STAT gene transcriptions: Difference between revisions
m (→See also) |
|||
| Line 63: | Line 63: | ||
STAT5 consensus sequence is TTCXXXGAA, where X = A, C, or G.<ref name=Gardmo/> Or, X = G or T.<ref name=Silva/> | STAT5 consensus sequence is TTCXXXGAA, where X = A, C, or G.<ref name=Gardmo/> Or, X = G or T.<ref name=Silva/> | ||
==STAT5 samplings== | |||
{{main|Sampling models|Samplings}} | |||
For the Basic programs (starting with SuccessablesSTAT5.bas) written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), the programs are, are looking for, and found: | |||
# negative strand in the negative direction (from ZSCAN22 to A1BG) is SuccessablesSTAT5--.bas, looking for 3'-TTCNNNGAA-5', 0, | |||
# negative strand in the positive direction (from ZNF497 to A1BG) is SuccessablesSTAT5-+.bas, looking for 3'-TTCNNNGAA-5', 1, 3'-TTCCGGGAA-5', 4247, | |||
# positive strand in the negative direction is SuccessablesSTAT5+-.bas, looking for 3'-TTCNNNGAA-5', 2, 3'-TTCGTTGAA-5', 3506, 3'-TTCCCTGAA-5', 3782, | |||
# positive strand in the positive direction is SuccessablesSTAT5++.bas, looking for 3'-TTCNNNGAA-5', 1, 3'-TTCCATGAA-5', 128, | |||
# complement, negative strand, negative direction is SuccessablesSTAT5c--.bas, looking for 3'-AAGNNNCTT-5', 2, 3'-AAGCAACTT-5', 3506, 3'-AAGGGACTT-5', 3782, | |||
# complement, negative strand, positive direction is SuccessablesSTAT5c-+.bas, looking for 3'-AAGNNNCTT-5', 1, 3'-AAGGTACTT-5', 128, | |||
# complement, positive strand, negative direction is SuccessablesSTAT5c+-.bas, looking for 3'-AAGNNNCTT-5', 0, | |||
# complement, positive strand, positive direction is SuccessablesSTAT5c++.bas, looking for 3'-AAGNNNCTT-5', 1, 3'-AAGGCCCTT-5', 4247, | |||
# inverse complement, negative strand, negative direction is SuccessablesSTAT5ci--.bas, looking for 3'-TTCNNNGAA-5', 0, | |||
# inverse complement, negative strand, positive direction is SuccessablesSTAT5ci-+.bas, looking for 3'-TTCNNNGAA-5', 1, 3'-TTCCGGGAA-5', 4247, | |||
# inverse complement, positive strand, negative direction is SuccessablesSTAT5ci+-.bas, looking for 3'-TTCNNNGAA-5', 2, 3'-TTCGTTGAA-5', 3506, 3'-TTCCCTGAA-5', 3782, | |||
# inverse complement, positive strand, positive direction is SuccessablesSTAT5ci++.bas, looking for 3'-TTCNNNGAA-5', 1, 3'-TTCCATGAA-5', 128, | |||
# inverse, negative strand, negative direction, is SuccessablesSTAT5i--.bas, looking for 3'-AAGNNNCTT-5', 2, 3'-AAGCAACTT-5', 3506, 3'-AAGGGACTT-5', 3782, | |||
# inverse, negative strand, positive direction, is SuccessablesSTAT5i-+.bas, looking for 3'-AAGNNNCTT-5', 1, 3'-AAGGTACTT-5', 128, | |||
# inverse, positive strand, negative direction, is SuccessablesSTAT5i+-.bas, looking for 3'-AAGNNNCTT-5', 0, | |||
# inverse, positive strand, positive direction, is SuccessablesSTAT5i++.bas, looking for 3'-AAGNNNCTT-5', 1, 3'-AAGGCCCTT-5', 4247. | |||
===STAT proximal promoters=== | |||
{{main|Proximal promoter gene transcriptions}} | |||
Negative strand, positive direction: TTCCGGGAA at 4247, and complement. | |||
===STAT distal promoters=== | |||
{{main|Distal promoter gene transcriptions}} | |||
Positive strand, negative direction: TTCGTTGAA at 3506, TTCCCTGAA at 3782, and complements. | |||
Positive strand, positive direction: TTCCATGAA at 128, and complement. | |||
==Human genes== | ==Human genes== | ||
Revision as of 22:55, 5 January 2021
Editor-In-Chief: Henry A. Hoff

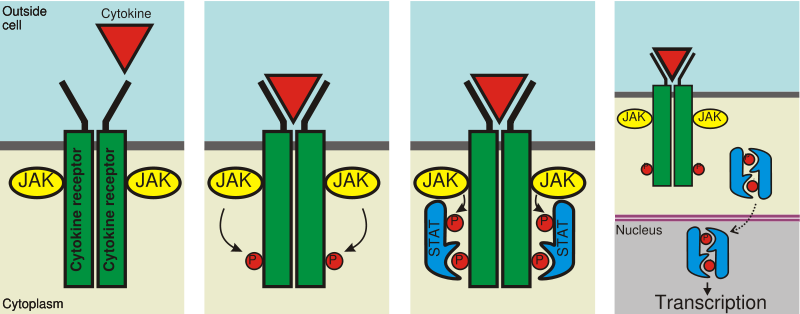
{{free media}}{{free media}}In the diagram on the right, STAT5 may be involved with an erythropoiesis receptor, or Epo Receptor. Murine, members of the subfamily Murinae, Epo Receptor truncations and known functions are included. Erythroid differentiation depends on transcriptional regulator GATA1, zinc finger DNA binding domain binds specifically to DNA consensus sequence [AT]GATA[AG] promoter elements. In erythropoiesis, EpoR is best known for inducing survival of progenitors.
The second down diagram on the right apparently describes the activation of STAT5.
The two live cell videos demonstrate that methylsulfonylmethane (MSM) suppresses breast cancer growth by down regulating STAT3 and STAT5b pathways.
A1BGs
{{fairuse}}Both "the 2.3 kb and the 160 bp proximal parts of the a1bg promoter direct sex-specific expression of the reporter gene, and that a negative regulatory element resides in the −1 kb to −160 bp region."[1]
"Computer analysis of the 2.3 kb rat a1bg promoter fragment revealed two putative Stat5 sites and one [hepatic nuclear factor 6] HNF6/HNF3 binding site at −2077/−2069, −69/−61 and −137/−128 respectively [...]."[1]
The "GH-dependent sexually dimorphic expression conveyed by the 2.3 kb a1bg promoter is enhanced by the HNF6/HNF3 site and, if anything, reduced by the proximal Stat5 site in that the impact of the 3′Stat5 mutation was more pronounced in males."[1]
The "binding of Stat5 and HNF6 to the respective site by electromobility shift analysis (EMSA) [was verified] using female-derived [rat] liver nuclear extracts. [...] Stat5 bound to the a1bg proximal Stat5 site, 3′Stat5 and the mutated oligonucleotide was unable to compete for the binding. Similarly, HNF6 bound to the a1bg HNF6 oligonucleotide, but in this case, the mutated oligonucleotide was able to compete for binding when added in large excess [...]. However, [...] the HNF6 binding capacity of the mutated oligonucleotide was clearly reduced. A 20 molar excess of the mutated oligonucleotide had only a marginal effect on the binding of HNF6 [...], whereas a 20 molar excess of unlabelled probe [...] completely abolished binding. Supershift analysis with an HNF6 antibody revealed a complex with a slightly lower mobility than the HNF6 complex [...]. By extending the electrophoresis run and including nuclear extract from hypophysectomized rats, devoid of GH and thereby lacking HNF6 (Lahuna et al. 1997), the two different complexes were clearly visualized. The complex with the lower mobility is most probably due to the binding of HNF3, in analogy with what was shown by Lahuna et al. for the CYP2C12 HNF6 binding site; HNF3 can bind to the site in the absence of HNF6 (Lahuna et al. 1997). To summarize the EMSA results, Stat5 and HNF6 could bind to their respective site in the a1bg promoter in vitro, and the mutations introduced in respective site abolished binding of the corresponding factor."[1]
The "expression of a −116/−89 deletion construct in which also the HNF6 site was mutated, (−116/−89) delmutHNF6-Luc, [...] the generated luciferase activities were reduced in both sexes [...]. This is in contrast to that mutation/deletion of the sites separately only affected the expression in female livers."[1]
The "−116/−89 region contains a site(s) of importance for the GH-dependent and female-specific expression of the a1bg gene, and that the impact of this region together with the HNF6 site is more complex than mere enhancement of the expression in females."[1]
The "Stat5 site conveys expression of a1bg to higher extent in male than in female livers, thereby reducing the sex difference. [...] On the other hand, HNF6 is expressed at higher levels in female than in male rat liver (Lahuna et al. 1997). Indeed, following mutation of the HNF6-binding element, mutHNF6-Luc, the sex-differentiated expression was attenuated due to reduced expression in females. Thus, for a1bg, the sex-related difference in amount of HNF6 is likely to contribute to the sex-differentiated and female characteristic expression."[1]
Nuclear "proteins binding to the a1bg −116/−89 region [are] members of the [nuclear factor 1] NF1 and the [octamer transcription factor] Oct families of transcription factors. NF1 genes are expressed in most adult tissues (Osada et al. 1999). It is not known how NF1 modulates transcriptional activity, and both activation and repression of transcription have been reported (Gronostajski 2000). Cofactors such as CBP/p300 and HDAC have been shown to interact with NF1 proteins suggesting modulation of chromatin structure (Chaudhry et al. 1999). NF1 factors have also been shown to interact directly with the basal transcription machinery as well as with other transcription factors, including Stat5 (Kim & Roeder 1994, Mukhopadhyay et al. 2001) and synergistic effects with HNF4 have been reported (Ulvila et al. 2004). In addition to the HNF6, Stat5 and NF1/Oct sites, the a1bg promoter harbours an imperfect HNF4 site at −51/−39 with two mismatches compared with the HNF4 consensus site. HNF4 is clearly important for the expression of CYP2C12 (Sasaki et al. 1999), however, the −51/−39 region in a1bg was not protected in the footprinting analysis and was therefore not analysed further. Like NF1, Oct proteins have been reported to be involved in activation as well as repression of gene expression (Phillips & Luisi 2000). Stat5 has been shown to form a stable complex with Oct-1 (Magne et al. 2003). Moreover, NF1 and Oct-1 have been shown to, reciprocally, facilitate each other’s binding (O’Connor & Bernard 1995, Belikov et al. 2004)."[1]
In the diagram on the right is liver "expression of a1bg-luciferase constructs. (A) Stat5 and HNF6 consensus sequences and corresponding sites in the 2.3 kb a1bg promoter alongside with the used mutations. (B) Female (black bars) and male (open bars) rats [results]."[1]
Consensus sequences
"STATs [signal transducers and activators of transcription] bind through their DNA-binding domain (DBD) to consensus elements (TTCTTGGAA, STAT5 consensus), resulting in gene transcription."[2]
STAT5 consensus sequence is TTCXXXGAA, where X = A, C, or G.[1] Or, X = G or T.[2]
STAT5 samplings
For the Basic programs (starting with SuccessablesSTAT5.bas) written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), the programs are, are looking for, and found:
- negative strand in the negative direction (from ZSCAN22 to A1BG) is SuccessablesSTAT5--.bas, looking for 3'-TTCNNNGAA-5', 0,
- negative strand in the positive direction (from ZNF497 to A1BG) is SuccessablesSTAT5-+.bas, looking for 3'-TTCNNNGAA-5', 1, 3'-TTCCGGGAA-5', 4247,
- positive strand in the negative direction is SuccessablesSTAT5+-.bas, looking for 3'-TTCNNNGAA-5', 2, 3'-TTCGTTGAA-5', 3506, 3'-TTCCCTGAA-5', 3782,
- positive strand in the positive direction is SuccessablesSTAT5++.bas, looking for 3'-TTCNNNGAA-5', 1, 3'-TTCCATGAA-5', 128,
- complement, negative strand, negative direction is SuccessablesSTAT5c--.bas, looking for 3'-AAGNNNCTT-5', 2, 3'-AAGCAACTT-5', 3506, 3'-AAGGGACTT-5', 3782,
- complement, negative strand, positive direction is SuccessablesSTAT5c-+.bas, looking for 3'-AAGNNNCTT-5', 1, 3'-AAGGTACTT-5', 128,
- complement, positive strand, negative direction is SuccessablesSTAT5c+-.bas, looking for 3'-AAGNNNCTT-5', 0,
- complement, positive strand, positive direction is SuccessablesSTAT5c++.bas, looking for 3'-AAGNNNCTT-5', 1, 3'-AAGGCCCTT-5', 4247,
- inverse complement, negative strand, negative direction is SuccessablesSTAT5ci--.bas, looking for 3'-TTCNNNGAA-5', 0,
- inverse complement, negative strand, positive direction is SuccessablesSTAT5ci-+.bas, looking for 3'-TTCNNNGAA-5', 1, 3'-TTCCGGGAA-5', 4247,
- inverse complement, positive strand, negative direction is SuccessablesSTAT5ci+-.bas, looking for 3'-TTCNNNGAA-5', 2, 3'-TTCGTTGAA-5', 3506, 3'-TTCCCTGAA-5', 3782,
- inverse complement, positive strand, positive direction is SuccessablesSTAT5ci++.bas, looking for 3'-TTCNNNGAA-5', 1, 3'-TTCCATGAA-5', 128,
- inverse, negative strand, negative direction, is SuccessablesSTAT5i--.bas, looking for 3'-AAGNNNCTT-5', 2, 3'-AAGCAACTT-5', 3506, 3'-AAGGGACTT-5', 3782,
- inverse, negative strand, positive direction, is SuccessablesSTAT5i-+.bas, looking for 3'-AAGNNNCTT-5', 1, 3'-AAGGTACTT-5', 128,
- inverse, positive strand, negative direction, is SuccessablesSTAT5i+-.bas, looking for 3'-AAGNNNCTT-5', 0,
- inverse, positive strand, positive direction, is SuccessablesSTAT5i++.bas, looking for 3'-AAGNNNCTT-5', 1, 3'-AAGGCCCTT-5', 4247.
STAT proximal promoters
Negative strand, positive direction: TTCCGGGAA at 4247, and complement.
STAT distal promoters
Positive strand, negative direction: TTCGTTGAA at 3506, TTCCCTGAA at 3782, and complements.
Positive strand, positive direction: TTCCATGAA at 128, and complement.
Human genes
GeneID: 6772 is STAT1 signal transducer and activator of transcription 1 (aka STAT91). "The protein encoded by this gene is a member of the STAT protein family. In response to cytokines and growth factors, STAT family members are phosphorylated by the receptor associated kinases, and then form homo- or heterodimers that translocate to the cell nucleus where they act as transcription activators. This protein can be activated by various ligands including interferon-alpha, interferon-gamma, EGF, PDGF and IL6. This protein mediates the expression of a variety of genes, which is thought to be important for cell viability in response to different cell stimuli and pathogens. Two alternatively spliced transcript variants encoding distinct isoforms have been described."[3]
- NP_009330.1 signal transducer and activator of transcription 1-alpha/beta isoform alpha.
- NP_644671.1 signal transducer and activator of transcription 1-alpha/beta isoform beta.
- XP_016860272.1 signal transducer and activator of transcription 1-alpha/beta isoform X1.
- XP_006712781.1 signal transducer and activator of transcription 1-alpha/beta isoform X2.
- XR_001738914.2 Homo sapiens signal transducer and activator of transcription 1 (STAT1), transcript variant X3, misc_RNA.
- XR_001738915.2 Homo sapiens signal transducer and activator of transcription 1 (STAT1), transcript variant X4, misc_RNA.
GeneID: 6773 is STAT2 signal transducer and activator of transcription 2 (aka STAT113). "The protein encoded by this gene is a member of the STAT protein family. In response to cytokines and growth factors, STAT family members are phosphorylated by the receptor associated kinases, and then form homo- or heterodimers that translocate to the cell nucleus where they act as transcription activators. In response to interferon (IFN), this protein forms a complex with STAT1 and IFN regulatory factor family protein p48 (ISGF3G), in which this protein acts as a transactivator, but lacks the ability to bind DNA directly. Transcription adaptor P300/CBP (EP300/CREBBP) has been shown to interact specifically with this protein, which is thought to be involved in the process of blocking IFN-alpha response by adenovirus. Multiple transcript variants encoding different isoforms have been found for this gene."[4]
- NP_005410.1 signal transducer and activator of transcription 2 isoform 1.
- NP_938146.1 signal transducer and activator of transcription 2 isoform 2.
- XP_011536999.1 signal transducer and activator of transcription 2 isoform X1.
- XP_011537000.1 signal transducer and activator of transcription 2 isoform X2.
- XP_011537001.1 signal transducer and activator of transcription 2 isoform X3.
- XP_011537002.1 signal transducer and activator of transcription 2 isoform X4.
- XP_016875393.1 signal transducer and activator of transcription 2 isoform X5.
- XR_001748856.1 Homo sapiens signal transducer and activator of transcription 2 (STAT2), transcript variant X6, misc_RNA.
- XR_002957375.1 Homo sapiens signal transducer and activator of transcription 2 (STAT2), transcript variant X7, misc_RNA.
- XR_001748857.1 Homo sapiens signal transducer and activator of transcription 2 (STAT2), transcript variant X8, misc_RNA.
- XR_245953.3 Homo sapiens signal transducer and activator of transcription 2 (STAT2), transcript variant X9, misc_RNA.
- XR_001748858.2 Homo sapiens signal transducer and activator of transcription 2 (STAT2), transcript variant X10, misc_RNA.
- XR_002957376.1 Homo sapiens signal transducer and activator of transcription 2 (STAT2), transcript variant X11, misc_RNA.
GeneID: 6774 is STAT3 signal transducer and activator of transcription 3. "The protein encoded by this gene is a member of the STAT protein family. In response to cytokines and growth factors, STAT family members are phosphorylated by the receptor associated kinases, and then form homo- or heterodimers that translocate to the cell nucleus where they act as transcription activators. This protein is activated through phosphorylation in response to various cytokines and growth factors including IFNs, EGF, IL5, IL6, HGF, LIF and BMP2. This protein mediates the expression of a variety of genes in response to cell stimuli, and thus plays a key role in many cellular processes such as cell growth and apoptosis. The small GTPase Rac1 has been shown to bind and regulate the activity of this protein. PIAS3 protein is a specific inhibitor of this protein. Mutations in this gene are associated with infantile-onset multisystem autoimmune disease and hyper-immunoglobulin E syndrome. Alternative splicing results in multiple transcript variants encoding distinct isoforms."[5]
- NP_644805.1 signal transducer and activator of transcription 3 isoform 1.
- NP_003141.2 signal transducer and activator of transcription 3 isoform 2.
- NP_998827.1 signal transducer and activator of transcription 3 isoform 3.
- XP_011523447.1 signal transducer and activator of transcription 3 isoform X1.
- XP_005257674.2 signal transducer and activator of transcription 3 isoform X1.
- XP_005257673.2 signal transducer and activator of transcription 3 isoform X2.
- XP_016880464.1 signal transducer and activator of transcription 3 isoform X2.
- XP_011523448.1 signal transducer and activator of transcription 3 isoform X3.
- XP_016880461.1 signal transducer and activator of transcription 3 isoform X3.
- XP_016880462.1 signal transducer and activator of transcription 3 isoform X4.
- XP_016880463.1 signal transducer and activator of transcription 3 isoform X4.
- XP_016880465.1 signal transducer and activator of transcription 3 isoform X4.
- XP_024306664.1 signal transducer and activator of transcription 3 isoform X5.
GeneID: 6775 is STAT4 signal transducer and activator of transcription 4. "The protein encoded by this gene is a member of the STAT family of transcription factors. In response to cytokines and growth factors, STAT family members are phosphorylated by the receptor associated kinases, and then form homo- or heterodimers that translocate to the cell nucleus where they act as transcription activators. This protein is essential for mediating responses to IL12 in lymphocytes, and regulating the differentiation of T helper cells. Mutations in this gene may be associated with systemic lupus erythematosus and rheumatoid arthritis. Alternate splicing results in multiple transcript variants that encode the same protein."[6]
- NP_003142.1 signal transducer and activator of transcription 4 (variant 1).
- NP_001230764.1 signal transducer and activator of transcription 4 (variant 2).
- XP_011510007.1 signal transducer and activator of transcription 4 isoform X1.
- XP_006712782.1 signal transducer and activator of transcription 4 isoform X1.
- XP_016860273.1 signal transducer and activator of transcription 4 isoform X2.
GeneID: ID: 6776 is STAT5A signal transducer and activator of transcription 5A (aka STAT5). "The protein encoded by this gene is a member of the STAT family of transcription factors. In response to cytokines and growth factors, STAT family members are phosphorylated by the receptor associated kinases, and then form homo- or heterodimers that translocate to the cell nucleus where they act as transcription activators. This protein is activated by, and mediates the responses of many cell ligands, such as IL2, IL3, IL7 GM-CSF, erythropoietin, thrombopoietin, and different growth hormones. Activation of this protein in myeloma and lymphoma associated with a TEL/JAK2 gene fusion is independent of cell stimulus and has been shown to be essential for tumorigenesis. The mouse counterpart of this gene is found to induce the expression of BCL2L1/BCL-X(L), which suggests the antiapoptotic function of this gene in cells. Alternatively spliced transcript variants have been found for this gene."[7]
- NP_001275647.1 signal transducer and activator of transcription 5A isoform 1 (variant 1).
- NP_003143.2 signal transducer and activator of transcription 5A isoform 1 (variant 2).
- NP_001275648.1 signal transducer and activator of transcription 5A isoform 2 (variant 3).
- NP_001275649.1 signal transducer and activator of transcription 5A isoform 3 (variant 4).
- XP_005257681.1 signal transducer and activator of transcription 5A isoform X1.
GeneID: ID: 6777 is STAT5B signal transducer and activator of transcription 5B (aka STAT5). "The protein encoded by this gene is a member of the STAT family of transcription factors. In response to cytokines and growth factors, STAT family members are phosphorylated by the receptor associated kinases, and then form homo- or heterodimers that translocate to the cell nucleus where they act as transcription activators. This protein mediates the signal transduction triggered by various cell ligands, such as IL2, IL4, CSF1, and different growth hormones. It has been shown to be involved in diverse biological processes, such as TCR signaling, apoptosis, adult mammary gland development, and sexual dimorphism of liver gene expression. This gene was found to fuse to retinoic acid receptor-alpha (RARA) gene in a small subset of acute promyelocytic leukemias (APLL). The dysregulation of the signaling pathways mediated by this protein may be the cause of the APLL."[8]
- NP_036580.2 signal transducer and activator of transcription 5B.
- XP_024306665.1 signal transducer and activator of transcription 5B isoform X1.
- XP_024306666.1 signal transducer and activator of transcription 5B isoform X1.
- XP_016880466.1 signal transducer and activator of transcription 5B isoform X2.
- XP_005257683.1 signal transducer and activator of transcription 5B isoform X3.
Signal transducer and activator of transcription 5 (STAT5) actually consists of STAT5A (GeneID: ID: 6776) and STAT5B (GeneID: ID: 6777).
GeneID: ID: 6778 is STAT6 signal transducer and activator of transcription 6 (aka STAT6B, STAT6C). "The protein encoded by this gene is a member of the STAT family of transcription factors. In response to cytokines and growth factors, STAT family members are phosphorylated by the receptor associated kinases, and then form homo- or heterodimers that translocate to the cell nucleus where they act as transcription activators. This protein plays a central role in exerting IL4 mediated biological responses. It is found to induce the expression of BCL2L1/BCL-X(L), which is responsible for the anti-apoptotic activity of IL4. Knockout studies in mice suggested the roles of this gene in differentiation of T helper 2 (Th2) cells, expression of cell surface markers, and class switch of immunoglobulins. Alternative splicing results in multiple transcript variants."[9]
- NP_001171549.1 signal transducer and activator of transcription 6 isoform 1 (variant 1).
- NP_003144.3 signal transducer and activator of transcription 6 isoform 1 (variant 2).
- NP_001171550.1 signal transducer and activator of transcription 6 isoform 1 (variant 3).
- NP_001171551.1 signal transducer and activator of transcription 6 isoform 2 (variant 4).
- NP_001171552.1 signal transducer and activator of transcription 6 isoform 2 (variant 5).
- NR_033659.1 Homo sapiens signal transducer and activator of transcription 6 (STAT6), transcript variant 6, non-coding RNA.
- XP_011537005.1 signal transducer and activator of transcription 6 isoform X1.
- XP_011537007.1 signal transducer and activator of transcription 6 isoform X1.
- XP_011537009.1 signal transducer and activator of transcription 6 isoform X1.
- XP_011537010.1 signal transducer and activator of transcription 6 isoform X2.
- XP_011537006.1 signal transducer and activator of transcription 6 isoform X1.
Acknowledgements
The content on this page was first contributed by: Henry A. Hoff.
Initial content for this page in some instances came from Wikiversity.
See also
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 1.9 Cissi Gardmo and Agneta Mode (1 December 2006). "In vivo transfection of rat liver discloses binding sites conveying GH-dependent and female-specific gene expression". Journal of Molecular Endocrinology. 37 (3): 433–441. doi:10.1677/jme.1.02116. Retrieved 2017-09-01.
- ↑ 2.0 2.1 Corinne M. Silva (2004). "Role of STATs as downstream signal transducers in Src family kinase-mediated tumorigenesis" (PDF). Oncogene. 23: 8017–8023. Retrieved 2017-09-02.
- ↑ RefSeq (July 2008). STAT1 signal transducer and activator of transcription 1 [ Homo sapiens (human) ]. 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 14 November 2018.
- ↑ RefSeq (March 2010). STAT2 signal transducer and activator of transcription 2 [ Homo sapiens (human) ]. 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 14 November 2018.
- ↑ RefSeq (September 2015). STAT3 signal transducer and activator of transcription 3 [ Homo sapiens (human) ]. 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 14 November 2018.
- ↑ RefSeq (August 2011). STAT4 signal transducer and activator of transcription 4 [ Homo sapiens (human) ]. 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 14 November 2018.
- ↑ RefSeq (December 2013). STAT5A signal transducer and activator of transcription 5A [ Homo sapiens (human) ]. 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 14 November 2018.
- ↑ RefSeq (July 2008). STAT5B signal transducer and activator of transcription 5B [ Homo sapiens (human) ]. 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 14 November 2018.
- ↑ RefSeq (May 2010). STAT6 signal transducer and activator of transcription 6 [ Homo sapiens (human) ]. 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 14 November 2018.