Electron transport chain
Overview
An electron transport chain associates electron carriers (such as NADH and FADH2) and mediating biochemical reactions that produce adenosine triphosphate (ATP), which is a major energy intermediate in living organisms. Only two sources of energy are available to biosynthesize organic molecules and maintain biochemical and kinetic processes in living organisms: oxidation-reduction (redox) reactions and some forms of radiation, such as sunlight (used for photosynthesis). Organisms that use redox reactions to produce ATP are called chemotrophs. Organisms that use sunlight are called phototrophs. Both chemotrophs and phototrophs use electron transport chains to convert energy into ATP. This is achieved through a three-step process:
Background
The Electron Transport Chain is also called the ETC. ATP is made by an enzyme called ATP Synthase. The structure of this enzyme and its underlying genetic code is remarkably conserved in all known forms of life.
ATP synthase is powered by a transmembrane electrochemical potential gradient, usually in the form of a proton gradient. The function of the electron transport chain is to produce this gradient. In all living organisms, a series of redox reactions is used to produce a transmembrane electrochemical potential gradient.
Redox reactions are chemical reactions in which electrons are transferred from a donor molecule to an acceptor molecule. The underlying force driving these reactions is the Gibbs free energy of the reactants and products. The Gibbs free energy is the energy available ("free") to do work. Any reaction that decreases the overall Gibbs free energy of a system will proceed spontaneously.
The transfer of electrons from a high-energy molecule (the donor) to a lower-energy molecule (the acceptor) can be spatially separated into a series of intermediate redox reactions. This is an electron transport chain.
The fact that a reaction is thermodynamically possible does not mean that it will actually occur; for example, a mixture of hydrogen gas and oxygen gas does not spontaneously ignite. It is necessary either to supply an activation energy or to lower the intrinsic activation energy of the system, in order to make most biochemical reactions proceed at a useful rate. Living systems use complex macromolecular structures (enzymes) to lower the activation energies of biochemical reactions.
It is possible to couple a thermodynamically favorable reaction (a transition from a high-energy state to a lower-energy state) to a thermodynamically unfavorable reaction (such as a separation of charges, or the creation of an osmotic gradient), in such a way that the overall free energy of the system decreases (making it thermodynamically possible), while useful work is done at the same time. Biological macromolecules that catalyze a thermodynamically unfavorable reaction if and only if a thermodynamically favorable reaction occurs simultaneously underlie all known forms of life.
Electron transport chains capture energy in the form of a transmembrane electrochemical potential gradient. This energy can then be harnessed to do useful work. The gradient can be used to transport molecules across membranes. It can be used to do mechanical work, such as rotating bacterial flagella. It can be used to produce ATP high-energy molecules that are necessary for growth.
A small amount of ATP is available from substrate-level phosphorylation (for example, in glycolysis). Some organisms can obtain ATP exclusively by fermentation. In most organisms, however, the majority of ATP is generated by electron transport chains.
Electron transport chains in mitochondria
The cells of all eukaryotes (all animals, plants, fungi, algae, protozoa – in other words, all living things except bacteria and archaea) contain intracellular organelles called mitochondria, which produce ATP. Energy sources such as glucose are initially metabolized in the cytoplasm. The products are imported into mitochondria. Mitochondria continue the process of catabolism using metabolic pathways including the Krebs cycle, fatty acid oxidation, and amino acid oxidation.
The end result of these pathways is the production of two kinds of energy-rich electron donors, NADH and FADH2. Electrons from these donors are passed through an electron transport chain to oxygen, which is reduced to water. This is a multi-step redox process that occurs on the mitochondrial inner membrane. The enzymes that catalyze these reactions have the remarkable ability to simultaneously create a proton gradient across the membrane, producing a thermodynamically unlikely high-energy state with the potential to do work. Although electron transport occurs with great efficiency, a small percentage of electrons are prematurely leaked to oxygen, resulting in the formation of the toxic free-radical superoxide.
The similarity between intracellular mitochondria and free-living bacteria is striking. The known structural, functional, and DNA similarities between mitochondria and bacteria provide strong evidence that mitochondria evolved from intracellular prokaryotic symbionts that took up residence in primitive eukaryotic cells.
Mitochondrial redox carriers
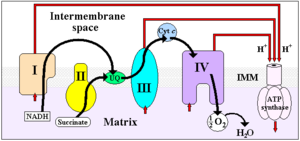
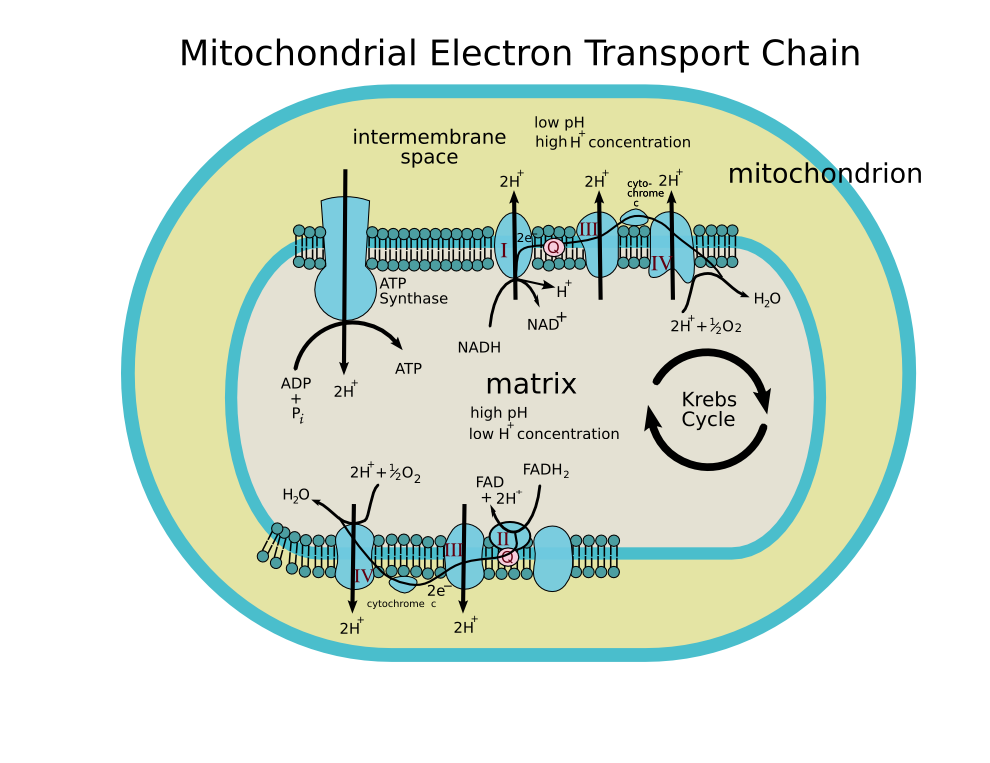
Four membrane-bound complexes have been identified in mitochondria. Each is an extremely complex transmembrane structure that is embedded in the inner membrane. Three of them are proton pumps. The structures are electrically connected by lipid-soluble electron carriers and water-soluble electron carriers. The overall electron transport chain
NADH → Complex I → Q → Complex III → cytochrome c → Complex IV → O2
↑
Complex II
Complex I
Complex I (NADH dehydrogenase, also called NADH:ubiquinone oxidoreductase; EC 1.6.5.3) removes two electrons from NADH and transfers them to a lipid-soluble carrier, ubiquinone (Q). The reduced product, ubiquinol (QH2) is free to diffuse within the membrane. At the same time, Complex I moves four protons (H+) across the membrane, producing a proton gradient. Complex I is one of the main sites at which premature electron leakage to oxygen occurs, thus being one of main sites of production of a harmful free radical called superoxide.
The pathway of electrons occurs as follows:
NADH is oxidized to NAD+, reducing Flavin mononucleotide to FMNH2 in one two-electron step. The next electron carrier is a Fe-S cluster, which can only accept one electron at a time to reduce the ferric ion into a ferrous ion. In a convenient manner, FMNH2 can be oxidized in only two one-electron steps, through a semiquinone intermediate. The electron thus travels from the FMNH2 to the Fe-S cluster, then from the Fe-S cluster to the oxidized Q to give the free-radical (semiquinone) form of Q. This happens again to reduce the semiquinone form to the ubiquinol form, QH2. During this process, four protons are translocated across the inner mitochondrial membrane, from the matrix to the intermembrane space. This creates a proton gradient that will be later used to generate ATP through oxidative phosphorylation.
Complex II
Complex II (succinate dehydrogenase; EC 1.3.5.1) is not a proton pump. It serves to funnel additional electrons into the quinone pool (Q) by removing electrons from succinate and transferring them (via FAD) to Q. Complex II consists of four protein subunits: SDHA,SDHB,SDHC, and SDHD. Other electron donors (e.g., fatty acids and glycerol 3-phosphate) also funnel electrons into Q (via FAD), again without producing a proton gradient.
Complex III
Complex III (cytochrome bc1 complex; EC 1.10.2.2) removes in a stepwise fashion two electrons from QH2 and transfers them to two molecules of cytochrome c, a water-soluble electron carrier located within the intermembrane space. At the same time, it moves two protons across the membrane, producing a proton gradient (in total 4 protons: 2 protons are translocated and 2 protons are released from ubiquinol). When electron transfer is hindered (by a high membrane potential, point mutations or respiratory inhibitors such as antimycin A), Complex III may leak electrons to oxygen resulting in the formation of superoxide, a highly-toxic species, which is thought to contribute to the pathology of a number of diseases, including aging.
Complex IV
Complex IV (cytochrome c oxidase; EC 1.9.3.1) removes four electrons from four molecules of cytochrome c and transfers them to molecular oxygen (O2), producing two molecules of water (H2O). At the same time, it moves four protons across the membrane, producing a proton gradient.
Coupling with oxidative phosphorylation
The chemiosmotic coupling hypothesis, as proposed by Nobel Prize in Chemistry winner Peter D. Mitchell, explains that the electron transport chain and oxidative phosphorylation are coupled by a proton gradient across the inner mitochondrial membrane. The efflux of protons creates both a pH gradient and an electrochemical gradient. This proton gradient is used by the FOF1 ATP synthase complex to make ATP via oxidative phosphorylation. ATP synthase is sometimes regarded as complex V of the electron transport chain. The FO component of ATP synthase acts as an ion channel for return of protons back to mitochondrial matrix. During their return, the free energy produced during the generation of the oxidized forms of the electron carriers (NAD+ and FAD) is released. This energy is used to drive ATP synthesis, catalyzed by the F1 component of the complex.
Coupling with oxidative phosphorylation is a key step for ATP production. However, in certain cases, uncoupling may be biologically useful. The inner mitochondrial membrane of brown adipose tissue contains a large amount of thermogenin (an uncoupling protein), which acts as uncoupler by forming an alternative pathway for the flow of protons back to matrix. This results in consumption of energy in thermogenesis rather than ATP production. This may be useful in cases when heat production is required, for example in colds or during arise of hibernating animals. Synthetic uncouplers (e.g., 2,4-dinitrophenol) also exist, and, at high doses, are lethal.
Summary
The mitochondrial electron transport chain removes electrons from an electron donor (NADH or FADH2) and passes them to a terminal electron acceptor (O2) via a series of redox reactions. These reactions are coupled to the creation of a proton gradient across the mitochondrial inner membrane. There are three proton pumps: I, III, and IV. The resulting transmembrane proton gradient is used to make ATP via ATP synthase.
The reactions catalyzed by Complex I and Complex III exist roughly at equilibrium. This means that these reactions are readily reversible, simply by increasing the concentration of the products relative to the concentration of the reactants (for example, by increasing the proton gradient). ATP synthase is also readily reversible. Thus ATP can be used to make a proton gradient, which in turn can be used to make NADH. This process of reverse electron transport is important in many prokaryotic electron transport chains.
Electron transport chains in bacteria
In eukaryotes, NADH is the most important electron donor. The associated electron transport chain is
NADH → Complex I → Q → Complex III → cytochrome c → Complex IV → O2 where Complexes I, III and IV are proton pumps, while Q and cytochrome c are mobile electron carriers. The electron acceptor is molecular oxygen.
In prokaryotes (bacteria and archaea) the situation is more complicated, because there is a number of different electron donors and a number of different electron acceptors. The generalized electron transport chain in bacteria is:
Donor Donor Donor
↓ ↓ ↓
dehydrogenase → quinone → bc1 → cytochrome
↓ ↓
oxidase(reductase) oxidase(reductase)
↓ ↓
Acceptor Acceptor
Note that electrons can enter the chain at three levels: at the level of a dehydrogenase, at the level of the quinone pool, or at the level of a mobile cytochrome electron carrier. These levels correspond to successively more positive redox potentials, or to successively decreased potential differences relative to the terminal electron acceptor. In other words, they correspond to successively smaller Gibbs free energy changes for the overall redox reaction Donor → Acceptor.
Individual bacteria use multiple electron transport chains, often simultaneously. Bacteria can use a number of different electron donors, a number of different dehydrogenases, a number of different oxidases and reductases, and a number of different electron acceptors. For example, E. coli (when growing aerobically using glucose as an energy source) uses two different NADH dehydrogenases and two different quinol oxidases, for a total of four different electron transport chains operating simultaneously.
A common feature of all electron transport chains is the presence of a proton pump to create a transmembrane proton gradient. Bacterial electron transport chains may contain as many as three proton pumps, like mitochondria, or they may contain only one or two. They always contain at least one proton pump.
Electron donors
In the present day biosphere, the most common electron donors are organic molecules. Organisms that use organic molecules as an energy source are called organotrophs. Organotrophs (animals, fungi, protists) and phototrophs (plants and algae) constitute the vast majority of all familiar life forms.
Some prokaryotes can use inorganic matter as an energy source. Such organisms are called lithotrophs ("rock-eaters"). Inorganic electron donors include hydrogen, carbon monoxide, ammonia, nitrite, sulfur, sulfide, and ferrous iron. Lithotrophs have been found growing in rock formations thousands of meters below the surface of Earth. Because of their volume of distribution, lithotrophs may actually outnumber organotrophs and phototrophs in our biosphere.
The use of inorganic electron donors as an energy source is of particular interest in the study of evolution. This type of metabolism must logically have preceded the use of organic molecules as an energy source.
Dehydrogenases
Bacteria can use a number of different electron donors. When organic matter is the energy source, the donor may be NADH or succinate, in which case electrons enter the electron transport chain via NADH dehydrogenase (similar to Complex I in mitochondria) or succinate dehydrogenase (similar to Complex II). Other dehydrogenases may be used to process different energy sources: formate dehydrogenase, lactate dehydrogenase, glyceraldehyde-3-phosphate dehydrogenase, H2 dehydrogenase (hydrogenase), etc. Some dehydrogenases are also proton pumps; others simply funnel electrons into the quinone pool.
Most dehydrogenases are synthesized only when needed. Depending on the environment in which they find themselves, bacteria select different enzymes from their DNA library and synthesize only those that are needed for growth. Enzymes that are synthesized only when needed are said to be inducible.
Quinone carriers
Quinones are mobile, lipid-soluble carriers that shuttles electrons (and protons) between large, relatively immobile macromolecular complexes imbedded in the membrane. Bacteria use ubiquinone (the same quinone that mitochondria use) and related quinones such as menaquinone.
Proton pumps
A proton pump is any process that creates a proton gradient across a membrane. Protons can be physically moved across a membrane; this is seen in mitochondrial Complexes I and IV. The same effect can be produced by moving electrons in the opposite direction. The result is the disappearance of a proton from the cytoplasm and the appearance of a proton in the periplasm. Mitochondrial Complex III uses this second type of proton pump, which is mediated by a quinone (the Q cycle).
Some dehydrogenases are proton pumps; others are not. Most oxidases and reductases are proton pumps, but some are not. Cytochrome bc1 is a proton pump found in many, but not all, bacteria (it is not found in E. coli). As the name implies, bacterial bc1 is similar to mitochondrial bc1 (Complex III).
Proton pumps are the heart of the electron transport process. They produce the transmembrane electrochemical gradient that supplies energy to the cell.
Cytochrome electron carriers
Cytochromes are pigments that contain iron. They are found in two very different environments.
Some cytochromes are water-soluble carriers that shuttle electrons to and from large, immobile macromolecular structures imbedded in the membrane. The mobile cytochrome electron carrier in mitochondria is cytochrome c. Bacteria use a number of different mobile cytochrome electron carriers.
Other cytochromes are found within macromolecules such as Complex III and Complex IV. They also function as electron carriers, but in a very different, intramolecular, solid-state environment.
Electrons may enter an electron transport chain at the level of a mobile cytochrome or quinone carrier. For example, electrons from inorganic electron donors (nitrite, ferrous iron, etc.) enter the electron transport chain at the cytochrome level. When electrons enter at a redox level greater than NADH, the electron transport chain must operate in reverse to produce this necessary, higher-energy molecule.
Terminal oxidases and reductases
When bacteria grow in aerobic environments, the terminal electron acceptor (O2) is reduced to water by an enzyme called an oxidase. When bacteria grow in anaerobic environments, the terminal electron acceptor is reduced by an enzyme called a reductase.
In mitochondria the terminal membrane complex (Complex IV) is cytochrome oxidase. Aerobic bacteria use a number of different terminal oxidases. For example, E. coli does not have a cytochrome oxidase or a bc1 complex. Under aerobic conditions, it uses two different terminal quinol oxidases (both proton pumps) to reduce oxygen to water.
Anaerobic bacteria, which do not use oxygen as a terminal electron acceptor, have terminal reductases individualized to their terminal acceptor. For example, E. coli can use fumarate reductase, nitrate reductase, nitrite reductase, DMSO reductase, or trimethylamine-N-oxide reductase, depending on the availability of these acceptors in the environment.
Most terminal oxidases and reductases are inducible. They are synthesized by the organism as needed, in response to specific environmental conditions.
Electron acceptors
Just as there are a number of different electron donors (organic matter in organotrophs, inorganic matter in lithotrophs), there are a number of different electron acceptors, both organic and inorganic. If oxygen is available, it is invariably used as the terminal electron acceptor, because it generates the greatest Gibbs free energy change and produces the most energy.
In anaerobic environments, different electron acceptors are used, including nitrate, nitrite, ferric iron, sulfate, carbon dioxide, and small organic molecules such as fumarate.
Since electron transport chains are redox processes, they can be described as the sum of two redox pairs. For example, the mitochondrial electron transport chain can be described as the sum of the NAD+/NADH redox pair and the O2/H2O redox pair. NADH is the electron donor and O2 is the electron acceptor.
Not every donor-acceptor combination is thermodynamically possible. The redox potential of the acceptor must be more positive than the redox potential of the donor. Furthermore, actual environmental conditions may be far different from standard conditions (1 molar concentrations, 1 atm partial pressures, pH = 7), which apply to standard redox potentials. For example, hydrogen-evolving bacteria grow at an ambient partial pressure of hydrogen gas of 10-4 atm. The associated redox reaction, which is thermodynamically favorable in nature, is thermodynamically impossible under “standard” conditions.
Summary
Bacterial electron transport pathways are, in general, inducible. Depending on their environment, bacteria can synthesize different transmembrane complexes and produce different electron transport chains in their cell membranes. Bacteria select their electron transport chains from a DNA library containing multiple possible dehydrogenases, terminal oxidases and terminal reductases. The situation is often summarized by saying that electron transport chains in bacteria are branched, modular, and inducible.
Photosynthetic electron transport chains
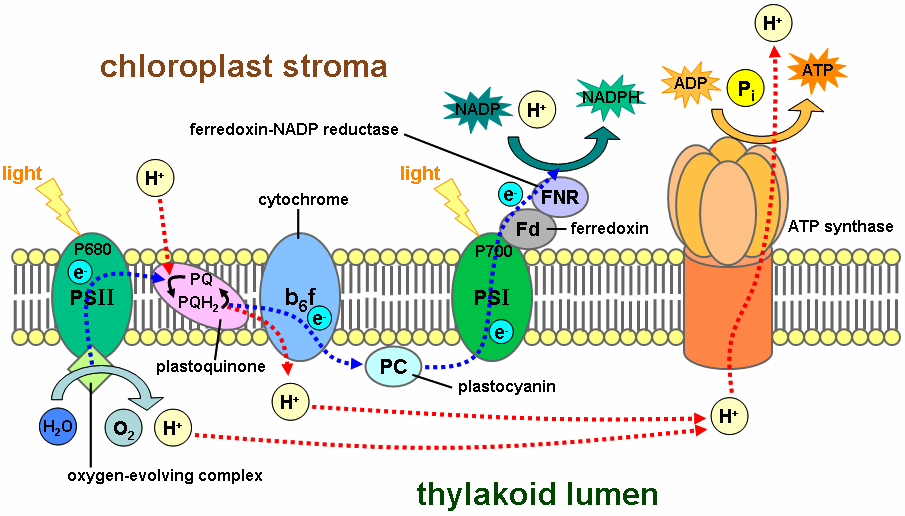
In oxidative phosphorylation, electrons are transferred from a high-energy electron donor (e.g., NADH) to an electron acceptor (e.g., O2) through an electron transport chain. In photophosphorylation, the energy of sunlight is used to create a high-energy electron donor and an electron acceptor. Electrons are then transferred from the donor to the acceptor through another electron transport chain.
Photosynthetic electron transport chains have many similarities to the oxidative chains discussed above. They use mobile, lipid-soluble carriers (quinones) and mobile, water-soluble carriers (cytochromes, etc.). They also contain a proton pump. It is remarkable that the proton pump in all photosynthetic chains resembles mitochondrial Complex III.
Photosynthetic electron transport chains are discussed in greater detail in the articles Photophosphorylation, Photosynthesis, Photosynthetic reaction center and Light-dependent reaction.
Summary
Electron transport chains are the source of energy for all known forms of life. They are redox reactions that transfer electrons from an electron donor to an electron acceptor. The transfer of electrons is coupled to the translocation of protons across a membrane, producing a proton gradient. The proton gradient is used to produce useful work.
The coupling of thermodynamically favorable to thermodynamically unfavorable biochemical reactions by biological macromolecules is an example of an emergent property – a property that could not have been predicted, even given full knowledge of the primitive geochemical systems from which these macromolecules evolved. It is an open question whether such emergent properties evolve only by chance, or whether they necessarily evolve in any large biogeochemical system, given the underlying laws of physics.
References
- Fenchel T (2006). Bacterial Biogeochemistry: The Ecophysiology of Mineral Cycling (2nd ed. ed.). Elsevier. ISBN 978-0121034559. Unknown parameter
|coauthors=ignored (help); Unknown parameter|month=ignored (help) - Lengeler JW (1999). Biology of the Prokaryotes. Blackwell Science. ISBN 978-0632053575. Unknown parameter
|month=ignored (help); Unknown parameter|coauthors=ignored (help) - Nelson DL (2005). Lehninger Principles of Biochemistry (4th ed ed.). W. H. Freeman. ISBN 978-0716743392. Unknown parameter
|coauthors=ignored (help); Unknown parameter|month=ignored (help) - Nicholls DG (2002). Bioenergetics 3. Academic Press. ISBN 978-0125181211. Unknown parameter
|coauthors=ignored (help); Unknown parameter|month=ignored (help) - Stumm W (1996). Aquatic Chemistry (3rd ed ed.). Wiley. ISBN 978-0471511854. Unknown parameter
|coauthor=ignored (help) - Thauer RK (1977). "Energy conservation in chemotrophic anaerobic bacteria". Bacteriol Rev. 41 (1): 100–80. PMID 860983. Unknown parameter
|coauthors=ignored (help); Unknown parameter|month=ignored (help) - White D. (1999). The Physiology and Biochemistry of Prokaryotes (2nd ed. ed.). Oxford University Press. ISBN 978-0195125795. Unknown parameter
|month=ignored (help) - Voet D (2004). Biochemistry (3rd ed ed.). Wiley. ISBN 978-0471586517. Unknown parameter
|coauthors=ignored (help); Unknown parameter|month=ignored (help)
External links
- Electron+Transport+Chain+Complex+Proteins at the US National Library of Medicine Medical Subject Headings (MeSH)
- UMich Orientation of Proteins in Membranes families/superfamily-3 - Complexes with cytochrome b-like domains
- UMich Orientation of Proteins in Membranes families/superfamily-4 - Bacterial and mitochondrial cytochrome c oxidases
- UMich Orientation of Proteins in Membranes families/superfamily-2 - Photosynthetic reaction centers and photosystems
- UMich Orientation of Proteins in Membranes families/superfamily-78 - Cytochrome c family
- UMich Orientation of Proteins in Membranes families/superfamily-101 - Cupredoxins
- UMich Orientation of Proteins in Membranes protein/pdbid-1e6e - Adrenodoxin reductase
- UMich Orientation of Proteins in Membranes families/superfamily-130 - Electron transfer flavoproteins
ar:سلسلة نقل الإلكترون ca:Cadena respiratòria cs:Dýchací řetězec da:Elektrontransportkæde de:Atmungskette it:Catena di trasporto degli elettroni mk:Респираторна низа sl:Dihalna veriga fi:Elektroninsiirtoketju sv:Elektrontransportkedja uk:Електронтранспортний ланцюг