Actin
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]
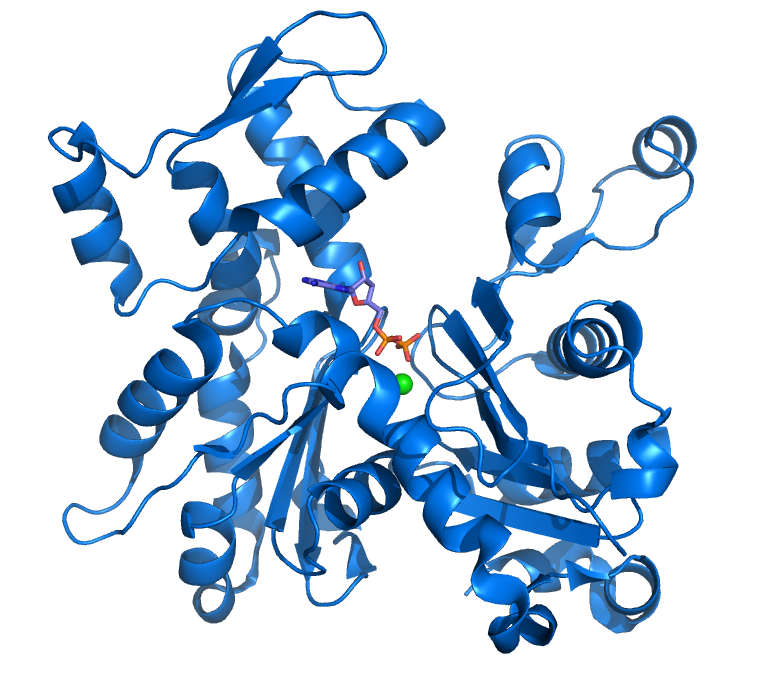
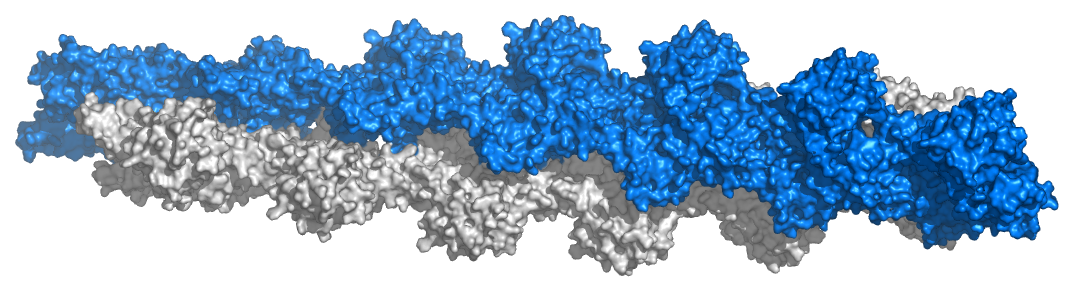
Actin is a globular, roughly 42-kDa protein found in all eukaryotic cells (except for nematode sperm) where it may be present at concentrations of over 100 μM. It is also one of the most highly-conserved proteins, differing by no more than 20% in species as diverse as algae and humans. It is the monomeric subunit of microfilaments, one of the three major components of the cytoskeleton, and of thin filaments, which are part of the contractile apparatus in muscle cells. Thus, actin participates in many important cellular functions, including muscle contraction, cell motility, cell division and cytokinesis, vesicle and organelle movement, cell signaling, and the establishment and maintenance of cell junctions and cell shape.
Formation of thin filament
Genetics
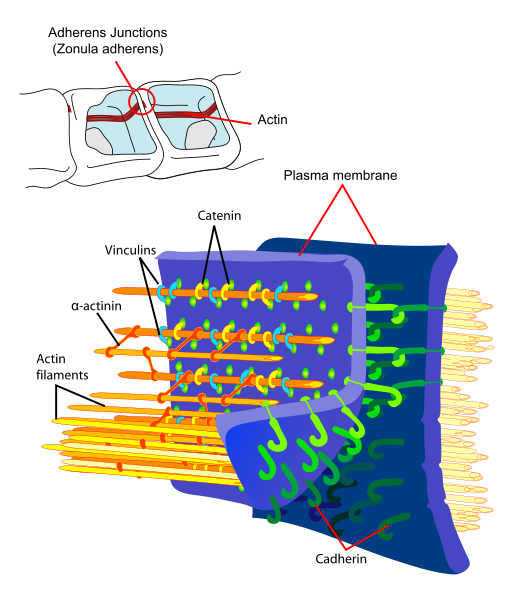
Principal interactions of structural proteins at cadherin-based adherens junction. Actin filaments are linked to α-actinin and to membrane through vinculin. The head domain of vinculin associates to E-cadherin via α-, β-, and γ-catenins. The tail domain of vinculin binds to membrane lipids and to actin filaments.
The protein actin is one of the most highly conserved throughout evolution because it interacts with a large number of other proteins, with 80.2% sequence conservation at the gene level between Homo sapiens and Saccharomyces cerevisiae (a species of yeast), and 95% conservation of the primary structure of the protein product.
Although most yeasts have only a single actin gene, higher eukaryotes, in general, express several isoforms of actin encoded by a family of related genes. Mammals have at least six actin isoforms coded by separate genes,[1] which are divided into three classes (alpha, beta and gamma) according to their isoelectric point. In general, alpha actins are found in muscle (α-skeletal, α-aortic smooth, α-cardiac, and γ2-enteric smooth), whereas beta and gamma isoforms are prominent in non-muscle cells (β- and γ1-cytoplasmic). Although the amino acid sequences and in vitro properties of the isoforms are highly similar, these isoforms cannot completely substitute for one another in vivo.[2]
The typical actin gene has an approximately 100-nucleotide 5' UTR, a 1200-nucleotide translated region, and a 200-nucleotide 3' UTR. The majority of actin genes are interrupted by introns, with up to 6 introns in any of 19 well-characterised locations. The high conservation of the family makes actin the favoured model for studies comparing the introns-early and introns-late models of intron evolution.
All non-spherical prokaryotes appear to possess genes such as MreB, which encode homologues of actin; these genes are required for the cell's shape to be maintained. The plasmid-derived gene ParM encodes an actin-like protein whose polymerised form is dynamically unstable, and appears to partition the plasmid DNA into the daughter cells during cell division by a mechanism analogous to that employed by microtubules in eukaryotic mitosis.[3] Actin is found in both smooth and rough endoplasmic reticulums.
Functions
Actin has three main functions in cells :
- To form the most dynamic one of the three subclasses of the cytoskeleton, which gives mechanical support to cells, and hardwires the cytoplasm with the surroundings to support signal transduction.
- To allow cell motility (see Actoclampin molecular motors).
- In muscle cells as well as non-muscle cells, to generate force together with myosin proteins to support muscle contraction, vesicle movement, and other transport processes.
Individual subunits of actin are known as globular actin (G-actin). G-actin subunits assemble into long filamentous polymers called F-actin. Two parallel F-actin strands twist around each other in a helical formation, giving rise to microfilaments of the cytoskeleton. Microfilaments measure approximately 7 nm in diameter with a loop of the helix repeating every 37 nm.
Polarity
The polarity of an actin filament can be determined by decorating the microfilament with myosin "S1" fragments, creating barbed (+) and pointed (-) ends on the filament. An S1 fragment is composed of the head and neck domains of myosin II.
Actomyosin filaments
In muscle, actin is the major component of thin filaments, which, together with the motor protein myosin (which forms thick filaments), are arranged into actomyosin myofibrils. These fibrils comprise the mechanism of muscle contraction. Using the hydrolysis of ATP for energy, myosin heads undergo a cycle during which they attach to thin filaments, exerting a tension, and then depending on the load, perform a power stroke that causes the thin filaments to slide past, shortening the muscle.
In contractile bundles, the actin-bundling protein alpha-actinin separates each thin filament by ~35 nm. This increase in distance allows thick filaments to fit in between and interact, enabling deformation or contraction. In deformation, one end of myosin is bound to the plasma membrane while the other end "walks" toward the plus end of the actin filament. This pulls the membrane into a different shape relative to the cell cortex. For contraction, the myosin molecule is usually bound to two separate filaments and both ends simultaneously "walk" toward their filament's plus end, sliding the actin filaments closer to each other. This results in the shortening, or contraction, of the actin bundle (but not the filament). This mechanism is responsible for muscle contraction and cytokinesis, the division of one cell into two.
Actin polymerization and depolymerization is necessary in chemotaxis and cytokinesis. Nucleating factors are necessary to stimulate actin polymerization. Also, Actin filaments themselves bind ATP, and hydrolysis of this ATP stimulates destabilization of the polymer.
History
Actin was first observed experimentally in 1887 by W.D. Halliburton, who extracted a protein from muscle that 'coagulated' preparations of myosin, and that he dubbed "myosin-ferment."[4] However, Halliburton was unable to further characterise his findings, and the discovery of actin is credited instead to Brúnó F. Straub, a young biochemist working in Albert Szent-Györgyi's laboratory at the Institute of Medical Chemistry at the University of Szeged, Hungary.
In 1942, Straub developed a novel technique for extracting muscle protein that allowed him to isolate substantial amounts of relatively-pure actin. Straub's method is essentially the same as that used in laboratories today. Szent-Gyorgyi had previously described the more viscous form of myosin produced by slow muscle extractions as 'activated' myosin, and, since Straub's protein produced the activating effect, it was dubbed actin. The hostilities of World War II meant that Szent-Gyorgyi and Straub were unable to publish the work in Western scientific journals; it became well-known in the West only in 1945, when it was published as a supplement to the Acta Physiologica Scandinavica.[5]
Straub continued to work on actin and in 1950 reported that actin contains bound ATP [6] and that, during polymerisation of the protein into microfilaments, the nucleotide is hydrolysed to ADP and inorganic phosphate (which remain bound in the microfilament). Straub suggested that the transformation of ATP-bound actin to ADP-bound actin played a role in muscular contraction. In fact, this is true only in smooth muscle, and was not supported through experimentation until 2001.[7]
The crystal structure of G-actin was solved in 1990 by Kabsch and colleagues.[8] In the same year a model for F-actin was proposed by Holmes and colleagues.[9] The model was derived by fitting a helix of G-actin structures according to low-resolution fiber diffraction data from the filament. Several models of the filament have been proposed since. However there is still no high-resolution X-ray structure of F-actin.
The Listeria bacteria use the cellular machinery to move around inside the host cell, by inducing directed polymerisation of actin by the ActA transmembrane protein, thus pushing the bacterial cell around.
See also
- MreB - an actin homologue in bacteria
- Motor protein
- ACTA1 - alpha actin 1
- ACTB - beta actin
- ACTG1 - gamma actin 1
References
- ↑ Vandekerckhove J. and Weber K. (1978) At least six different actins are expressed in a higher mammal: an analysis based on the amino acid sequence of the amino-terminal tryptic peptide. J Mol Biol 126:783–802 Template:Entrez Pubmed
- ↑ Khaitlina SY (2001) Functional specificity of actin isoforms. Int Rev Cytol 202:35-98 Template:Entrez Pubmed
- ↑ Garner EC et al (2007) Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science 315:1270-1274 Template:Entrez Pubmed
- ↑ Halliburton, W.D. (1887) On muscle plasma. J. Physiol. 8, 133
- ↑ Szent-Gyorgyi, A. (1945) Studies on muscle. Acta Physiol Scandinav 9 (suppl. 25)
- ↑ Straub, F.B. and Feuer, G. (1950) Adenosinetriphosphate the functional group of actin. Biochim. Biophys. Acta. 4, 455-470 Template:Entrez Pubmed
- ↑ Bárány, M., Barron, J.T., Gu, L., and Bárány, K. (2001) Exchange of the actin-bound nucleotide in intact arterial smooth muscle. J. Biol. Chem., 276, 48398-48403 Template:Entrez Pubmed
- ↑ Kabsch, W., Mannherz, E.G., Suck, D., Pai, E.F., and Holmes, K.C. (1990) Atomic structure of the actin:DNase I complex. Nature, 347, 37-44 Template:Entrez Pubmed
- ↑ Holmes KC, Popp D, Gebhard W, Kabsch W. (1990) Atomic model of the actin filament. Nature, 347, 21-2 Template:Entrez Pubmed
bg:Актин cs:Aktin da:Actin de:Aktin et:Aktiin gl:Actina it:Actina he:אקטין mk:Актин nl:Actine sr:Актински филаменти fi:Aktiini sv:Aktin tl:Actin uk:Актин