(C/T)C-5', 2, 3'-GTTTTTTTTC-5', 1399, 3'-GTAATTTTCC-5', 3441,
- complement, negative strand, positive direction is SuccessablesCArGc-+.bas, looking for 3'-G(A/G/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(C/T)C-5', 0,
- complement, positive strand, negative direction is SuccessablesCArGc+-.bas, looking for 3'-G(A/G/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(C/T)C-5', 0,
- complement, positive strand, positive direction is SuccessablesCArGc++.bas, looking for 3'-G(A/G/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(C/T)C-5', 0,
- inverse complement, negative strand, negative direction is SuccessablesCArGci--.bas, looking for 3'-C(C/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/G/T)G-5', 0,
- inverse complement, negative strand, positive direction is SuccessablesCArGci-+.bas, looking for 3'-C(C/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/G/T)G-5', 0,
- inverse complement, positive strand, negative direction is SuccessablesCArGci+-.bas, looking for 3'-C(C/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/G/T)G-5', 0,
- inverse complement, positive strand, positive direction is SuccessablesCArGci++.bas, looking for 3'-C(C/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(A/G/T)G-5', 0,
- inverse, negative strand, negative direction, is SuccessablesCArGi--.bas, looking for 3'-G(A/G)(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(C/A/T)C-5', 0,
- inverse, negative strand, positive direction, is SuccessablesCArGi-+.bas, looking for 3'-G(A/G)(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(C/A/T)C-5', 0,
- inverse, positive strand, negative direction, is SuccessablesCArGi+-.bas, looking for 3'-G(A/G)(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(C/A/T)C-5', 0,
- inverse, positive strand, positive direction, is SuccessablesCArGi++.bas, looking for 3'-G(A/G)(A/T)(A/T)(A/T)(A/T)(A/T)(A/T)(C/A/T)C-5', 0.
HY boxes
The hypertrophy region HY box is between -89 and -60 nucleotides (nts) upstream from the transcription start site.[41]
For the Basic programs (starting with SuccessablesHY.bas) written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), including extending the number of nts from 958 to 4445, the programs are, are looking for, and found:
- negative strand in the negative direction is SuccessablesHY--.bas, looking for 3'-TG(A/T)GGG-5', 1, 3'-TGTGGG-5' at 749,
- negative strand in the positive direction is SuccessablesHY-+.bas, looking for 3'-TG(A/T)GGG-5', 4, 3'-TGTGGG-5' at 11, 3'-TGAGGG-5' at 40, 3'-TGAGGG-5' at 440, 3'-TGTGGG-5' at 956,
- positive strand in the negative direction is SuccessablesHY+-.bas, looking for 3'-TG(A/T)GGG-5', 5, 3'-TGAGGG-5' at 88, 3'-TGAGGG-5' at 2699, 3'-TGAGGG-5' at 3652, 3'-TGTGGG-5' at 3712, 3'-TGAGGG-5' at 4558,
- positive strand in the positive direction is SuccessablesHY++.bas, looking for 3'-TG(A/T)GGG-5', 1, 3'-TGTGGG-5' at 94,
- complement, negative strand, negative direction is SuccessablesHYc--.bas, looking for 3'-AC(A/T)CCC-5', 0,
- complement, negative strand, positive direction is SuccessablesHYc-+.bas, looking for 3'-AC(A/T)CCC-5', 1, 3'-ACACCC-5', 94,
- complement, positive strand, negative direction is SuccessablesHYc+-.bas, looking for 3'-AC(A/T)CCC-5', 1 , 3'-ACACCC-5', 749,
- complement, positive strand, positive direction is SuccessablesHYc++.bas, looking for 3'-AC(A/T)CCC-5', 4, 3'-ACACCC-5', 11, 3'-ACTCCC-5', 40, 3'-ACTCCC-5', 440, 3'-ACACCC-5', 956,
- inverse complement, negative strand, negative direction is SuccessablesHYci--.bas, looking for 3'-CCC(A/T)CA-5', 4, 3'-CCCTCA-5', 2702, 3'-CCCACA-5', 3184, 3'-CCCTCA-5', 3889, 3'-CCCTCA-5', 4498,
- inverse complement, negative strand, positive direction is SuccessablesHYci-+.bas, looking for 3'-CCC(A/T)CA-5', 1, 3'-CCCTCA-5', 64,
- inverse complement, positive strand, negative direction is SuccessablesHYci+-.bas, looking for 3'-CCC(A/T)CA-5', 0,
- inverse complement, positive strand, positive direction is SuccessablesHYci++.bas, looking for 3'-CCC(A/T)CA-5', 0,
- inverse, negative strand, negative direction, is SuccessablesHYi--.bas, looking for 3'-GGG(A/T)GT-5', 0,
- inverse, negative strand, positive direction, is SuccessablesHYi-+.bas, looking for 3'-GGG(A/T)GT-5', 0,
- inverse, positive strand, negative direction, is SuccessablesHYi+-.bas, looking for 3'-GGG(A/T)GT-5', 4, 3'-GGGAGT-5', 2702, 3'-GGGTGT-5', 3184, 3'-GGGAGT-5', 3889, 3'-GGGAGT-5', 4498,
- inverse, positive strand, positive direction, is SuccessablesHYi++.bas, looking for 3'-GGG(A/T)GT-5', 1, 3'-GGGAGT-5', 64.
HNF6
Consensus sequence for HNF6 is DWRTCMATXD, but TTATTGATTA found.[9] D = A, G, or T, W = A or T, R = A or G, M = A or C; however, the fourth T from the left should be a C and the next letter should be A or C, not G.
The "HNF-6 binding sequence [consensus sequence is] DHWATTGAYTWWD (where W = A or T, Y = T or C, H is not G, and D is not C)".[42] The more open consensus[9] is (A/G/T)(A/T)(A/G)T(C/T)(A/C/G)AT(A/C/G/T)(A/G/T).
For the Basic programs (starting with SuccessablesHNF6.bas) written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), including extending the number of nts from 958 to 4445, the programs are, are looking for, and found:
- negative strand in the negative direction (from ZSCAN22 to A1BG) is SuccessablesHNF6--.bas, looking for 3'-(A/G/T)(A/T)(A/G)T(C/T)(A/C/G)AT(A/C/G/T)(A/G/T)-5', 3, 3'-GTGTTAATAA-5', 1725, 3'-TAGTTGATAA-5', 3527, 3'-TTATTAATCG-5', 4229,
- negative strand in the positive direction (from ZNF497 to A1BG) is SuccessablesHNF6-+.bas, looking for 3'-(A/G/T)(A/T)(A/G)T(C/T)(A/C/G)AT(A/C/G/T)(A/G/T)-5', 3, 3'-ATGTCCATGG-5', 3581, 3'-TTATTAATCA-5', 4147, 3'-TTATTGATTA-5', 4164,
- positive strand in the negative direction is SuccessablesHNF6+-.bas, looking for 3'-(A/G/T)(A/T)(A/G)T(C/T)(A/C/G)AT(A/C/G/T)(A/G/T)-5', 1, 3'-AAATTGATAA-5', 3361,
- positive strand in the positive direction is SuccessablesHNF6++.bas, looking for 3'-(A/G/T)(A/T)(A/G)T(C/T)(A/C/G)AT(A/C/G/T)(A/G/T)-5', 1, 3'-GAGTCCATTG-5', 3732,
- complement, negative strand, negative direction is SuccessablesHNF6c--.bas, looking for 3'-(A/C/T)(A/T)(C/T)A(A/G)(C/G/T)TA(A/C/G/T)(A/C/T)-5', 1, 3'-TTTAACTATT-5', 3361,
- complement, negative strand, positive direction is SuccessablesHNF6c-+.bas, looking for 3'-(A/C/T)(A/T)(C/T)A(A/G)(C/G/T)TA(A/C/G/T)(A/C/T)-5', 1, 3'-CTCAGGTAAC-5', 3732,
- complement, positive strand, negative direction is SuccessablesHNF6c+-.bas, looking for 3'-(A/C/T)(A/T)(C/T)A(A/G)(C/G/T)TA(A/C/G/T)(A/C/T)-5', 3, 3'-CACAATTATT-5', 1725, 3'-ATCAACTATT-5', 3527, 3'-AATAATTAGC-5', 4229,
- complement, positive strand, positive direction is SuccessablesHNF6c++.bas, looking for 3'-(A/C/T)(A/T)(C/T)A(A/G)(C/G/T)TA(A/C/G/T)(A/C/T)-5', 3, 3'-TACAGGTACC-5', 3581, 3'-AATAATTAGT-5', 4147, 3'-AATAACTAAT-5', 4164,
- inverse complement, negative strand, negative direction is SuccessablesHNF6ci--.bas, looking for 3'-(A/C/T)(A/C/G/T)AT(C/G/T)(A/G)A(C/T)(A/T)(A/C/T)-5', 2, 3'-ACATGGACAT-5', 802, 3'-TAATGAACTT-5', 1301,
- inverse complement, negative strand, positive direction is SuccessablesHNF6ci-+.bas, looking for 3'-(A/C/T)(A/C/G/T)AT(C/G/T)(A/G)A(C/T)(A/T)(A/C/T)-5', 1, 3'-TTATTGATTA-5', 4164,
- inverse complement, positive strand, negative direction is SuccessablesHNF6ci+-.bas, looking for 3'-(A/C/T)(A/C/G/T)AT(C/G/T)(A/G)A(C/T)(A/T)(A/C/T)-5', 3, 3'-AAATTGATAA-5', 3361, 3'-TCATCAACTA-5', 3525, 3'-TTATTAATTC-5', 4542,
- inverse complement, positive strand, positive direction is SuccessablesHNF6ci++.bas, looking for 3'-(A/C/T)(A/C/G/T)AT(C/G/T)(A/G)A(C/T)(A/T)(A/C/T)-5', 2, 3'-CCATTGACTC-5', 3736, 3'-ATATTAACAA-5', 4172,
- inverse, negative strand, negative direction, is SuccessablesHNF6i--.bas, looking for 3'-(A/G/T)(A/C/G/T)TA(A/C/G)(C/T)T(A/G)(A/T)(A/G/T)-5', 3, 3'-TTTAACTATT-5', 3361, 3'-AGTAGTTGAT-5', 3525, 3'-AATAATTAAG-5', 4542,
- inverse, negative strand, positive direction, is SuccessablesHNF6i-+.bas, looking for 3'-(A/G/T)(A/C/G/T)TA(A/C/G)(C/T)T(A/G)(A/T)(A/G/T)-5', 2, 3'-GGTAACTGAG-5', 3736, 3'-TATAATTGTT-5', 4172,
- inverse, positive strand, negative direction, is SuccessablesHNF6i+-.bas, looking for 3'-(A/G/T)(A/C/G/T)TA(A/C/G)(C/T)T(A/G)(A/T)(A/G/T)-5', 2, 3'-TGTACCTGTA-5', 802, 3'-ATTACTTGAA-5', 1301,
- inverse, positive strand, positive direction, is SuccessablesHNF6i++.bas, looking for 3'-(A/G/T)(A/C/G/T)TA(A/C/G)(C/T)T(A/G)(A/T)(A/G/T)-5', 1, 3'-AATAACTAAT-5', 4164.
Metal responsive elements
"[T]hree potential metal response elements (MREs) [overlap] the E-boxes in the repeats, (TGCACGT with TGCRCNC being the consensus sequence; 17,18)."[43]
The reproducible consensus sequence seems to be 3'-TGCRCNC-5', specifically 3'-TGC-(A/G)-C-(A/C/G/T)-C-5'.
For the Basic programs (starting with SuccessablesMRE.bas) written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), including extending the number of nts from 958 to 4445, the programs are, are looking for, and found:
- negative strand in the negative direction is SuccessablesMRE--.bas, looking for 3'-T-G-C-(A/G)-C-(A/C/G/T)-C-5', 0,
- negative strand in the positive direction is SuccessablesMRE-+.bas, looking for 3'-T-G-C-(A/G)-C-(A/C/G/T)-C-5', 11, 3'-TGCGCCC-5', 453, 3'-TGCACAC-5', 549, 3'-TGCACAC-5', 1221, 3'-TGCGCCC-5', 1247, 3'-TGCACTC-5', 1373, 3'-TGCGCCC-5', 1399, 3'-TGCACTC-5', 1473, 3'-TGCGCCC-5', 1499, 3'-TGCGCCC-5', 1657, 3'-TGCACAC-5', 2963, 3'-TGCACCC-5', 3323,
- positive strand in the negative direction is SuccessablesMRE+-.bas, looking for 3'-T-G-C-(A/G)-C-(A/C/G/T)-C-5', 7, 3'-TGCGCTC-5', 891, 3'-TGCACTC-5', 1348, 3'-TGCACTC-5', 2001, 3'-TGCACTC-5', 2427, 3'-TGCACCC-5', 2762, 3'-TGCACTC-5', 3290, 3'-TGCACTC-5', 4341,
- positive strand in the positive direction is SuccessablesMRE++.bas, looking for 3'-T-G-C-(A/G)-C-(A/C/G/T)-C-5', 2, 3'-TGCGCCC-5', 872, 3'-TGCGCCC-5', 972,
- complement, negative strand, negative direction is SuccessablesMREc--.bas, looking for 3'-A-C-G-(T/C)-G-(A/C/G/T)-G-5', 7, 3'-ACGCGAG-5', 891, 3'-ACGTGAG-5', 1348, 3'-ACGTGAG-5', 2001, 3'-ACGTGAG-5', 2427, 3'-ACGTGGG-5', 2762, 3'-ACGTGAG-5', 3290, 3'-ACGTGAG-5', 4341,
- complement, negative strand, positive direction is SuccessablesMREc-+.bas, looking for 3'-A-C-G-(T/C)-G-(A/C/G/T)-G-5', 2, 3'-ACGCGGG-5', 872, 3'-ACGCGGG-5', 972,
- complement, positive strand, negative direction is SuccessablesMREc+-.bas, looking for 3'-A-C-G-(T/C)-G-(A/C/G/T)-G-5', 0,
- complement, positive strand, negative direction is SuccessablesMREc++.bas, looking for 3'-A-C-G-(T/C)-G-(A/C/G/T)-G-5', 11, 3'-ACGCGGG-5', 453, 3'-ACGTGTG-5', 549, 3'-ACGTGTG-5', 1221, 3'-ACGCGGG-5', 1247, 3'-ACGTGAG-5', 1373, 3'-ACGCGGG-5', 1399, 3'-ACGTGAG-5', 1473, 3'-ACGCGGG-5', 1499, 3'-ACGCGGG-5', 1657, 3'-ACGTGTG-5', 2963, 3'-ACGTGGG-5', 3323,
- inverse complement, negative strand, negative direction is SuccessablesMREci--.bas, looking for 3'-G-(A/C/G/T)-G-(T/C)-G-C-A-5', 2, 3'-GTGTGCA-5', 531, 3'-GAGTGCA-5', 1772,
- inverse complement, negative strand, positive direction is SuccessablesMREci-+.bas, looking for 3'-G-(A/C/G/T)-G-(T/C)-G-C-A-5', 10, 3'-GCGTGCA-5', 546, 3'-GCGCGCA-5', 684, 3'-GGGCGCA-5', 876, 3'-GGGCGCA-5', 976, 3'-GCGTGCA-5', 1218, 3'-GTGCGCA-5', 1523, 3'-GAGTGCA-5', 1786, 3'-GAGTGCA-5', 2326, 3'-GGGTGCA-5', 2800, 3'-GGGTGCA-5', 3883,
- inverse complement, positive strand, negative direction is SuccessablesMREci+-.bas, looking for 3'-G-(A/C/G/T)-G-(T/C)-G-C-A-5', 2, 3'-GAGTGCA-5', 1470, 3'-GTGTGCA-5', 2863,
- inverse complement, positive strand, positive direction is SuccessablesMREci++.bas, looking for 3'-G-(A/C/G/T)-G-(T/C)-G-C-A-5', 0,
- inverse, negative strand, negative direction, is SuccessablesMREi--.bas, looking for 3'-C-(A/C/G/T)-C-(A/G)-C-G-T-5', 2, 3'-CTCACGT-5', 1470, 3'-CACACGT-5', 2863,
- inverse, negative strand, positive direction, is SuccessablesMREi-+.bas, looking for 3'-C-(A/C/G/T)-C-(A/G)-C-G-T-5', 0,
- inverse, positive strand, negative direction, is SuccessablesMREi+-.bas, looking for 3'-C-(A/C/G/T)-C-(A/G)-C-G-T-5', 2, 3'-CACACGT-5', 531, 3'-CTCACGT-5', 1772,
- inverse, positive strand, positive direction, is SuccessablesMREi++.bas, looking for 3'-C-(A/C/G/T)-C-(A/G)-C-G-T-5', 10, 3'-CGCACGT-5', 546, 3'-CGCGCGT-5', 684, 3'-CCCGCGT-5', 876, 3'-CCCGCGT-5', 976, 3'-CGCACGT-5', 1218, 3'-CACGCGT-5', 1523, 3'-CTCACGT-5', 1786, 3'-CTCACGT-5', 2326, 3'-CCCACGT-5', 2800, 3'-CCCACGT-5', 3883.
Nuclear factors
STAT5
"STATs [signal transducers and activators of transcription] bind through their DNA-binding domain (DBD) to consensus elements (TTCTTGGAA, STAT5 consensus), resulting in gene transcription."[44]
STAT5 consensus sequence is TTCXXXGAA, where X = A, C, or G.[9] Or, X = G or T.[44]
For the Basic programs (starting with SuccessablesSTAT5.bas) written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), including extending the number of nts from 958 to 4445, the programs are, are looking for, and found:
- negative strand in the negative direction (from ZSCAN22 to A1BG) is SuccessablesSTAT5--.bas, looking for 3'-TTCNNNGAA-5', 0,
- negative strand in the positive direction (from ZNF497 to A1BG) is SuccessablesSTAT5-+.bas, looking for 3'-TTCNNNGAA-5', 1, 3'-TTCCGGGAA-5', 808,
- positive strand in the negative direction is SuccessablesSTAT5+-.bas, looking for 3'-TTCNNNGAA-5', 2, 3'-TTCGTTGAA-5', 3506, 3'-TTCCCTGAA-5', 3782,
- positive strand in the positive direction is SuccessablesSTAT5++.bas, looking for 3'-TTCNNNGAA-5', 0,
- complement, negative strand, negative direction is SuccessablesSTAT5c--.bas, looking for 3'-AAGNNNCTT-5', 2, 3'-AAGCAACTT-5', 3506, 3'-AAGGGACTT-5', 3782,
- complement, negative strand, positive direction is SuccessablesSTAT5c-+.bas, looking for 3'-AAGNNNCTT-5', 0,
- complement, positive strand, negative direction is SuccessablesSTAT5c+-.bas, looking for 3'-AAGNNNCTT-5', 0,
- complement, positive strand, positive direction is SuccessablesSTAT5c++.bas, looking for 3'-AAGNNNCTT-5', 1, 3'-AAGGCCCTT-5', 808,
- inverse complement, negative strand, negative direction is SuccessablesSTAT5ci--.bas, looking for 3'-TTCNNNGAA-5', 0,
- inverse complement, negative strand, positive direction is SuccessablesSTAT5ci-+.bas, looking for 3'-TTCNNNGAA-5', 1, 3'-TTCCGGGAA-5', 808,
- inverse complement, positive strand, negative direction is SuccessablesSTAT5ci+-.bas, looking for 3'-TTCNNNGAA-5', 2, 3'-TTCGTTGAA-5', 3506, 3'-TTCCCTGAA-5', 3782,
- inverse complement, positive strand, positive direction is SuccessablesSTAT5ci++.bas, looking for 3'-TTCNNNGAA-5', 0,
- inverse, negative strand, negative direction, is SuccessablesSTAT5i--.bas, looking for 3'-AAGNNNCTT-5', 2, 3'-AAGCAACTT-5', 3506, 3'-AAGGGACTT-5', 3782,
- inverse, negative strand, positive direction, is SuccessablesSTAT5i-+.bas, looking for 3'-AAGNNNCTT-5', 0,
- inverse, positive strand, negative direction, is SuccessablesSTAT5i+-.bas, looking for 3'-AAGNNNCTT-5', 0,
- inverse, positive strand, positive direction, is SuccessablesSTAT5i++.bas, looking for 3'-AAGNNNCTT-5', 1, 3'-AAGGCCCTT-5', 808.
Core promoters
Nucleotides
A1BG is on chromosome 19 between the genes for ZSCAN22 and ZNF497. Before each untranslated region are nucleotides between the genes.
Between ZNF497 and A1BG are 1006 nts before the TSS from this direction. Between ZSCAN22 and A1BG there are 4618 nts.
Depending on which direction the RNA polymerase II holoenzyme transcribes from is the exact nt of the transcription start site (TSS). For transcription from the ZNF497 direction the TSS sits among the local nts as follows: TTGG+1GGC. For transcription from the ZSCAN22 direction, the TSS sits among TGAA+1ACT.
NCBI at the National Institutes of Health includes these in between nucleotides as well as those for each gene. NCBI has predetermined whether the strand is the coding strand (positive strand) or the template strand (negative strand).
The nucleotides between ZNF497 and A1BG as A1BG is approached from ZNF497 on the negative strand are (in modification 108 3'-58354441:) in modification 107 3'-58865723:
GCTCCAGGCTCCAGAGGGCATTGGGCATCCAGGGACCTGGAGGAGAAGAGTGGGGGATGACAGCCCA CCCACCGTCGTGTCCCCCTCAGGCCCTCTGTCTGGTGTTCTTTGTATTCTCTAAGGTACTTGCCTTT TCTCTCCTGTTTTAGGTCACAGGCGCGAGATGGAGTCCCCAAGAGGGTGGACCCTGCAGGTGGCCCC AGAGGAAGGCCAGGTCCTCTGCAATGTGAAGACTGCCACGAGGGGCCTCTCTGAGGGGGCTGTGTCT GGAGGCTGGGGGGCCTGGGAAAACTCCACGGAGGTTCCGAGGGAGGCAGGGGACGGCCAGCGGCAGC AAGCCACACTGGGGGCGGCGGACGAACAGGGAGGCCCCGGCAGGGAGCTGGGCCCCGCAGACGGTGG GCGGGACGGGGCTGGGCCCAGGAGCGAGCCTGCAGACCGGGCGTTGCGCCCTTCGCCTCTCCCAGAG GAGCCGGGCTGCCGGTGCGGGGAGTGCGGCAAGGCGTTCAGCCAGGGCTCTTACTTGCTGCAGCATC GGCGCGTGCACACAGGCGAGAAGCCGTACACGTGCCCCGAGTGCGGCAAGGCCTTCGCCTGGAGCTC CAACCTCAGCCAGCACCAGCGCATCCACAGCGGCGAGAAGCCCTACGCTTGCAGGGAGTGCGGCAAG GCCTTCCGCGCGCACTCGCAGCTCATCCACCACCAGGAGACACACAGCGGCCTGAAGCCCTTCCGCT GCCCGGACTGCGGCAAGTCCTTCGGCCGAAGCACCACGCTGGTGCAGCACCGACGCACGCACACGCC TGCCGGGACTGTGGCAAGGCCTTCAGCCAGAGCTCCAACCTGGCCGAGCACCTGAAGATCCACGCGG GCGCACGGCCACACGCCTGTCCCGACTGCCGCCTGCCGGGACTGTGGCAAGGCCTTCAGCCAGAGCT CCAACCTGGCCGAGCACCTGAAGATCCACGCGGGCGCACGGCCACACGCCTGTCCCGACTGCGGCAA GGCCTTCGTGCGTGTGGCGGGGCTGCGGCAGCACCGGCGCACGCACAGCAGCGAGAAGCCCTTCCCC TGCGCCGAGTGCGGAAAGGCTTTCCGCGAGAGCTCGCAGCTCCTGCAGCACCAGCGCACGCACACTG GTGAGCGGCCCTTCGAGTGCGCCGAGTGCGGCCAGGCTTTCGTCATGGGCTCCTACCTGGCGGAGCA CCGGCGCGTGCACACGGGCGAGAAGCCTCATGCGTGCGCCCAGTGCGGCAAGGCCTTCAGCCAGCGC TCCAACCTACTGAGCCACCGGCGCACGCCTTCGCCTGCGCAGAATGCGGCAAGGCCTTCCGCGGCAG CTCCGAGCTGCGCCAGCACCAGCGCCTGCACTCTGGCGAGAGGCCGTTCGTCTGCGCCCACCTTCGC CTGCGCAGAATGCGGCAAGGCCTTCCGCGGCAGCTCCGAGCTGCGCCAGCACCAGCGCCTGCACTCT GGCGAGAGGCCGTTCGTCTGCGCCCACTGCAGCAAGGCCTTCGTGCGCAAGTCGGAGCTCTTAAGCC ACCGGCGCACGCACACGGGCGAGAGGCCCTACGCTTGCGGCGAGTGCGGGAAGCCTTTCAGCCACCG TTGCAACCTCAACGAGCACCAGAAGCGGCACGGGGGCCGCGCTGCGCCCTGACCCGAGGACGCCCTG AGCGGGAGGTCGCGGACACACGGCATTGCGGGGTCTCGGGCGTGAGTGCGCTGTCTGCTGGCCCAGA CTTTTTCGGGCCGCCGGTGCGGGCGCCCTCCTGCTGGGAGTGCAGGGGCGGCCTTGGGTGTGGAGAA CCCTGGCTGCACAGTCCCTTTGACGATAGTCCACCGGCCACCCAGGCCTGTCTGGGGACATGTAGGA TGGGCTCTTACCCCAGGGAGGGCGGCAGGCTCCACTTCGGCGAGAGGTTCGTCCATGCAGAGGTGGG CAAGAACTGGGGTCTCCGACAGGTGTGGCTATTTCTTTGAGTTCTCTGGCACTGTCAAAAGCAGCCA ACCCACCCCCCAGTCCACATGGTCACCACTGCTGCTACCAGCTGCTCAGTGCAGTGGCCACTGTGTC TCCTAAGGTGCTCGCTTCAGTCAGCACTTCATCTCAGGCAACCACAGGTGACAGTTAAACATGATGA AACCGCATGCTATGGCTTTCTAGTGTCTCATATTCTGTTGGCAAGAAGCTCAGCACTGCATTCCTGA CCGAGGTCAGAACCAGATCAATCTCAGAATCTCACCTGTGTAGGTCTGTTTCATGGGACTTTTCTTT TTTGGGGGAGGGGGCAGGGTTTCACTCTGTCACCCAGGCTGGAGTGCAGTGGTGCAATCACTGTTTA TTGCAGCCGCGACTTCTTAGGCTCAGGTGATCCTCCCACCTCAGCCTCCCAAGCAGCTGGGATGACA GACTTGCGTCACTACACCTGGCTAATTTTAAAATTTTTTGTAAGGACAGTGTCTCACCATGTTGCCT AGGCGGGTCTCAAAACTCCTGGGCTCAAGTGATCCTCCTGCCTCAGCCTCCCAAAGCGTTGGGATTA TAGGCGTGAGCCACCGCACCTGGCCAGGGAACCTTCTTTATACCAAGTACCACACCAGTCAGAGTCA AGTCAGGAAATAGAAACCACACTAGGTATTTCAAACAGAGGGGATATAAGTTAGGGACTGATTGTGC AGGTTTTGCAAGGCTCAGAGAGCAAAAGTGGATGTTGCAGAATCTCAGAGCTGGATACTTGCAGGAA GCTGCTACCACCCTTAGGGCTGGAGAACCCAGGGAAGCTAAGAGGAGGGTGCAACGAGGCTGGTGTT GGGACTGCCAAAGGAAACACAGAATGACAGCTCCATCTCTGCAGACCTCAGATTTGACCAGGCCTCT GGCTGCCTGGGCAGCCTCATTGCCAATGGGCTGAAAGGTTCTGTCACTGTTCAGTGGTCTGACTTCT GAGTTCTGTGCACACCCTGAGCTCTGCTCCTCTGGCCTGGCCACCAGTCCTTGTCTGAGTGCCCCAG GTCTGGTTATGACTTCAGGCCTCAGCACCTGGTTGTCTCTTCTGCAAAGAATGCATTCTCCCCCAGT CCACCCAGAACACACAGCCTCCATCCAGGTACTGGCTCTGAGCAGGACAGAGAGCATGCAGCAGGTG CTCAGCAATATGGAAATGGGACCAAACAGAGGGAGTCTCCACAGCTCCCCGTCCCTCACAGCAGAAG CCAGAGCCGCTGCAGTGCCCAGCTGGTCTTCATCACGTCTATGAGCTCTGCAATCGCTCTGCAGTCG CCTCAACTCCTGGTCTCCTCTGCTCCTTCTCACTGCACCCGCATCTCCCATGTTTGCACTGGCTGTT CCCTCTGCCTGGAATGCTCCTTGCCCAGTTATCCCACTGTCTTCTTTGCATCTGGCACACAGTAGAT GCTCAATAAATGCCTGTGGAATGAATGAGTGGGGAGGATGCAGTGCAGGGGGCAGATGAGGGCTAGG CGGTTGCCCTGGGCCCTCACACTCGTAGCGGAGCTAGGCTGGGACACCCAGGGTGGGGACCAGACCT CCCCGGGTGGGAATGACAGGATGTCCATGGAGGCTGAGTGTGAAAGCACCACGGTCTCACCCCTGTC CTGTTCCATCCCAACAGGCTGTGGTGAGGAGAGGGGGAGGCAGGGGAAGCGGGAGGCCTGGCCTCCA GGCAGCAGGCTATAGCCACATGAGTGACCACCAGCAGCTCAGGTAACTGAGCACATGTCACGGGTAG GCCTGGGAAGCGCAGGTCTCAGCTGAATGACCTGGGTGGAAATCCGACTCCAGAGCCGTGGTGGGTC ACACCATGCAGAATGAACCAGTGATGGAGAAGGAACCACAGTCCTCAGGAAGAGTGAGGGTGCACCT CCAGACAGCCCATGTGAGGGCAACCGCAGAAAGTCTGAAAAGAGGTGAACCCCACCTTTGGTGTCAC ATGTGCAGTGTGGTGTGACAGGGAGGGGCTCGCTGGGCTTCAGCCCCGGCACTCTCCACTTGACCTC AGCAGCTCCAGGTAGAGTGGGGAGAACTCAGCGTCTCCTTCTAGAACAGGTTCTAGGATCCATCACT GAAATGAGGATGAGGTGGTTTTAACATCATTTTATCACTCTTGATTTAGTTTATTAATCATACATGA TTATTGATTATAATTGTTGCTGGGCATCCTGAGGCCTCAGAAGTTCACCCTTTGCCCTGACCCCATG GGGGCCCTGCCCCCGCCTTCCGGGAAGGACAAACACGGGAAGAGGTCAGTGCCCGAGCCACCCCACC GCCCTCCCTTGG+1 :58864973-5'(107) or 58350139-5' (108).
The nucleotides between ZSCAN22 and A1BG as A1BG is approached from ZSCAN22 on the negative strand are 3'-58853715:
TTTAAGATTGTCTGACTTAAAGAAAAACCTGGTCGGTATACAAGAAATCTTTTCTATGTGGATTTTG TTCCTATACCCTTCAACTCCCGTTTCCCTATCTTTTCCTATATGTTCCATCCGTACTTTGACTTCTTT CGACTACCTCGACGTAATCAGCGCGTTTTGTCTGTAATTCCATATTTTCGTAAAATTCTTAGTACCC CAGTGATACAAAAATATTTTCTTTGTTAGATAATCCTTCTACATTATCAAGTTTTGGTCCGTGTATT ATACTCAGAACATCTTGTCTTAGTGTCACTCTTTGACTGCTTTGGAATTCACATGAACCTTCACGAT TGTGCAAGAAATACTATCTTTTGTAAACTTCTCCGGCCCACGCCACCGAGGGTGGACATTAGGGTC GTGAAACCCTCCGGTTCTGTCCGCCTAGTGCTCCAGTCCTCACGCTCCGTTCGGACCCGTTGTATC ACTTTGACAGATGTTTTTTATGCTTTTAATCGGTCGGACCCGGCCCGTGTCACCTAGTGTGCATTAA GGTCGTGAAACCCTCCGGTTCTGTCCGTCCAGTGCTCCAGTCCTCTAACTCTAGTAGGACCGATTA TACCACACATTGGGGAAGAGATGATTTTTATGTTTTTTAACCGGTCCGTGCCACCGAGTGCGGACA TTAGGGTCGTGAAACCCTCCGGTTCCGTCCGCCTAGTGCTCCAGTCCTCAAGCTCTGGTCGGACTG GTCGCACCACTGTGGGGCAGAGATGATTTTTATGTTTTTTAATCGACCTACACCACCACACATGGAC ATTAGGGTCGATGATTCCTCCGACTCCGTCCTCTTAGCGAACTTGGGTCCTCCGCCTCCAACGCCA CTCGGTTCTAGTGTGGTAACGCGAGGTCGGACCCGTTGTCTCGCTCTGAGACAGAGTTTTTTTTTTT TTTTTTTTTCGGTCCGTACCGCCGTGTGCGGACATCTAGGTCGATTAGTCCTCCGACTCCGTCCTCT TAACGAACTTGGACCCTCCGTCTCCAACGTCACTCGGCTCTAACCCACTGAAGTGAGGTCGGAGCT GTTGTCTCACTCTGAGACAGAGTTTTTTTTTTTTTTTTTCCGACCCGTGTCACCGAGTGTGGACATT AGGGTCGTGAAACCCTGCGGCTCCACCCACCTAGTGGACTCAAGTGCTCAAACGCTGGTCGGACCG GTTGTACCACTTTGGGGCACAGATGTTTTTTAATCGGCCCGCACCACCGCCCACGGACATTAGGGT CGATGAGTCCTCCGACTCCGTCCTCTTAATGAACTTGGATCCTCCGTCTCCAACGTCACTCGGCTC TAACGTGGTAACGTGAGGTCAGACCCGTTATTCTCGTTTTGAGGTAAAGTTTTTGTTTGTTTTTTTT CTGAGTTGGGTCTTAAGAAAAAAAAAAAAAAAAACTCTACCTCAGAGCGAGACAACGGGTCCGACC TCACGTCACCACACTAGAGTCGAGTGACGTTCGAGGCGGAGGGCAGTTGGGTCTTAAGATATAGGT CACGTTTGTGTGAATTTCTTGCTTCCGTTTTATGTCTGTAGGAGTTTACTTTGTTTGGATTCTATATA TAAAGAACGGTCGTCTGAACGGACTTTTCTTTACGGTTTCCTTTGAGAACCATCTTCCCTTTACTAT GGTCTCCCTTTCGCCCTTGAAACCCTTAATTTACTATCATTATCTCTAACGTGTTAATAAAATAGAA ATTTTATACCGACCTCCGCCGACCCGCGCCACCGAGTGCAGACATTAGGGTCGTGAAACCCTCCGA CTCCACCCGCCTAGGGTTCCCGTCCTCTACCTCTGGTAGGACCGATTGTACAACTTTGGGGTAGAG ATGATTTTTATGTTTTTTTAATCGACCCGCACCACCACCCGAGGACATTAGGGTCGATGAACCCTCT GACTCCGTCCTCTTACCGTACTTGGACCCTCCGTCTCGAACGTCACTCGGCTCTAGTGCGGTGACG TGAGGTCGGACCCGCTGTCTCGTTCTGAGACAGAGTTTTTTTTTTTTTTTTTTTCTTTTTTTTACACC GACCTCCGGTCCACGTCACATTAGGATCGTGAAACCCTCCGACTCCACCTGTCTGGTGAACTCGAG TCCTCAAACTCTGGTTGTACCGCTTTATGACAGAGATGATTTCTATGTTTTTTACTGGCCCACGCCA CCGAGTGCGGTCATGAAACCCTCCGACTCCGCCAACCCTAGTGTTCCAGTCCTCAAACTCTGGTCG GACCTGTCGTACCACTTTGGGGTAGAGATGATTTTTATGTTTTTTGATCGGCCCGTACGACCACCCA CGGACATCATGGTCGATGAGGCCTCCGACTCCGTCCTCTTAACGAACTTGGACCGTCCGCCTCCAA CGTCACTCGGCTCTAGTGTGGTGACGTGAGGTCGGACCCGTTGTCTCACTCTAAGCCCTTTTTTTTT TTTTTTCGTTTTCGTTTGTTTGTTTTGAGTTATCATTCTTTTGTTTGTCCGGTCCGTGCCACCGAGTA CGGACATCAGGGTTGTGAAACCTTCCGACTCCGTCCACCTAGTGAACTCCAGTCCTCAACTTCTGG TCACACCGGTTGTACCACTTTGGGGGAGAGGTGATTTATATGTTTTTAGTCGGTCACACCACCGTG TACGGACATTAGGGTCGATGTGTCCTCCGACTCCCTCAACTTAGCGAACTTGGACCCGCCGCCTTC AACGTCACTCGACTTTAGTACGGTGACGTGGGGTCGGACCCGTTGTCTCGTACTGAGAGAGTTTTT CTTTCTTTTCTCTTCTTTTTTCTTTTCTCTTCTTTTTTCTTTTGTTGGGTTATAAAATTTCACACGTTT TATATATTTGTCTGTAAAGTAGTTTCTACGATATATCTACCGTTGATTCGTGAACCCCTTTTTTACGA ATTGTAGTAGTCTGTAATTCCTTTGCGTTCCTTTTGGTGATAATCTATACAGATGTATGGATAATCT TACCGATTTTATTTGAAAAATTTTTTGATTCGACCCCGACCCGCACCACCGAGTGGGGACATTAGGG TCGTGAAACCCTCCGACTCCGCCCACCTAGTGAACTCGAGTCCTCAACCTCTGGTCGGACCGGTGG GTTGTTCCACTTTGGGGTAGAGATGATTTTTGTATTTTTAATCGACCCACACCACCACCCACGGACA GTCGGGTCGATGAGTCCTCCGACTCCGTGCTCTTAGTGAACTTGGGTCCTCCATCTCCAACGACAC TCGGTTCTAGTGCGGTAACGTGAGGTCGGACCCGTTGTTCTCGTTTTAAGACAGAGTTTTTGTTTAT TTGTTCTTTTCCAAAATTTTTAACTATTTGGTGATCGTTCTTTCTCTCTCTCCTGTGTAGTCCGAACT CTCTTCACTGTAGTAGTATCTGACACGTCTGTAATTTTCCGTATCCGTATAATCCTTGATCTATAAT GATTCCGTCATTTGGTCTAGTGATCGTTTAAGCAACTTTTTGTGTTAAGTAGTTGATAACCAACTAT TATGACTATGCGTAGAAGACACACGGTCCCGGAACTCCGGGACGAGGTCCGTCTTTGAGCCGTCAC CAACCCTTCCGTAGTCTACAGTCTACCGTGTTCTCCTGAGTCTCGACTCCCTATTACCCTTTCTTGT GTCTCCTTAGGTCGGTAAAGGTGTCGCAGGTCGAGACGACACCCTCCGACCCTTGTCGGGTCGTGA TGGTGGGACCTGACCCTCCTGTTCTGGTGTTTTACGTCGAAGGGACTTGGAGGAGAACCACTACCC CAACTACACCAGCTCCATCTCCACTCATACAGACCTCGGAGTTCGTTGACGGGTATGGACGACCCT GGTCCGGCTCCACGGGTCCCTCATTCTCCGTCGTAGGACCTTCTCGTGTCTACTTTCTCCGGGACT CTTCACAACCAACCGGTCCACGACACCGAGTGTGGACATTAGGGTTGTGAAACCCTCCGACTCCGC CCTCCTAGTGAACTCGGGTTCTCAAGTTCTGGTCGGACCCGTTGTATCACTCTGAGTAGAGATGTT TTTTATTTATTATCTTTCTTTTTACACTCAACCGGTCCGTACCACCGAGTACGGACATCAGGGTCGA TGAGTCCTCCGACTCCACCCTCCTAGTGAATTCCGGTCCTCAAGTTCTGTTCGAACCCGTTGTGTCA CTCTGGGACAGATGTTTTTTATTATTAATCGGTCTGCACCGCTACGTACGGGTCCGAGGGTCGATG AACCCTCCGACTCCGTCCTCCTAGCGAACTCGGACCCTCCAGTTTTGACGTCACTCGGCCTGACGT TGTGACGTGAGGTCGGACCCACTGTCACACTCTGGGACAGAGTTTTTTCTTTTTTCTTTTCTTTTGA CACGAGAATTCTCGGTCAAGAGGTGAGGAGATGGAGTCCTCGGTGGGGTCTTGGGTAGGTGAA+1 :58858175-5'.
The nucleotides between ZNF497 and A1BG as A1BG is approached from ZNF497 on the positive strand are 3'-58865723:
CGAGGTCCGAGGTCTCCCGTAACCCGTAGGTCCCTGGACCTCCTCTTCTCACCCCCTACTGTCGGGT GGGTGGCAGCACAGGGGGAGTCCGGGAGACAGACCACAAGAAACATAAGAGATTCCATGAACGGAAA AGAGAGGACAAAATCCAGTGTCCGCGCTCTACCTCAGGGGTTCTCCCACCTGGGACGTCCACCGGGG TCTCCTTCCGGTCCAGGAGACGTTACACTTCTGACGGTGCTCCCCGGAGAGACTCCCCCGACACAGA CCTCCGACCCCCCGGACCCTTTTGAGGTGCCTCCAAGGCTCCCTCCGTCCCCTGCCGGTCGCCGTCG TTCGGTGTGACCCCCGCCGCCTGCTTGTCCCTCCGGGGCCGTCCCTCGACCCGGGGCGTCTGCCACC CGCCCTGCCCCGACCCGGGTCCTCGCTCGGACGTCTGGCCCGCAACGCGGGAAGCGGAGAGGGTCTC CTCGGCCCGACGGCCACGCCCCTCACGCCGTTCCGCAAGTCGGTCCCGAGAATGAACGACGTCGTAG CCGCGCACGTGTGTCCGCTCTTCGGCATGTGCACGGGGCTCACGCCGTTCCGGAAGCGGACCTCGAG GTTGGAGTCGGTCGTGGTCGCGTAGGTGTCGCCGCTCTTCGGGATGCGAACGTCCCTCACGCCGTTC CGGAAGGCGCGCGTGAGCGTCGAGTAGGTGGTGGTCCTCTGTGTGTCGCCGGACTTCGGGAAGGCGA CGGGCCTGACGCCGTTCAGGAAGCCGGCTTCGTGGTGCGACCACGTCGTGGCTGCGTGCGTGTGCGG ACGGCCCTGACACCGTTCCGGAAGTCGGTCTCGAGGTTGGACCGGCTCGTGGACTTCTAGGTGCGCC CGCGTGCCGGTGTGCGGACAGGGCTGACGGCGGACGGCCCTGACACCGTTCCGGAAGTCGGTCTCGA GGTTGGACCGGCTCGTGGACTTCTAGGTGCGCCCGCGTGCCGGTGTGCGGACAGGGCTGACGCCGTT CCGGAAGCACGCACACCGCCCCGACGCCGTCGTGGCCGCGTGCGTGTCGTCGCTCTTCGGGAAGGGG ACGCGGCTCACGCCTTTCCGAAAGGCGCTCTCGAGCGTCGAGGACGTCGTGGTCGCGTGCGTGTGAC CACTCGCCGGGAAGCTCACGCGGCTCACGCCGGTCCGAAAGCAGTACCCGAGGATGGACCGCCTCGT GGCCGCGCACGTGTGCCCGCTCTTCGGAGTACGCACGCGGGTCACGCCGTTCCGGAAGTCGGTCGCG AGGTTGGATGACTCGGTGGCCGCGTGCGGAAGCGGACGCGTCTTACGCCGTTCCGGAAGGCGCCGTC GAGGCTCGACGCGGTCGTGGTCGCGGACGTGAGACCGCTCTCCGGCAAGCAGACGCGGGTGGAAGCG GACGCGTCTTACGCCGTTCCGGAAGGCGCCGTCGAGGCTCGACGCGGTCGTGGTCGCGGACGTGAGA CCGCTCTCCGGCAAGCAGACGCGGGTGACGTCGTTCCGGAAGCACGCGTTCAGCCTCGAGAATTCGG TGGCCGCGTGCGTGTGCCCGCTCTCCGGGATGCGAACGCCGCTCACGCCCTTCGGAAAGTCGGTGGC AACGTTGGAGTTGCTCGTGGTCTTCGCCGTGCCCCCGGCGCGACGCGGGACTGGGCTCCTGCGGGAC TCGCCCTCCAGCGCCTGTGTGCCGTAACGCCCCAGAGCCCGCACTCACGCGACAGACGACCGGGTCT GAAAAAGCCCGGCGGCCACGCCCGCGGGAGGACGACCCTCACGTCCCCGCCGGAACCCACACCTCTT GGGACCGACGTGTCAGGGAAACTGCTATCAGGTGGCCGGTGGGTCCGGACAGACCCCTGTACATCCT ACCCGAGAATGGGGTCCCTCCCGCCGTCCGAGGTGAAGCCGCTCTCCAAGCAGGTACGTCTCCACCC GTTCTTGACCCCAGAGGCTGTCCACACCGATAAAGAAACTCAAGAGACCGTGACAGTTTTCGTCGGT TGGGTGGGGGGTCAGGTGTACCAGTGGTGACGACGATGGTCGACGAGTCACGTCACCGGTGACACAG AGGATTCCACGAGCGAAGTCAGTCGTGAAGTAGAGTCCGTTGGTGTCCACTGTCAATTTGTACTACT TTGGCGTACGATACCGAAAGATCACAGAGTATAAGACAACCGTTCTTCGAGTCGTGACGTAAGGACT GGCTCCAGTCTTGGTCTAGTTAGAGTCTTAGAGTGGACACATCCAGACAAAGTACCCTGAAAAGAAA AAACCCCCTCCCCCGTCCCAAAGTGAGACAGTGGGTCCGACCTCACGTCACCACGTTAGTGACAAAT AACGTCGGCGCTGAAGAATCCGAGTCCACTAGGAGGGTGGAGTCGGAGGGTTCGTCGACCCTACTGT CTGAACGCAGTGATGTGGACCGATTAAAATTTTAAAAAACATTCCTGTCACAGAGTGGTACAACGGA TCCGCCCAGAGTTTTGAGGACCCGAGTTCACTAGGAGGACGGAGTCGGAGGGTTTCGCAACCCTAAT ATCCGCACTCGGTGGCGTGGACCGGTCCCTTGGAAGAAATATGGTTCATGGTGTGGTCAGTCTCAGT TCAGTCCTTTATCTTTGGTGTGATCCATAAAGTTTGTCTCCCCTATATTCAATCCCTGACTAACACG TCCAAAACGTTCCGAGTCTCTCGTTTTCACCTACAACGTCTTAGAGTCTCGACCTATGAACGTCCTT CGACGATGGTGGGAATCCCGACCTCTTGGGTCCCTTCGATTCTCCTCCCACGTTGCTCCGACCACAA CCCTGACGGTTTCCTTTGTGTCTTACTGTCGAGGTAGAGACGTCTGGAGTCTAAACTGGTCCGGAGA CCGACGGACCCGTCGGAGTAACGGTTACCCGACTTTCCAAGACAGTGACAAGTCACCAGACTGAAGA CTCAAGACACGTGTGGGACTCGAGACGAGGAGACCGGACCGGTGGTCAGGAACAGACTCACGGGGTC CAGACCAATACTGAAGTCCGGAGTCGTGGACCAACAGAGAAGACGTTTCTTACGTAAGAGGGGGTCA GGTGGGTCTTGTGTGTCGGAGGTAGGTCCATGACCGAGACTCGTCCTGTCTCTCGTACGTCGTCCAC GAGTCGTTATACCTTTACCCTGGTTTGTCTCCCTCAGAGGTGTCGAGGGGCAGGGAGTGTCGTCTTC GGTCTCGGCGACGTCACGGGTCGACCAGAAGTAGTGCAGATACTCGAGACGTTAGCGAGACGTCAGC GGAGTTGAGGACCAGAGGAGACGAGGAAGAGTGACGTGGGCGTAGAGGGTACAAACGTGACCGACAA GGGAGACGGACCTTACGAGGAACGGGTCAATAGGGTGACAGAAGAAACGTAGACCGTGTGTCATCTA CGAGTTATTTACGGACACCTTACTTACTCACCCCTCCTACGTCACGTCCCCCGTCTACTCCCGATCC GCCAACGGGACCCGGGAGTGTGAGCATCGCCTCGATCCGACCCTGTGGGTCCCACCCCTGGTCTGGA GGGGCCCACCCTTACTGTCCTACAGGTACCTCCGACTCACACTTTCGTGGTGCCAGAGTGGGGACAG GACAAGGTAGGGTTGTCCGACACCACTCCTCTCCCCCTCCGTCCCCTTCGCCCTCCGGACCGGAGGT CCGTCGTCCGATATCGGTGTACTCACTGGTGGTCGTCGAGTCCATTGACTCGTGTACAGTGCCCATC CGGACCCTTCGCGTCCAGAGTCGACTTACTGGACCCACCTTTAGGCTGAGGTCTCGGCACCACCCAG TGTGGTACGTCTTACTTGGTCACTACCTCTTCCTTGGTGTCAGGAGTCCTTCTCACTCCCACGTGGA GGTCTGTCGGGTACACTCCCGTTGGCGTCTTTCAGACTTTTCTCCACTTGGGGTGGAAACCACAGTG TACACGTCACACCACACTGTCCCTCCCCGAGCGACCCGAAGTCGGGGCCGTGAGAGGTGAACTGGAG TCGTCGAGGTCCATCTCACCCCTCTTGAGTCGCAGAGGAAGATCTTGTCCAAGATCCTAGGTAGTGA CTTTACTCCTACTCCACCAAAATTGTAGTAAAATAGTGAGAACTAAATCAAATAATTAGTATGTACT AATAACTAATATTAACAACGACCCGTAGGACTCCGGAGTCTTCAAGTGGGAAACGGGACTGGGGTAC CCCCGGGACGGGGGCGGAAGGCCCTTCCTGTTTGTGCCCTTCTCCAGTCACGGGCTCGGTGGGGTGG CGGGAGGGAACC+1 :58864973-5'.
The nucleotides between ZSCAN22 and A1BG as A1BG is approached from ZSCAN22 on the positive strand are 3'-58853715:
AAATTCTAACAGACTGAATTTCTTTTTGGACCAGCCATATGTTCTTTAGAAAAGATACACCTAAAAC AAGGATATGGGAAGTTGAGGGCAAAGGGATAGAAAAGGATATACAAGGTAGGCATGAAACTGAAGA AAGCTGATGGAGCTGCATTAGTCGCGCAAAACAGACATTAAGGTATAAAAGCATTTTAAGAATCAT GGGGTCACTATGTTTTTATAAAAGAAACAATCTATTAGGAAGATGTAATAGTTCAAAACCAGGCACA TAATATGAGTCTTGTAGAACAGAATCACAGTGAGAAACTGACGAAACCTTAAGTGTACTTGGAAGTG CTAACACGTTCTTTATGATAGAAAACATTTGAAGAGGCCGGGTGCGGTGGCTCCCACCTGTAATCC CAGCACTTTGGGAGGCCAAGACAGGCGGATCACGAGGTCAGGAGTGCGAGGCAAGCCTGGGCAAC ATAGTGAAACTGTCTACAAAAAATACGAAAATTAGCCAGCCTGGGCCGGGCACAGTGGATCACACG TAATTCCAGCACTTTGGGAGGCCAAGACAGGCAGGTCACGAGGTCAGGAGATTGAGATCATCCTGG CTAATATGGTGTGTAACCCCTTCTCTACTAAAAATACAAAAAATTGGCCAGGCACGGTGGCTCACGC CTGTAATCCCAGCACTTTGGGAGGCCAAGGCAGGCGGATCACGAGGTCAGGAGTTCGAGACCAGCC TGACCAGCGTGGTGACACCCCGTCTCTACTAAAAATACAAAAAATTAGCTGGATGTGGTGGTGTGT ACCTGTAATCCCAGCTACTAAGGAGGCTGAGGCAGGAGAATCGCTTGAACCCAGGAGGCGGAGGTT GCGGTGAGCCAAGATCACACCATTGCGCTCCAGCCTGGGCAACAGAGCGAGACTCTGTCTCAAAAA AAAAAAAAAAAAAAAGCCAGGCATGGCGGCACACGCCTGTAGATCCAGCTAATCAGGAGGCTGAGG CAGGAGAATTGCTTGAACCTGGGAGGCAGAGGTTGCAGTGAGCCGAGATTGGGTGACTTCACTCCA GCCTCGACAACAGAGTGAGACTCTGTCTCAAAAAAAAAAAAAAAAAGGCTGGGCACAGTGGCTCAC ACCTGTAATCCCAGCACTTTGGGACGCCGAGGTGGGTGGATCACCTGAGTTCACGAGTTTGCGACC AGCCTGGCCAACATGGTGAAACCCCGTGTCTACAAAAAATTAGCCGGGCGTGGTGGCGGGTGCCTG TAATCCCAGCTACTCAGGAGGCTGAGGCAGGAGAATTACTTGAACCTAGGAGGCAGAGGTTGCAGT GAGCCGAGATTGCACCATTGCACTCCAGTCTGGGCAATAAGAGCAAAACTCCATTTCAAAAACAAA CAAAAAAAAGACTCAACCCAGAATTCTTTTTTTTTTTTTTTTTGAGATGGAGTCTCGCTCTGTTGCCC AGGCTGGAGTGCAGTGGTGTGATCTCAGCTCACTGCAAGCTCCGCCTCCCGTCAACCCAGAATTCT ATATCCAGTGCAAACACACTTAAAGAACGAAGGCAAAATACAGACATCCTCAAATGAAACAAACCT AAGATATATATTTCTTGCCAGCAGACTTGCCTGAAAAGAAATGCCAAAGGAAACTCTTGGTAGAAGG GAAATGATACCAGAGGGAAAGCGGGAACTTTGGGAATTAAATGATAGTAATAGAGATTGCACAATT ATTTTATCTTTAAAATATGGCTGGAGGCGGCTGGGCGCGGTGGCTCACGTCTGTAATCCCAGCACT TTGGGAGGCTGAGGTGGGCGGATCCCAAGGGCAGGAGATGGAGACCATCCTGGCTAACATGTTGAA ACCCCATCTCTACTAAAAATACAAAAAAATTAGCTGGGCGTGGTGGTGGGCTCCTGTAATCCCAGC TACTTGGGAGACTGAGGCAGGAGAATGGCATGAACCTGGGAGGCAGAGCTTGCAGTGAGCCGAGA TCACGCCACTGCACTCCAGCCTGGGCGACAGAGCAAGACTCTGTCTCAAAAAAAAAAAAAAAAAAA GAAAAAAAATGTGGCTGGAGGCCAGGTGCAGTGTAATCCTAGCACTTTGGGAGGCTGAGGTGGACA GACCACTTGAGCTCAGGAGTTTGAGACCAACATGGCGAAATACTGTCTCTACTAAAGATACAAAAA ATGACCGGGTGCGGTGGCTCACGCCAGTACTTTGGGAGGCTGAGGCGGTTGGGATCACAAGGTCAG GAGTTTGAGACCAGCCTGGACAGCATGGTGAAACCCCATCTCTACTAAAAATACAAAAAACTAGCC GGGCATGCTGGTGGGTGCCTGTAGTACCAGCTACTCCGGAGGCTGAGGCAGGAGAATTGCTTGAAC CTGGCAGGCGGAGGTTGCAGTGAGCCGAGATCACACCACTGCACTCCAGCCTGGGCAACAGAGTG AGATTCGGGAAAAAAAAAAAAAAAGCAAAAGCAAACAAACAAAACTCAATAGTAAGAAAACAAACA GGCCAGGCACGGTGGCTCATGCCTGTAGTCCCAACACTTTGGAAGGCTGAGGCAGGTGGATCACTT GAGGTCAGGAGTTGAAGACCAGTGTGGCCAACATGGTGAAACCCCCTCTCCACTAAATATACAAAA ATCAGCCAGTGTGGTGGCACATGCCTGTAATCCCAGCTACACAGGAGGCTGAGGGAGTTGAATCGC TTGAACCTGGGCGGCGGAAGTTGCAGTGAGCTGAAATCATGCCACTGCACCCCAGCCTGGGCAACA GAGCATGACTCTCTCAAAAAGAAAGAAAAGAGAAGAAAAAAGAAAAGAGAAGAAAAAAGAAAACAA CCCAATATTTTAAAGTGTGCAAAATATATAAACAGACATTTCATCAAAGATGCTATATAGATGGCAA CTAAGCACTTGGGGAAAAAATGCTTAACATCATCAGACATTAAGGAAACGCAAGGAAAACCACTAT TAGATATGTCTACATACCTATTAGAATGGCTAAAATAAACTTTTTAAAAAACTAAGCTGGGGCTGGG CGTGGTGGCTCACCCCTGTAATCCCAGCACTTTGGGAGGCTGAGGCGGGTGGATCACTTGAGCTCA GGAGTTGGAGACCAGCCTGGCCACCCAACAAGGTGAAACCCCATCTCTACTAAAAACATAAAAATT AGCTGGGTGTGGTGGTGGGTGCCTGTCAGCCCAGCTACTCAGGAGGCTGAGGCACGAGAATCACTT GAACCCAGGAGGTAGAGGTTGCTGTGAGCCAAGATCACGCCATTGCACTCCAGCCTGGGCAACAAG AGCAAAATTCTGTCTCAAAAACAAATAAACAAGAAAAGGTTTTAAAAATTGATAAACCACTAGCAAG AAAGAGAGAGAGGACACATCAGGCTTGAGAGAAGTGACATCATCATAGACTGTGCAGACATTAAAA GGCATAGGCATATTAGGAACTAGATATTACTAAGGCAGTAAACCAGATCACTAGCAAATTCGTTGAA AAACACAATTCATCAACTATTGGTTGATAATACTGATACGCATCTTCTGTGTGCCAGGGCCTTGAGG CCCTGCTCCAGGCAGAAACTCGGCAGTGGTTGGGAAGGCATCAGATGTCAGATGGCACAAGAGGAC TCAGAGCTGAGGGATAATGGGAAAGAACACAGAGGAATCCAGCCATTTCCACAGCGTCCAGCTCTG CTGTGGGAGGCTGGGAACAGCCCAGCACTACCACCCTGGACTGGGAGGACAAGACCACAAAATGCA GCTTCCCTGAACCTCCTCTTGGTGATGGGGTTGATGTGGTCGAGGTAGAGGTGAGTATGTCTGGAG CCTCAAGCAACTGCCCATACCTGCTGGGACCAGGCCGAGGTGCCCAGGGAGTAAGAGGCAGCATC CTGGAAGAGCACAGATGAAAGAGGCCCTGAGAAGTGTTGGTTGGCCAGGTGCTGTGGCTCACACCT GTAATCCCAACACTTTGGGAGGCTGAGGCGGGAGGATCACTTGAGCCCAAGAGTTCAAGACCAGCC TGGGCAACATAGTGAGACTCATCTCTACAAAAAATAAATAATAGAAAGAAAAATGTGAGTTGGCCA GGCATGGTGGCTCATGCCTGTAGTCCCAGCTACTCAGGAGGCTGAGGTGGGAGGATCACTTAAGGC CAGGAGTTCAAGACAAGCTTGGGCAACACAGTGAGACCCTGTCTACAAAAAATAATAATTAGCCAG ACGTGGCGATGCATGCCCAGGCTCCCAGCTACTTGGGAGGCTGAGGCAGGAGGATCGCTTGAGCCT GGGAGGTCAAAACTGCAGTGAGCCGGACTGCAACACTGCACTCCAGCCTGGGTGACAGTGTGAGAC CCTGTCTCAAAAAAGAAAAAAGAAAAGAAAACTGTGCTCTTAAGAGCCAGTTCTCCACTCCTCTACC TCAGGAGCCACCCCAGAACCCATCCACTT+1 :58858175-5'.
According to Michael David Winther, Leah Christine Knickle, Martin Haardt, Stephen John Allen, Andre Ponton, Roberto Justo De Antueno, Kenneth Jenkins, Solomon O. Nwaka, and Y. Paul Goldberg, Fat Regulated Genes, Uses Thereof and Compounds for Mudulating Same at https://patents.google.com/patent/US20040146872A1/en, A1BG is transcribed from the direction of ZNF497: 58350056: CGAGCCACCCCACCGCCCTCCCTTGGGGCCTCATTGCTGCAGACGCTCACCCCAG ACACTCACTGCACCGGAGTGAGCGCGACCATCA+1TG : 58351767.
To obtain the nts between genes as well as the first several nucleotides within the first UTR, input the GeneID followed by [uid]. Under "Genomic context" in parentheses is (58858172..58864865, complement). These are the nucleotide numbers on the chromosome. The nearest neighboring gene ZNF497 has (58865723..58874214), where 58864801 to 58866601 (see above) are the nucleotides between the genes that contain the promoter for A1BG.
Transcription start sites
Notation: let the symbol PPP denote a promoter prediction program.
Notation: let the symbol CpG stand for cytosine - phosphodiester bond - guanine, which indicates that C and G are next to each other on the same DNA strand connected by phosphate.
"One important question is what the different PPPs are actually trying to predict. Some programs aim to predict the exact location of the promoter region of known protein-coding genes, while others focus on finding the transcription start site (TSS)."[45] "Recent research has shown that there is often no single TSS, but rather a whole transcription start region (TSR) containing multiple TSSs that are used at different frequencies (Frith et al., 2008)."[45] "The most recent large-scale validation of PPPs included more programs than any of the earlier studies and introduced for the first time an evaluation based on all experimentally determined TSSs in the human genome (Abeel et al., 2008a, 2008b)."[45] "[T]he current state-of-the-art in promoter prediction is biased toward housekeeping genes that contain CpG islands."[45]
Each of the currently described promoter elements is tested for possible occurrences between ZSCAN22 and A1BG on both the negative and positive strands, and between ZNF497 and A1BG on both strands, going from the neighboring gene toward A1BG.
E boxes
A1BG has an E-box: 3'-CACATG-5' ending at -2118 nt (G) in the distal promoter region.
MREs
On the negative strand going from ZSCAN22 to A1BG, there are no MREs.
eIF4Es
A1BG does not have an eIF4E basal element on the template strand between ZSCAN22 and A1BG TSS.
HY boxes
The only HY box upstream or downstream from A1BG ends (G) at -3711 nts (3'-TGTGGG-5') upstream from the TSS.
CAAT boxes
A1BG does not have a CAAT box in the transcription direction along the negative strand from ZSCAN22 to A1BG.
GC boxes
A1BG does not have a GC box in the transcription direction on either the positive or negative strand (ZSCAN22 to A1BG).
BREs
There are three B recognition elements (BREs) going along the negative strand from ZSCAN22 to A1BG: 3'-CCACGCC-5' out 380 nts from the last nt of the ending untranslated region for ZSCAN22, 3'-CCGCGCC-5' out 1762, and 3'-CCACGCC-5' out 2197 nts.
"The position in nucleotides (nts) relative to the transcription start site (TSS, +1)" is -35 for the BRE."[46] None of the three BREs located are anywhere near the TSS at some 4600 nts out from ZSCAN22.
TATA boxes
On the positive strand, in the nucleotide region between gene ZSCAN22 (NCBI GeneID: 342945) and A1BG (NCBI GeneID: 1) are 211 TATA box-like 8 nt long sequences. Of these,
- TATAAAAG occurs at 58853713 + 183 nts and
- TATAAAAG at 58853713 + 222. This is a TATA box found with some genes.[47] But, the optimal TBP recognition sequence 3'-TATATAAG-5',[48] does not occur.
- TATATAAA occurs only once at 2874 nts from the end of ZSCAN22. TBP is bound to this sequence and TATAAAAG above.[49][50]
- TATAAA occurs seven times, with the closest one at 2874 nts from the end of ZSCAN22. "In virtually every RNA polymerase II-transcribed gene examined, the sequence TATAAA was present 25 to 30 nts upstream of the transcription start site."[25]
A1BG does not have a TATA box in the core promoter region. There is the sequence 3'-TGCTATATAGATGGCAACTAAGCACTTGGGGAAAAAA-5' for which the first nt (T) is number 58856598 or 1574 nt upstream from the beginning of the 3'-UTR at 58858172. Unless another variant exists, -1574 nt from the beginning of the 3'-UTR is a large number of nts away from the TSS.
The closest TATA box-like sequence is 3'-CTCTTAAG-5' on the template strand at 4408 nts from the end of ZSCAN22, which is upstream from the core promoter.
The extra TATA boxes between ZSCAN22 and A1BG strongly suggest that there is at least one gene (or pseudogene) between ZSCAN22 and A1BG not currently in the NCBI database.
On the negative strand between ZNF497 and A1BG, there are no TATA boxes of the form 3’-TATA-A/T-A-A/T-A/G-5’.
For the negative strand going from ZSCAN22 to A1BG there are two TATA boxes: 3'-TATATATA-5' at 1600 nts and 3'-TATATAAA-5' at 1602 nts. These are way too far from the possible TSS in this direction.
These two TATA boxs in the distal promoter at approximately -2860 nts from the TSS, which suggests that there may be a short gene between ZSCAN22 and A1BG.
dBREs
A1BG does not have a TATA box. The closest 7 nucleotide dBRE is at -450 nts which is outside the core promoter in the distal promoter. The closest 6 nt dBRE of consensus sequence 3'-A/G-T-A/G/T-G/T-G/T-G/T-5' is also at -451 nts.
Consensus sequence 3'-T-A/G/T-G/T-G/T-G/T-G/T-5' has an expression at -108 nts with 3'-TGGGTG-5' in the proximal promoter.
Consensus sequence 3'-A/G-T-A/G/T-G/T-G/T-5' has one dBRE at -99 nts with 3'-GTGTG-5' in the proximal promoter.
3'-T-A/G/T-G/T-G/T-G/T-5' is another 5 nts consensus sequence that has a dBRE at -109 nts with 3'-TGGGT-5'.
Another 5 nts consensus sequence that has the same dBRE at -99 nts is 3'-GTGTG-5' in the proximal promoter.
A 4 nts consensus sequence has a dBRE at -100 nts is 3'-GTGT-5' also in the proximal promoter.
A second set of 4 nts consensus sequence dBREs are at -59 nts and +2 nts. The dBRE has not been reported to include the TSS.
A third 4 nts consensus sequence dBRE occurs at -43 nts, roughly just outside the core promoter, with another including the TSS and none in between.
XCPE 1s
There is no X core promoter element 1 between ZSCAN22 and A1BG on the template strand.
MTEs
There is no motif ten element between ZSCAN22 and A1BG on the template strand.
GAACs
There are two GAAC elements in the distal promoter between ZSCAN22 and A1BG.
A1BG does not have a GAAC element within 2-7 nucleotides of the TSS. The closest GAAC element, 3'-GAACT-5', is -999 nts from the TSS.
Inrs
Along the negative strand from ZSCAN22 to A1BG there are at least 43 initiator elements, with the closest one 3'-TCACACT-5' ending at 4361 nts rather than including the TSS of A+1 at 4460 nts.
In the sequence along the positive strand of nucleotides from genes ZSCAN22 and A1BG, the sequence 3'-CCATCCACT-5' occurs only once, just before the transcription start site (TSS) of the A1BG gene. The DCE 3'-CTT-5' contains the TSS at its 5' end.
The TSS for A1BG has the following nts around it: 3'-CCATCCACTT+1TGAGGACAC-5'. Most genes studied early on contained an adenosine (A+1) at the TSS, a cytosine (C-1), and a few pyrimidines (Pys) surrounding these nts.[51]
Usually the Inr contains the TSS. The sequence 3'-CCACTT+1T-5' does contain the TSS but not at A+1. The nearest other Inr ends -24 nt upstream from the TSS.
There are at least 15 Inrs between ZSCAN22 and A1BG.
The sequence 3'-CCACTT+1T-5' is also an Inr, where the TSS is indicated. But, the only DPE nearby 3'-GGACA-5' begins on the fourth nt (+5) after the TSS (+1), not precisely +28 to +32 relative to the TSS nucleotide. No other DPE is even close to this +28 to +32 window.
AGCE1s
A1BG contains 3'-CTT+1-5'. This is an angiotensinogen core promoter element 1 (AGCE1).
The AGCE1 occurs at 3'-CTT+1-5' and 3'-ATC-5' which ends 5 nts upstream from the first and is the only AGCE1 within -25 and -1 nts of the TSS.
DCEs
The downstream core element (DCE) SI 3'-CTT+1-5' contains the TSS but is not downstream of the TSS. Of the DCE SI elements, the closest is some 60 nts downstream from the TSS (3'-CTTC-5' ending at +69).
Of the DCE SII elements such as 3'-CTGT-5', there are none within +1 to +100 nts downstream. Element 3'-CTG-5' occurs starting at +32 nts, which is some +11 nts further downstream than expected and falls within the range for SIII. The element 3’-TGT-5’ has no occurrence within the TSS and +100 nts downstream.
DCE SIII elements (3’-AGC-5’) occur at +19 and +28 nts. The second DCE SIII is close but overlaps any likely nts for DCE SII and is supposed to be after any DCE SII. The DCE SII is unlikely to be used but the second DCE SIII may be close enough to act alone.
Along the negative strand from ZSCAN22 to A1BG and past the TSS, there are no DCE SIs of the type 3'-CTTC-5' past the TSS. Of the type 3'-CTT-5', only one occurs at 4552 nts or 92 nts past the TSS. For type 3'-TTC-5' of DCE SI the closest past is 3'-TTC-5' ending at 4504, or 44 nts past the TSS.
Type SII 3'-CTGT-5' has the closest one ending at 4468 nts and the next ending at 4507 nts.
DPEs
Within the nucleotides of the negative strand going from gene ZSCAN22 to A1BG are at least 163 downstream promoter elements (DPEs), when using the minimal five-nucleotide consensus sequence. There are three DPEs near the required +28 (4487) to +32 (4491) nts from the TSS at 4460 nts from the end of ZSCAN22: 3'-GGTCG-5' at 4480, 3'-AGTCG-5' at 4489, and 3'-GGACC-5' at 4494 nts.
TAFs
Each of the foregoing core promoter elements does not appear to be involved in the transcription of A1BG. The Inr does not contain the known TSS. The DCE is not supposed to contain the TSS, nor is AGCE1 although it is one nt off by containing it. Currently, the only way to default transcribe A1BG is by directing the transcription program directly to the known TSS.
When there is no TATA box in the promoter, a TAF binds sequence specifically, and forces the TBP to bind non-sequence specifically.
5'-untranslated region
The 5' UTR for A1BG contains some 216 nts, depending upon the location of the TSS.
3’-T+1TGAGGACACGAGATCCCAGCCCACTCAGCCCTGGGAGTCCAAAGACATTTTAAACAGAGCCTCTCTTCACATTTA TTAATTCCTGGGAGGAATGAGGGAGGCTTCTCCAGCCCCCCAGAGACCCCGGCCTTGTGCTGCAACAGGAGGGGA GGGAGCCAGTCCAGAATCCCCGGCACTTCTGAGGACACCAACAGCACCCTGGGCCCGCGGCTGCA-5’
The five prime untranslated region (5' UTR), can contain elements for controlling gene expression by way of regulatory elements. It begins at the transcription start site and ends one nucleotide (nt) before the start codon (usually AUG) of the coding region. ... The 5' UTR has a median length of ~150 nt in eukaryotes, but can be as long as several thousand bases. ... Several regulatory sequences may be found in the 5' UTR:
- Binding sites for proteins, that may affect the mRNA's stability or translation, for example iron responsive elements, that regulate gene expression in response to iron.
- Riboswitches.
- Sequences that promote or inhibit translation initiation.
- Introns within 5' UTRs have been linked to regulation of gene expression and mRNA export.[52]
TBP binding
If TBP can bind to any variety of seven A/Ts before the TSS, then the sequence 3'-AAAAAATAATAATTA-5' is likely to be the 3'-"xod-ATAT"-5', rather than the traditional 3'-"TATA-box"-5'. The first (T)-5' is at 58858405, only 233 nts from TSS, but way outside the core promoter.
RNA polymerase II holoenzymes
RNA polymerase II ... is recruited to the promoters of protein-coding genes in living cells.[53] Or, transcription factories are present and the euchromatin is brought within the nearest transcription factory and A1BG messenger RNA (mRNA) is transcribed.
For those circumstances in which the holoenzyme is built onto the euchromatin, it is necessary to consider the holoenzyme components and the likely sequence of binding, RNA polymerase II entrance upon the scene and subsequent action.
RNA polymerase II (also called RNAP II and Pol II) ... catalyzes the transcription of DNA to synthesize precursors of mRNA and most snRNA and microRNA.[54] ... In humans RNAP II consists of seventeen protein molecules (gene products encoded by POLR2A-L, where the proteins synthesized from 2C-, E-, and F-form homodimers).
Hypotheses
- A1BG may be transcribed from either direction.
- A1BG may be transcribed from either strand.
- Each of the many ways to alter gene expression can be applied to the expression of A1BG.
Acknowledgements
The content on this page was first contributed by: Henry A. Hoff.
Initial content for this page in some instances came from Wikiversity.
See also
- A box gene transcriptions
- AGC box gene transcriptions
- AGCE gene transcriptions
- Angiotensinogen core promoter element gene transcriptions
- ATA box gene transcriptions
- B box gene transcriptions
- Bridge gene transcriptions
- Box gene transcriptions
- CAAT box gene transcriptions
- cAMP response element gene transcriptions
- C and D boxes gene transcriptions
- CARE gene transcriptions
- CArG box gene transcriptions
- CAT box gene transcriptions
- C box gene transcriptions
- CENP-B box gene transcriptions
- CGCG box gene transcriptions
- CRE box gene transcriptions
- D box gene transcriptions
- Downstream core element gene transcriptions
- Downstream promoter element gene transcriptions
- Downstream TFIIB recognition element gene transcriptions
- DREB box gene transcriptions
- E2 box gene transcriptions
- Element gene transcriptions
- E box gene transcriptions
- EIF4E basal element gene transcriptions
- Enhancer box gene transcriptions
- Factor II B recognition element gene transcriptions
- F box gene transcriptions
- Fur box gene transcriptions
- GAAC element gene transcriptions
- GARE gene transcriptions
- GA responsive complex gene transcriptions
- GATA gene transcriptions
- G box gene transcriptions
- GC box gene transcriptions
- GCC box gene transcriptions
- General transcription factor II A gene transcriptions
- General transcription factor II B gene transcriptions
- General transcription factor II D gene transcriptions
- General transcription factor II F gene transcriptions
- General transcription factor II H gene transcriptions
- General transcription factor gene transcriptions
- GLM box gene transcriptions
- H and ACA box gene transcriptions
- H box gene transcriptions
- HMG box gene transcriptions
- HNF gene transcriptions
- Homeobox gene transcriptions
- HY box gene transcriptions
- I box gene transcriptions
- Initiator element gene transcriptions
- Kruppel-associated box gene transcriptions
- Kruppel-like factor gene transcriptions
- L box gene transcriptions
- M35 box gene transcriptions
- MADS box gene transcriptions
- M box gene transcriptions
- Metal responsive element gene transcriptions
- Motif ten element gene transcriptions
- MYB recognition element gene transcriptions
- Nuclear factor gene transcriptions
- Nuclear factor of activated T cell gene transcriptions (NFAT)
- P box gene transcriptions
- Preinitiation complex
- Preinitiation complex gene transcriptions
- Pribnow box gene transcriptions
- Prolamin box gene transcriptions
- Pyrimidine box gene transcriptions
- RNA polymerase II holoenzyme complex
- SARE gene transcriptions
- Sp1 gene transcriptions
- STAT gene transcriptions
- TACTAAC box gene transcriptions
- TATA binding protein associated factor gene transcriptions
- TATA binding protein gene transcriptions
- TATA box gene transcriptions
- TAT box gene transcriptions
- TATC box gene transcriptions
- T box gene transcriptions
- TC element gene transcriptions
- TCT gene transcriptions
- Tetradecanoylphorbol-13-acetate response element gene transcriptions
- Transcription factor gene transcriptions
- Transthyration gene transcriptions
- Upstream response element gene transcriptions
- Upstream stimulatory factor gene transcriptions
- V and P box gene transcriptions
- V box gene transcriptions
- W box gene transcriptions
- X box gene transcriptions
- X core promoter element 1 gene transcriptions
- Y box gene transcriptions
- Z box gene transcriptions
References
- ↑ Han Wang, Wei Yan, Zuohua Feng, Yuan Gao, Liu Zhang, Xinxia Feng & Dean Tian (6 January 2020). "Plasma proteomic analysis of autoimmune hepatitis in an improved AIH mouse model". Journal of Translational Medicine 18:3. doi:10.1186/s12967-019-02180-3.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 Tamar Juven-Gershon and James T. Kadonaga (15 March 2010). "Regulation of gene expression via the core promoter and the basal transcriptional machinery". Developmental Biology. 339 (2): 225–9. doi:10.1016/j.ydbio.2009.08.009. Retrieved 2016-01-16.
- ↑ 3.0 3.1 3.2 Mei Tian, Ya-Zhou Cui, Guan-Hua Song, Mei-Juan Zong, Xiao-Yan Zhou, Yu Chen and Jin-Xiang Han (August 2008). "Proteomic analysis identifies MMP-9, DJ-1 and A1BG as overexpressed proteins in pancreatic juice from pancreatic ductal adenocarcinoma patients". BMC Cancer. 16 (8): 241. PMID 18706098. Retrieved 2011-11-28.
- ↑ 4.0 4.1 4.2 Rongming Miao, Bangmei Ding, Yingyi Zhang, Qian Xia, Yong Li, and Baoli Zhu (March 2016). "Proteomic profiling change during the early development of silicosis disease". Journal of Thoracic Discovery. 8 (3): 329–41. doi:10.21037/jtd.2016.02.46. PMID 27076927. Retrieved 2016-06-15.
- ↑ Norma Angélica Galicia Canales, Vicente Madrid Marina, Jorge Salmerón Castro, Alfredo Antúnez Jiménez, Guillermo Mendoza-Hernández, Elizabeth Langley McCarron, Margarita Bahena Roman, and Julieta Ivone Castro-Romero (August 2014). "A1BG and C3 are overexpressed in patients with cervical intraepithelial neoplasia III". Oncology Letters. 8 (2): 939–47. doi:10.3892/ol.2014.2195. PMID 25009667. Retrieved 2016-06-15.
- ↑ Jianling Liu, Dan Wang, Chaoqi Zhang, Zhen Zhang, Xinfeng Chen, Jingyao Lian, Jinbo Liu, Guixian Wang, Weitang Yuan, Zhenqiang Sun, Weijia Wang, Mengjia Song, Yaping Wang, Qian Wu, Ling Cao, Dong Wang, and Yi Zhang (December 2018) Identification of liver metastasis-associated genes in human colon carcinoma by mRNA profiling. Liver metastasis-associated genes in human colon carcinoma". Chinese Journal of Cancer Research 30 (6):633-646. doi: 10.21147/j.issn.1000-9604.2018.06.08.
- ↑ Jung-Mo Ahn, Hye-Jin Sung, Yeon-Hee Yoon, Byung-Gyu Kim, Won Suk Yang, Cheolju Lee, Hae-Min Park, Bum-Jin Kim, Byung-Gee Kim, Soo-Youn Lee, Hyun-Joo An and Je-Yoel Cho (January 2014) "Integrated Glycoproteomics Demonstrates Fucosylated Serum Paraoxonase 1 Alterations in Small Cell Lung Cancer". Molecular & Cellular Proteomics 13 (1): P30-48. doi:10.1074/mcp.M113.028621.
- ↑ Hong-Jun Song, Yan-Li Xue, Zhong-Ling Qiu, and Quan-Yong Luo (6 November 2013). "Comparative serum proteomic analysis identified afamin as a downregulated protein in papillary thyroid carcinoma patients with non-131I-avid lung metastases". Nuclear Medicine Communications. 34 (12): 1196–203. doi:10.1097/MNM.0000000000000001. Retrieved 2016-06-21.
- ↑ 9.00 9.01 9.02 9.03 9.04 9.05 9.06 9.07 9.08 9.09 9.10 9.11 9.12 9.13 9.14 9.15 9.16 9.17 9.18 9.19 9.20 Cissi Gardmo and Agneta Mode (1 December 2006). "In vivo transfection of rat liver discloses binding sites conveying GH-dependent and female-specific gene expression". Journal of Molecular Endocrinology. 37 (3): 433–441. doi:10.1677/jme.1.02116. Retrieved 2017-09-01.
- ↑ H Eiberg, ML Bisgaard, and J Mohr (1989). "Linkage between alpha 1B-glycoprotein (A1BG) and Lutheran (LU) red blood group system: assignment to chromosome 19: new genetic variants of A1BG". Clinical Genetics. 36 (6): 415–8. PMID 2591067. Retrieved 2016-06-17.
- ↑ JJ Catanese and LF Kress (21 January 1992). "Isolation from opossum serum of a metalloproteinase inhibitor homologous to human alpha 1B-glycoprotein". Biochemistry. 31 (2): 410–8. PMID 1731898. Retrieved 2016-06-15.
- ↑ 12.0 12.1 Surza L. G. Rocha, Bruno Lomonte, Ana G. C. Neves-Ferreira, Monique R. O. Trugilho, Inácio de L. M. Junqueira-de-Azevedo, Paulo L. Ho, Gilberto B. Domont, José M. Gutiérrez, and Jonas Perales (December 2002). "Functional analysis of DM64, an antimyotoxic protein with immunoglobulin-like structure from Didelphis marsupialis serum". The FEBS Journal. 269 (24): 6052–62. doi:10.1046/j.1432-1033.2002.03308.x. PMID 12473101. Retrieved 2016-06-15.
- ↑ Bryan McBournie (5 September 2012). "Human genome study could unlock the biology of disease, In: The Washington Post". Retrieved 2016-06-01.
- ↑ 14.0 14.1 Malcolm Ritter (6 September 2012). "Far from being mostly junk, human DNA is 'a jungle' of complex activity, huge project shows". The Washington Post. Retrieved 2016-06-01.
- ↑ T. Wolfsberg, J. McEntyre, and G. Schuler (2001). "Guide to the draft human genome" (PDF). Nature. 409 (6822): 824–6. doi:10.1038/35057000. PMID 11236998. Retrieved 2016-06-01.
- ↑ The ENCODE Project Consortium (2007). "Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project". Nature. 447 (7146): 799–816. Bibcode:2007Natur.447..799B. Retrieved 2016-06-01.
- ↑ D. Rearick, A. Prakash, A. McSweeny, S.S. Shepard, L. Fedorova, and A. Fedorov (March 2011). "Critical association of ncRNA with introns". Nucleic Acids Research. 39 (6): 2357–66. doi:10.1093/nar/gkq1080. PMID 21071396. Retrieved 2016-06-01.
- ↑ 18.0 18.1 18.2 Caitrin W. McDonough, Yan Gong, Sandosh Padmanabhan, Ben Burkley, Taimour Y. Langaee, Olle Melander, Carl J. Pepine, Anna F. Dominiczak, Rhonda M. Cooper-DeHoff, Julie A. Johnson (2013). "Pharmacogenomic Association of Nonsynonymous SNPs in SIGLEC12, A1BG, and the Selectin Region and Cardiovascular Outcomes". Hypertension. 62 (1): 48–52. doi:10.1161/HYPERTENSIONAHA.111.00823. Retrieved 2016-06-17.
- ↑ R.K. Juneja, N. Saha, J.S.H. Tay, P.S. Low & B. Gahne (1994). "Distribution of plasma alpha-1-B-glycoprotein (A1BG) polymorphism in several populations of the Indian subcontinent". Annals of Human Biology. 21 (5): 443–8. doi:10.1080/03014469400003462. Retrieved 2016-06-17.
- ↑ Bradley E. Bernstein, Alexander Meissner, and Eric S. Lander (23 February 2007). "The Mammalian Epigenome". Cell. 128 (4): 669–81. doi:10.1016/j.cell.2007.01.033. Retrieved 2016-06-01.
- ↑ Peter Baumann, Fiona E Benson, Stephen C West (15 November 1996). "Human Rad51 Protein Promotes ATP-Dependent Homologous Pairing and Strand Transfer Reactions In Vitro". Cell. 87 (4): 757–766. doi:10.1016/S0092-8674(00)81394-X. Retrieved 2017-01-25.
- ↑ Phil Green, Brent Ewing, Webb Miller, Pamela J. Thomas, & Eric D. Green (2003). "Transcription-associated mutational asymmetry in mammalian evolution". Nature Genetics. 33 (4): 514–7. doi:10.1038/ng1103. Retrieved 2017-01-25.
- ↑ A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis by Manabu Yoshikawa, Angela Peragine, Mee Yeon Park and R. Scott Poethig in Genes Dev. (2005) Volume 19, pages 2164–2175.
- ↑ Discover the rules of DNA base pairing with an online simulator The base pairing rules for RNA are similar.
- ↑ 25.0 25.1 25.2 25.3 Stephen T. Smale and James T. Kadonaga (July 2003). "The RNA Polymerase II Core Promoter" (PDF). Annual Review of Biochemistry. 72 (1): 449–79. doi:10.1146/annurev.biochem.72.121801.161520. PMID 12651739. Retrieved 2012-05-07.
- ↑ Bork P, Holm L, Sander C (September 1994). "The immunoglobulin fold. Structural classification, sequence patterns and common core". Journal of Molecular Biology. 242 (4): 309–20. doi:10.1006/jmbi.1994.1582. PMID 7932691.
- ↑ Brümmendorf T, Rathjen FG (1995). "Cell adhesion molecules 1: immunoglobulin superfamily". Protein Profile. 2 (9): 963–1108. PMID 8574878.
- ↑ 28.0 28.1 Edda Topfer-Petersen, Mahnaz Ekhlasi-Hundrieser, Christiane Kirchhoff, Tosso Leeb, Harald Sieme (October 2005). "The role of stallion seminal proteins in fertilisation". Animal Reproduction Science. 89 (1–4): 159–70. doi:10.1016/j.anireprosci.2005.06.018. Retrieved 2012-02-26.
- ↑ Lucy J. Schmidt, Kevin M. Regan, S. Keith Anderson, Zhifu Sun, Karla V. Ballman, Donald J. Tindall (December 2009). "Effects of the 5 alpha‐reductase inhibitor dutasteride on gene expression in prostate cancer xenografts" (PDF). The Prostate. 69 (16): 1730–43. doi:10.1002/pros.21022. PMID 19676081. Retrieved 2012-02-26.
- ↑ 30.0 30.1 30.2 30.3 Lene Udby, Ole E. Sørensen, Jesper Pass, Anders H. Johnsen, Niels Behrendt, Niels Borregaard and Lars Kjeldsen (October 2004). "Cysteine-Rich Secretory Protein 3 Is a Ligand of α1B-Glycoprotein in Human Plasma". Biochemistry. 43 (40): 12877–86. doi:10.1021/bi048823e. PMID 15461460. Retrieved 2011-11-28.
- ↑ "The Opossum: Our Marvelous Marsupial, The Social Loner". Wildlife Rescue League.
- ↑ Journal Of Venomous Animals And Toxins – Anti-Lethal Factor From Opossum Serum Is A Potent Antidote For Animal, Plant And Bacterial Toxins. Retrieved 2009-12-29.
- ↑ 33.0 33.1 B Haendler, J Krätzschmar, F Theuring and W D Schleuning (July 1993). "Transcripts for cysteine-rich secretory protein-1 (CRISP-1; DE/AEG) and the novel related CRISP-3 are expressed under androgen control in the mouse salivary gland". Endocrinology. 133 (1): 192–8. doi:10.1210/en.133.1.192. PMID 8319566. Retrieved 2012-02-20.
- ↑ 34.0 34.1 Junfeng Ye, Ludong Tan, Yu Fu, Hongji Xu, Lijia Wen, Yu Deng and Kai Liu (13 June 2019). "LncRNA SNHG15 promotes hepatocellular carcinoma progression by sponging miR‐141‐3p". Journal of Cellular Biochemistry. 120:19775-19775. doi: 10.1002/jcb.29283, https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/jcb.29283.
- ↑ Mehrdad Hashemi, Shima Hajimazdarany, Chakrabhavi Dhananjaya Mohan, Maryam Mohammadi, Shamin Rezaei, Yeganeh Olyae, Yeganeh Goldoost, Amin Ghorbani, Seyed Reza Mirmazloomi, Nazanin Gholinia, Amirabbas Kakavand, Shokooh Salimimoghadam, Yavuz Nuri Ertas, Kanchugarakoppal S. Rangappa, Afshin Taheriazam and Maliheh Entezari (30 October 2022). "Long non-coding RNA/epithelial-mesenchymal transition axis in human cancers: Tumorigenesis, chemoresistance, and radioresistance". Pharmacological Research 186: 106535-106542. doi:10.1016/j.phrs.2022.106535.
- ↑ 36.00 36.01 36.02 36.03 36.04 36.05 36.06 36.07 36.08 36.09 36.10 36.11 36.12 Bryan J. Matthews & David J. Waxman (17 July 2020). "Impact of 3D genome organization, guided by cohesin and CTCF looping, on sex-biased chromatin interactions and gene expression in mouse liver". Epigenetics & Chromatin. 13: 30. doi:10.1186/s13072-020-00350-y. Retrieved 8 May 2023.
- ↑ Collin Homer-Bouthiette, Yang Zhao, Lauren B. Shunkwiler, Benjamine Van Peel, Elizabeth Garrett-Mayer, Rachael C. Baird, Anna I. Rissman, Stephen T. Guest, Stephen P. Ethier, Manorama C. John, Patricia A. Powers, Jill D. Haag, Michael N. Gould & Bart M. G. Smits (10 December 2018). "Deletion of the murine ortholog of the 8q24 gene desert has anti-cancer effects in transgenic mammary cancer models". BMC Cancer 18:1233. doi:10.1186/s12885-018-5109-8.
- ↑ 38.0 38.1 Thomas Abeel, Yvan Saeys, Eric Bonnet, Pierre Rouzé, and Yves Van de Peer (February 2008). "Generic eukaryotic core promoter prediction using structural features of DNA". Genome Research. 18 (2): 310–23. doi:10.1101/gr.6991408. Retrieved 2012-04-04.
- ↑ Jaideep Chaudhary, Michael K. Skinner. "Basic Helix-Loop-Helix Proteins Can Act at the E-Box within the Serum Response Element of the c-fos Promoter to Influence Hormone-Induced Promoter Activation in Sertoli Cells". Molecular Endocrinology. 12 (5): 774–786.
- ↑ 40.0 40.1 40.2 40.3 40.4 Oliver G. McDonald, Brian R. Wamhoff, Mark H. Hoofnagle, and Gary K. Owens (January 4, 2006). "Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo". The Journal of Clinical Investigation. 116 (1): 36–48. Retrieved 2014-06-05.
- ↑ Akiro Higashikawa, Taku Saito, Toshiyuki Ikeda, Satoru Kamekura, Naohiro Kawamura, Akinori Kan, Yasushi Oshima, Shinsuke Ohba, Naoshi Ogata, Katsushi Takeshita, Kozo Nakamura, Ung-Il Chung, Hiroshi Kawaguchi (January 2009). "Identification of the core element responsive to runt-related transcription factor 2 in the promoter of human type x collagen gene". Arthritis & Rheumatism. 60 (1): 166–78. doi:10.1002/art.24243. PMID 19116917. Retrieved 2013-06-18.
- ↑ Uzma Samadani and Robert H. Costa (November 1996). "The transcriptional activator hepatocyte nuclear factor 6 regulates liver gene expression" (PDF). Molecular and Cellular Biology. 16 (11): 6273–84. Retrieved 2017-09-05.
- ↑ Barbara Levinson, Rebecca Conant, Rhonda Schnur, Soma Das, Seymour Packman and Jane Gitschier (1996). "A Repeated Element in the Regulatory Region of the MNK Gene and Its Deletion in A Patient With Occipital Horn Syndrome". Human Molecular Genetics. 5 (11): 1737–42. doi:10.1093/hmg/5.11.1737. Retrieved 2013-04-15.
- ↑ 44.0 44.1 Corinne M. Silva (2004). "Role of STATs as downstream signal transducers in Src family kinase-mediated tumorigenesis" (PDF). Oncogene. 23: 8017–8023. Retrieved 2017-09-02.
- ↑ 45.0 45.1 45.2 45.3 Thomas Abeel, Yves Van de Peer and Yvan Saeys (September 2009). "Toward a gold standard for promoter prediction evaluation". Bioinformatics. 25 (12): i313–20. doi:10.1093/bioinformatics/btp191. Retrieved 2012-04-04.
- ↑ Chuhu Yang, Eugene Bolotin, Tao Jiang, Frances M. Sladek, Ernest Martinez. (March 7, 2007). "Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters". Gene. 389 (1): 52–65. doi:10.1016/j.gene.2006.09.029. PMID 17123746.
- ↑ Georgia A. Patikoglou, Joseph L. Kim, Liping Sun, Sang-Hwa Yang, Thomas Kodadek, and Stephen K. Burley (1999). "TATA element recognition by the TATA box-binding protein has been conserved throughout evolution". Genes & Development. 13: 3217–30. Retrieved 2012-06-12.
- ↑ Jie Min Wong and Erik Bateman (25 May 1994). "TBP-DNA interactions in the minor groove discriminate between A: T and T: A base pairs". Nucleic Acids Research. 22 (10): 1890–96. doi:10.1093/nar/22.10.1890. Retrieved 2012-06-12.
- ↑ Joseph L. Kim, Dimitar B. Nikolov & Stephen K. Burley (7 October 1993). "Co-crystal structure of TBP recognizing the minor groove of a TATA element". Nature. 365: 520–7. doi:10.1038/365520a0. Retrieved 2012-06-12.
- ↑ Youngchang Kim, James. H. Geiger, Steven Hahn & Paul B. Sigler (7 October 1993). "Crystal structure of a yeast TBP/TATA-box complex". Nature. 365: 512–20. Retrieved 2012-06-12.
- ↑ J. Corden, B. Wasylyk, A. Buchwalder, P. Sassone-Corsi, C. Kedinger, P. Chambon (19 September 1980). "Promoter sequences of eukaryotic protein-coding genes". Science. 209 (4463): 1405–14. doi:10.1126/science.6251548. Retrieved 2012-06-12.
- ↑ Cenik, C (2011). "Genome analysis reveals interplay between 5' UTR introns and nuclear mRNA export for secretory and mitochondrial genes". PLoS Genetics. 7 (4). doi:10.1371/journal.pgen.1001366.
- ↑ Myer VE, Young RA (October 1998). "RNA polymerase II holoenzymes and subcomplexes" (PDF). J. Biol. Chem. 273 (43): 27757–60. doi:10.1074/jbc.273.43.27757. PMID 9774381.
- ↑ Kornberg R (1999). "Eukaryotic transcriptional control". Trends in Cell Biology. 9 (12): M46. doi:10.1016/S0962-8924(99)01679-7. PMID 10611681.
Further reading
- Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale ST (January 1994). "DNA Sequence Requirements for Transcriptional Initiator Activity in Mammalian Cells". Mol Cell Biol. 14 (1): 116–27. PMID 8264580.
- Liston DR, Johnson PJ (March 1999). "Analysis of a Ubiquitous Promoter Element in a Primitive Eukaryote: Early Evolution of the Initiator Element". Mol Cell Biol. 19 (3): 2380–8.
- Myer VE, Young RA (October 1998). "RNA polymerase II holoenzymes and subcomplexes" (PDF). J. Biol. Chem. 273 (43): 27757–60. doi:10.1074/jbc.273.43.27757. PMID 9774381.
External links
- GenomeNet KEGG database
- Home - Gene - NCBI
- NCBI All Databases Search
- NCBI Site Search
- PubChem Public Chemical Database
- Virtual Cell Animation Collection, Introducing Transcription