Immune system

|
WikiDoc Resources for Immune system |
|
Articles |
|---|
|
Most recent articles on Immune system Most cited articles on Immune system |
|
Media |
|
Powerpoint slides on Immune system |
|
Evidence Based Medicine |
|
Clinical Trials |
|
Ongoing Trials on Immune system at Clinical Trials.gov Trial results on Immune system Clinical Trials on Immune system at Google
|
|
Guidelines / Policies / Govt |
|
US National Guidelines Clearinghouse on Immune system NICE Guidance on Immune system
|
|
Books |
|
News |
|
Commentary |
|
Definitions |
|
Patient Resources / Community |
|
Patient resources on Immune system Discussion groups on Immune system Patient Handouts on Immune system Directions to Hospitals Treating Immune system Risk calculators and risk factors for Immune system
|
|
Healthcare Provider Resources |
|
Causes & Risk Factors for Immune system |
|
Continuing Medical Education (CME) |
|
International |
|
|
|
Business |
|
Experimental / Informatics |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]
An immune system is a collection of mechanisms within an organism that protects against disease by identifying and killing pathogens and tumor cells. It detects a wide variety of agents, from viruses to parasitic worms, and needs to distinguish them from the organism's own healthy cells and tissues in order to function properly. Detection is complicated as pathogens adapt and evolve new ways to successfully infect the host organism.
To survive this challenge, several mechanisms evolved that recognize and neutralize pathogens. Even simple unicellular organisms such as bacteria possess enzyme systems that protect against viral infections. Other basic immune mechanisms evolved in ancient eukaryotes and remain in their modern descendants, such as plants, fish, reptiles, and insects. These mechanisms include antimicrobial peptides called defensins, phagocytosis, and the complement system. More sophisticated mechanisms, however, developed relatively recently, with the evolution of vertebrates.[1] The immune systems of vertebrates such as humans consist of many types of proteins, cells, organs, and tissues, which interact in an elaborate and dynamic network. As part of this more complex immune response, the vertebrate system adapts over time to recognize particular pathogens more efficiently. The adaptation process creates immunological memories and allows even more effective protection during future encounters with these pathogens. This process of acquired immunity is the basis of vaccination.
Disorders in the immune system can cause disease. Immunodeficiency diseases occur when the immune system is less active than normal, resulting in recurring and life-threatening infections. Immunodeficiency can either be the result of a genetic disease, such as severe combined immunodeficiency, or be produced by pharmaceuticals or an infection, such as the acquired immune deficiency syndrome (AIDS) that is caused by the retrovirus HIV. In contrast, autoimmune diseases result from a hyperactive immune system attacking normal tissues as if they were foreign organisms. Common autoimmune diseases include rheumatoid arthritis, diabetes mellitus type 1 and lupus erythematosus. These critical roles of immunology in health and disease are areas of intense scientific study.
Layered defense in immunity
The immune system protects you from dying from infection with layered defenses of increasing specificity. Most simply, physical barriers prevent pathogens such as bacteria and viruses from entering the body. If a pathogen breaches these barriers, the innate immune system provides an immediate, but non-specific response. Innate immune systems are found in all plants and animals.[2] However, if pathogens successfully evade the innate response, vertebrates possess a third layer of protection, the adaptive immune system, which is activated by the innate response. Here, the immune system adapts its response during an infection to improve its recognition of the pathogen. This improved response is then retained after the pathogen has been eliminated, in the form of an immunological memory, and allows the adaptive immune system to mount faster and stronger attacks each time this pathogen is encountered.[3]
| Innate immune system | Adaptive immune system |
|---|---|
| Response is non-specific | Pathogen and antigen specific response |
| Exposure leads to immediate maximal response | Lag time between exposure and maximal response |
| Cell-mediated and humoral components | Cell-mediated and humoral components |
| No immunological memory | Exposure leads to immunological memory |
| Found in nearly all forms of life | Found only in jawed vertebrates |
Both innate and adaptive immunity depend on the ability of the immune system to distinguish between self and non-self molecules. In immunology, self molecules are those components of an organism's body that can be distinguished from foreign substances by the immune system.[4] Conversely, non-self molecules are those recognized as foreign molecules. One class of non-self molecules are called antigens (short for antibody generators) and are defined as substances that bind to specific immune receptors and elicit an immune response.[5]
Surface barriers
Several barriers protect organisms from infection, including mechanical, chemical and biological barriers. The waxy cuticle of many leaves, the exoskeleton of insects, the shells and membranes of externally deposited eggs, and skin are examples of the mechanical barriers that are the first line of defense against infection.[5] However, as organisms cannot be completely sealed against their environments, other systems act to protect body openings such as the lungs, intestines, and the genitourinary tract. In the lungs, coughing and sneezing mechanically eject pathogens and other irritants from the respiratory tract. The flushing action of tears and urine also mechanically expels pathogens, while mucus secreted by the respiratory and gastrointestinal tract serves to trap and entangle microorganisms.[6]
Chemical barriers also protect against infection. The skin and respiratory tract secrete antimicrobial peptides such as the β-defensins.[7] Enzymes such as lysozyme and phospholipase A2 in saliva, tears, and breast milk are also antibacterials.[8][9] Vaginal secretions serve as a chemical barrier following menarche, when they become slightly acidic, while semen contains defensins and zinc to kill pathogens.[10][11] In the stomach, gastric acid and proteases serve as powerful chemical defenses against ingested pathogens.
Within the genitourinary and gastrointestinal tracts, commensal flora serve as biological barriers by competing with pathogenic bacteria for food and space and, in some cases, by changing the conditions in their environment, such as pH or available iron.[12] This reduces the probability that pathogens will be able to reach sufficient numbers to cause illness. However, since most antibiotics non-specifically target bacteria and do not affect fungi, oral antibiotics can lead to an “overgrowth” of fungi and cause conditions such as a vaginal candidiasis (yeast infection).[13] There is good evidence that re-introduction of probiotic flora, such as pure cultures of the lactobacilli normally found in yoghurt, helps restore a healthy balance of microbial populations in intestinal infections in children and encouraging preliminary data in studies on bacterial gastroenteritis, inflammatory bowel diseases, urinary tract infection and post-surgical infections.[14][15][16]
Innate immunity
Microorganisms that successfully enter an organism will encounter the cells and mechanisms of the innate immune system. The innate response is usually triggered when microbes are identified by pattern recognition receptors, which recognize components that are conserved among broad groups of microorganisms.[17] Innate immune defenses are non-specific, meaning these systems respond to pathogens in a generic way.[5] This system does not confer long-lasting immunity against a pathogen. The innate immune system is the dominant system of host defense in most organisms.[2]
Humoral and chemical barriers
Inflammation
Inflammation is one of the first responses of the immune system to infection.[18] The symptoms of inflammation are redness and swelling, which are caused by increased blood flow into a tissue. Inflammation is produced by eicosanoids and cytokines, which are released by injured or infected cells. Eicosanoids include prostaglandins that produce fever and the dilation of blood vessels associated with inflammation, and leukotrienes that attract certain white blood cells (leukocytes).[19][20] Common cytokines include interleukins that are responsible for communication between white blood cells; chemokines that promote chemotaxis; and interferons that have anti-viral effects, such as shutting down protein synthesis in the host cell.[21] Growth factors and cytotoxic factors may also be released. These cytokines and other chemicals recruit immune cells to the site of infection and promote healing of any damaged tissue following the removal of pathogens.[22]
Complement system
The complement system is a biochemical cascade that attacks the surfaces of foreign cells. It contains over 20 different proteins and is named for its ability to “complement” the killing of pathogens by antibodies. Complement is the major humoral component of the innate immune response.[23][24] Many species have complement systems, including non-mammals like plants, fish, and some invertebrates.[25]
In humans, this response is activated by complement binding to antibodies that have attached to these microbes or the binding of complement proteins to carbohydrates on the surfaces of microbes. This recognition signal triggers a rapid killing response.[26] The speed of the response is a result of signal amplification that occurs following sequential proteolytic activation of complement molecules, which are also proteases. After complement proteins initially bind to the microbe, they activate their protease activity, which in turn activates other complement proteases, and so on. This produces a catalytic cascade that amplifies the initial signal by controlled positive feedback.[27] The cascade results in the production of peptides that attract immune cells, increase vascular permeability, and opsonize (coat) the surface of a pathogen, marking it for destruction. This deposition of complement can also kill cells directly by disrupting their plasma membrane.[23]
Cellular barriers of the innate system

Leukocytes (white blood cells) act like independent, single-celled organisms and are the second arm of the innate immune system.[5] The innate leukocytes include the phagocytes (macrophages, neutrophils, and dendritic cells), mast cells, eosinophils, basophils, and natural killer cells. These cells identify and eliminate pathogens, either by attacking larger pathogens through contact or by engulfing and then killing microorganisms.[25] Innate cells are also important mediators in the activation of the adaptive immune system.[3]
Phagocytosis is an important feature of cellular innate immunity performed by cells called 'phagocytes' that engulf, or eat, pathogens or particles. Phagocytes generally patrol the body searching for pathogens, but can be called to specific locations by cytokines.[5] Once a pathogen has been engulfed by a phagocyte, it becomes trapped in an intracellular vesicle called a phagosome, which subsequently fuses with another vesicle called a lysosome to form a phagolysosome. The pathogen is killed by the activity of digestive enzymes or following a respiratory burst that releases free radicals into the phagolysosome.[28][29] Phagocytosis evolved as a means of acquiring nutrients, but this role was extended in phagocytes to include engulfment of pathogens as a defense mechanism.[30] Phagocytosis probably represents the oldest form of host defense, as phagocytes have been identified in both vertebrate and invertebrate animals.[31]
Neutrophils and macrophages are phagocytes that travel throughout the body in pursuit of invading pathogens.[32] Neutrophils are normally found in the bloodstream and are the most abundant type of phagocyte, normally representing 50% to 60% of the total circulating leukocytes.[33] During the acute phase of inflammation, particularly as a result of bacterial infection, neutrophils migrate toward the site of inflammation in a process called chemotaxis, and are usually the first cells to arrive at the scene of infection. Macrophages are versatile cells that reside within tissues and produce a wide array of chemicals including enzymes, complement proteins, and regulatory factors such as interleukin 1.[34] Macrophages also act as scavengers, ridding the body of worn-out cells and other debris, and as antigen-presenting cells that activate the adaptive immune system.[3]
Dendritic cells (DC) are phagocytes in tissues that are in contact with the external environment; therefore, they are located mainly in the skin, nose, lungs, stomach, and intestines.[35] They are named for their resemblance to neuronal dendrites, as both have many spine-like projections, but dendritic cells are in no way connected to the nervous system. Dendritic cells serve as a link between the innate and adaptive immune systems, as they present antigen to T cells, one of the key cell types of the adaptive immune system.[35]
Mast cells reside in connective tissues and mucous membranes, and regulate the inflammatory response.[36] They are most often associated with allergy and anaphylaxis.[33] Basophils and eosinophils are related to neutrophils. They secrete chemical mediators that are involved in defending against parasites and play a role in allergic reactions, such as asthma.[37] Natural killer (NK cells) cells are leukocytes that attack and destroy tumor cells, or cells that have been infected by viruses.[38]
Adaptive immunity
The adaptive immune system evolved in early vertebrates and allows for a stronger immune response as well as immunological memory, where each pathogen is "remembered" by a signature antigen.[39] The adaptive immune response is antigen-specific and requires the recognition of specific “non-self” antigens during a process called antigen presentation. Antigen specificity allows for the generation of responses that are tailored to specific pathogens or pathogen-infected cells. The ability to mount these tailored responses is maintained in the body by "memory cells". Should a pathogen infect the body more than once, these specific memory cells are used to quickly eliminate it.
Lymphocytes
The cells of the adaptive immune system are special types of leukocytes, called lymphocytes. B cells and T cells are the major types of lymphocytes and are derived from hematopoietic stem cells in the bone marrow.[25] B cells are involved in the humoral immune response, whereas T cells are involved in cell-mediated immune response.
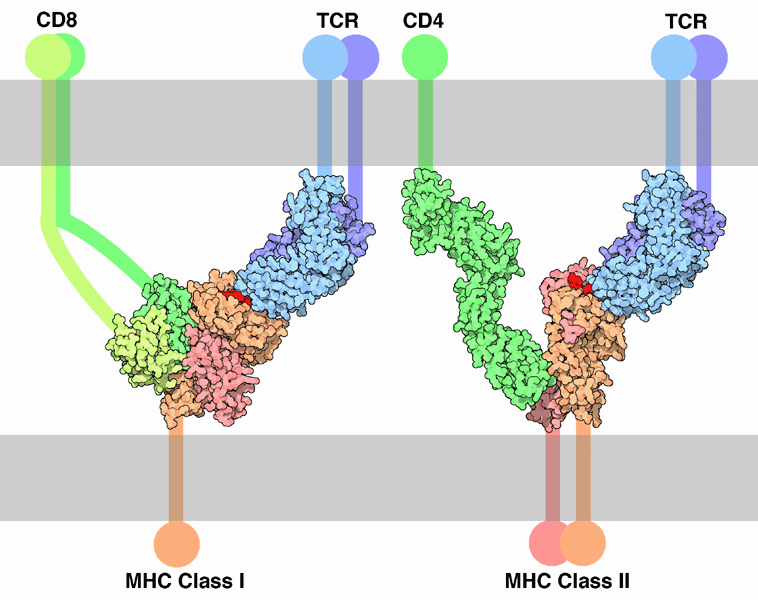
Both B cells and T cells carry receptor molecules that recognize specific targets. T cells recognize a “non-self” target, such as a pathogen, only after antigens (small fragments of the pathogen) have been processed and presented in combination with a “self” receptor called a major histocompatibility complex (MHC) molecule. There are two major subtypes of T cells: the killer T cell and the helper T cell. Killer T cells only recognize antigens coupled to Class I MHC molecules, while helper T cells only recognize antigens coupled to Class II MHC molecules. These two mechanisms of antigen presentation reflect the different roles of the two types of T cell. A third, minor subtype are the γδ T cells that recognize intact antigens that are not bound to MHC receptors.[40]
In contrast, the B cell antigen-specific receptor is an antibody molecule on the B cell surface, and recognizes whole pathogens without any need for antigen processing. Each lineage of B cell expresses a different antibody, so the complete set of B cell antigen receptors represent all the antibodies that the body can manufacture.[25]
Killer T cells

Killer T cell are a sub-group of T cells that kill cells infected with viruses (and other pathogens), or are otherwise damaged or dysfunctional.[42] As with B cells, each type of T cell recognises a different antigen. Killer T cells are activated when their T cell receptor (TCR) binds to this specific antigen in a complex with the MHC Class I receptor of another cell. Recognition of this MHC:antigen complex is aided by a co-receptor on the T cell, called CD8. The T cell then travels throughout the body in search of cells where the MHC I receptors bear this antigen. When an activated T cell contacts such cells, it releases cytotoxins, such as perforin, which form pores in the target cell's plasma membrane, allowing ions, water and toxins to enter. The entry of another toxin called granulysin (a protease) induces the target cell to undergo apoptosis.[43] T cell killing of host cells is particularly important in preventing the replication of viruses. T cell activation is tightly controlled and generally requires a very strong MHC/antigen activation signal, or additional activation signals provided by "helper" T cells (see below).[43]
Helper T cells
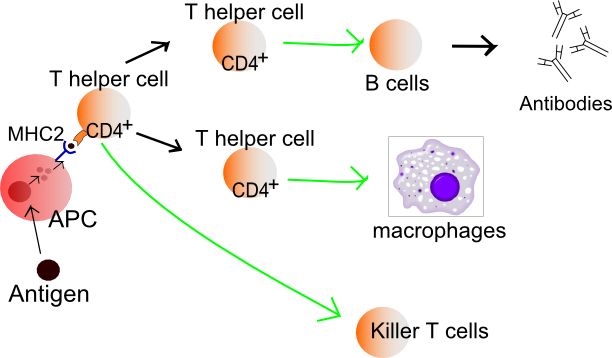
Helper T cells regulate both the innate and adaptive immune responses and help determine which types of immune responses the body will make to a particular pathogen.[44][45] These cells have no cytotoxic activity and do not kill infected cells or clear pathogens directly. They instead control the immune response by directing other cells to perform these tasks.
Helper T cells express T cell receptors (TCR) that recognize antigen bound to Class II MHC molecules. The MHC:antigen complex is also recognized by the helper cell's CD4 co-receptor, which recruits molecules inside the T cell (e.g. Lck) that are responsible for T cell's activation. Helper T cells have a weaker association with the MHC:antigen complex than observed for killer T cells, meaning many receptors (around 200–300) on the helper T cell must be bound by an MHC:antigen in order to activate the helper cell, while killer T cells can be activated by engagement of a single MHC:antigen molecule. Helper T cell activation also requires longer duration of engagement with an antigen-presenting cell.[46] The activation of a resting helper T cell causes it to release cytokines that influence the activity of many cell types. Cytokine signals produced by helper T cells enhance the microbicidal function of macrophages and the activity of killer T cells.[5] In addition, helper T cell activation causes an upregulation of molecules expressed on the T cell's surface, such as CD40 ligand (also called CD154), which provide extra stimulatory signals typically required to activate antibody-producing B cells.[47]
γδ T cells
γδ T cells possess an alternative T cell receptor (TCR) as opposed to CD4+ and CD8+ (αβ) T cells and share the characteristics of helper T cells, cytotoxic T cells and NK cells. The conditions that produce responses from γδ T cells are not fully understood. Like other 'unconventional' T cell subsets bearing invariant TCRs, such as CD1d-restricted Natural Killer T cells, γδ T cells straddle the border between innate and adaptive immunity.[48] On one hand, γδ T cells are a component of adaptive immunity as they rearrange TCR genes to produce receptor diversity and can also develop a memory phenotype. On the other hand, the various subsets are also part of the innate immune system, as restricted TCR or NK receptors may be used as pattern recognition receptors. For example, large numbers of human Vγ9/Vδ2 T cells respond within hours to common molecules produced by microbes, and highly restricted Vδ1+ T cells in epithelia will respond to stressed epithelial cells.[49]
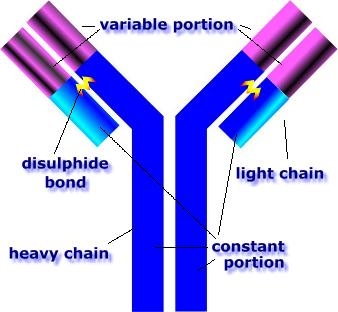
B lymphocytes and antibodies
A B cell identifies pathogens when antibodies on its surface bind to a specific foreign antigen.[50] This antigen/antibody complex is taken up by the B cell and processed by proteolysis into peptides. The B cell then displays these antigenic peptides on its surface MHC class II molecules. This combination of MHC and antigen attracts a matching helper T cell, which releases lymphokines and activates the B cell.[51] As the activated B cell then begins to divide, its offspring (plasma cells) secrete millions of copies of the antibody that recognizes this antigen. These antibodies circulate in blood plasma and lymph, bind to pathogens expressing the antigen and mark them for destruction by complement activation or for uptake and destruction by phagocytes. Antibodies can also neutralize challenges directly, by binding to bacterial toxins or by interfering with the receptors that viruses and bacteria use to infect cells.[52]CD20 antigen is also found on B lymphocytes.
Alternative adaptive immune system
Although the classical molecules of the adaptive immune system (e.g. antibodies and T cell receptors) exist only in jawed vertebrates, a distinct lymphocyte-derived molecule has been discovered in primitive jawless vertebrates, such as the lamprey and hagfish. These animals possess a large array of molecules called variable lymphocyte receptors (VLRs) that, like the antigen receptors of jawed vertebrates, are produced from only a small number (one or two) of genes. These molecules are believed to bind pathogenic antigens in a similar way to antibodies, and with the same degree of specificity.[53]
Immunological memory
When B cells and T cells are activated and begin to replicate, some of their offspring will become long-lived memory cells. Throughout the lifetime of an animal, these memory cells will remember each specific pathogen encountered and can mount a strong response if the pathogen is detected again. This is "adaptive" because it occurs during the lifetime of an individual as an adaptation to infection with that pathogen and prepares the immune system for future challenges. Immunological memory can either be in the form of passive short-term memory or active long-term memory.
Passive memory
Passive immunity is usually short-term, lasting between a few days and several months. Newborn infants have no prior exposure to microbes and are particularly vulnerable to infection. Several layers of passive protection are provided by the mother. During pregnancy, a particular type of antibody, called IgG, is transported from mother to baby directly across the placenta, so human babies have high levels of antibodies even at birth, with the same range of antigen specificities as their mother.[54] Breast milk also contains antibodies that are transferred to the gut of the infant and protect against bacterial infections until the newborn can synthesize its own antibodies.[55] This is passive immunity because the fetus does not actually make any memory cells or antibodies, it only borrows them. In medicine, protective passive immunity can also be transferred artificially from one individual to another via antibody-rich serum.[56]
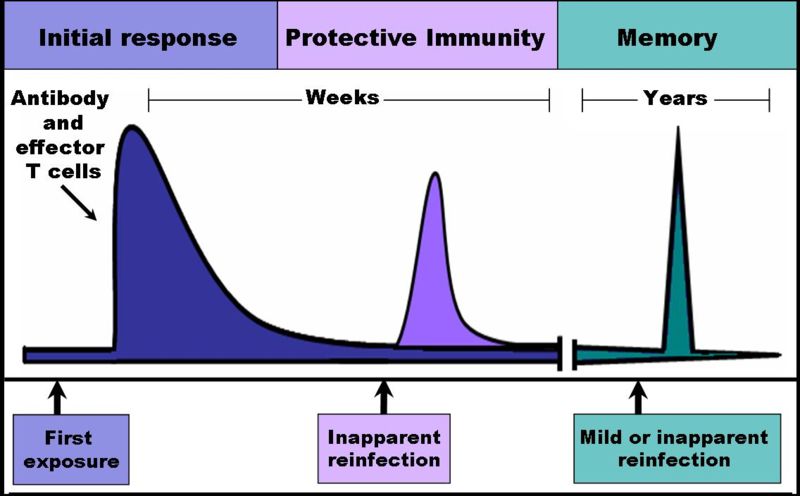
Active memory and immunization
Long-term active memory is acquired following infection by activation of B and T cells. Active immunity can also be generated artificially, through vaccination. The principle behind vaccination (also called immunization) is to introduce an antigen from a pathogen in order to stimulate the immune system and develop specific immunity against that particular pathogen without causing disease associated with that organism.[5] This deliberate induction of an immune response is successful because it exploits the natural specificity of the immune system, as well as its inducibility. With infectious disease remaining one of the leading causes of death in the human population, vaccination represents the most effective manipulation of the immune system mankind has developed.[57][25]
Most viral vaccines are based on live attenuated viruses, while many bacterial vaccines are based on acellular components of micro-organisms, including harmless toxin components.[5] Since many antigens derived from acellular vaccines do not strongly induce the adaptive response, most bacterial vaccines are provided with additional adjuvants that activate the antigen-presenting cells of the innate immune system and maximize immunogenicity.[58]
Disorders of human immunity
The immune system is a remarkably effective structure that incorporates specificity, inducibility and adaptation. Failures of host defense do occur, however, and fall into three broad categories: immunodeficiencies, autoimmunity, and hypersensitivities.
Immunodeficiencies
Immunodeficiencies occur when one or more of the components of the immune system are inactive. The ability of the immune system to respond to pathogens is diminished in both the young and the elderly, with immune responses beginning to decline at around 50 years of age due to immunosenescence.[59][60] In developed countries, obesity, alcoholism, and illegal drug abuse are common causes of poor immune function.[60] However, malnutrition is the most common cause of immunodeficiency in developing countries.[60] Diets lacking sufficient protein are associated with impaired cell-mediated immunity, complement activity, phagocyte function, IgA antibody concentrations, and cytokine production. Deficiency of single nutrients such as iron; copper; zinc; selenium; vitamins A, C, E, and B6; and folic acid (vitamin B9) also reduces immune responses.[60] Additionally, the loss of the thymus at an early age through genetic mutation or surgical removal results in severe immunodeficiency and a high susceptibility to infection.[61]
Immunodeficiencies can also be inherited or 'acquired'.[5] Chronic granulomatous disease, where phagocytes have a reduced ability to destroy pathogens, is an example of an inherited, or congenital, immunodeficiency. AIDS and some types of cancer cause acquired immunodeficiency.[62][63]
Autoimmunity
Overactive immune responses comprise the other end of immune dysfunction, particularly the autoimmune disorders. Here, the immune system fails to properly distinguish between self and non-self, and attacks part of the body. Under normal circumstances, many T cells and antibodies react with “self” peptides.[64] One of the functions of specialized cells (located in the thymus and bone marrow) is to present young lymphocytes with self antigens produced throughout the body and to eliminate those cells that recognize self-antigens, preventing autoimmunity.[50]
Hypersensitivity
Hypersensitivity is an immune response that damages the body's own tissues. They are divided into four classes (Type I – IV) based on the mechanisms involved and the time course of the hypersensitive reaction. Type I hypersensitivity is an immediate or anaphylactic reaction, often associated with allergy. Symptoms can range from mild discomfort to death. Type I hypersensitivity is mediated by IgE released from mast cells and basophils.[65] Type II hypersensitivity occurs when antibodies bind to antigens on the patient's own cells, marking them for destruction. This is also called antibody-dependent (or cytotoxic) hypersensitivity, and is mediated by IgG and IgM antibodies.[65] Immune complexes (aggregations of antigens, complement proteins, and IgG and IgM antibodies) deposited in various tissues trigger Type III hypersensitivity reactions.[65] Type IV hypersensitivity (also known as cell-mediated or delayed type hypersensitivity) usually takes between two and three days to develop. Type IV reactions are involved in many autoimmune and infectious diseases, but may also involve contact dermatitis (poison ivy). These reactions are mediated by T cells, monocytes, and macrophages.[65]
Other mechanisms of host defense
It is likely that a multicomponent, adaptive immune system arose with the first vertebrates, as invertebrates do not generate lymphocytes or an antibody-based humoral response.[1] Many species, however, utilize mechanisms that appear to be precursors of these aspects of vertebrate immunity. Immune systems appear even in the most structurally-simple forms of life, with bacteria using a unique defense mechanism, called the restriction modification system to protect themselves from viral pathogens, called bacteriophages.[66]
Pattern recognition receptors are proteins used by nearly all organisms to identify molecules associated with pathogens. Antimicrobial peptides called defensins are an evolutionarily conserved component of the innate immune response found in all animals and plants, and represent the main form of invertebrate systemic immunity.[1] The complement system and phagocytic cells are also used by most forms of invertebrate life. Ribonucleases and the RNA interference pathway are conserved across all eukaryotes, and are thought to play a role in the immune response to viruses.[67]
Unlike animals, plants lack phagocytic cells, and most plant immune responses involve systemic chemical signals that are sent through a plant.[68] When a part of a plant becomes infected, the plant produces a localized hypersensitive response, whereby cells at the site of infection undergo rapid apoptosis to prevent the spread of the disease to other parts of the plant. Systemic acquired resistance (SAR) is a type of defensive response used by plants that renders the entire plant resistant to a particular infectious agent.[68] RNA silencing mechanisms are particularly important in this systemic response as they can block virus replication.[69]
Tumor immunology
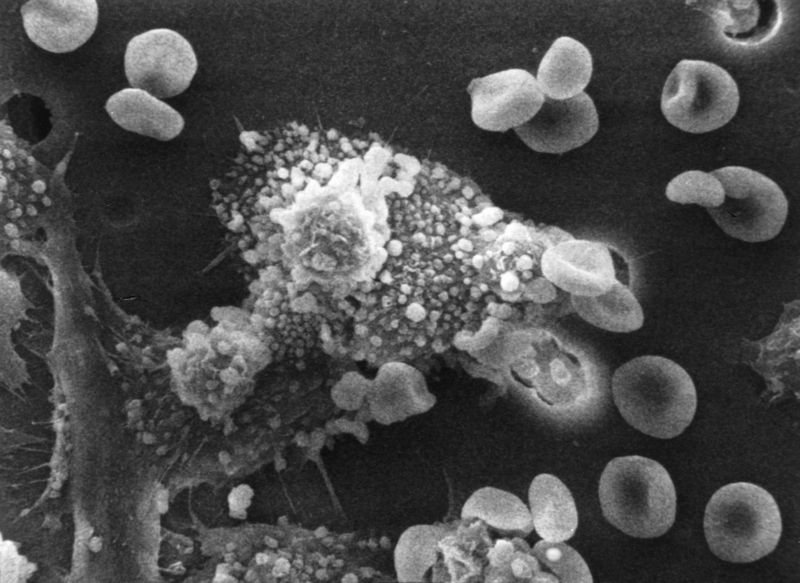
Another important role of the immune system is to identify and eliminate tumors. The transformed cells of tumors express antigens that are not found on normal cells. To the immune system, these antigens appear foreign, and their presence causes immune cells to attack the transformed tumor cells. The antigens expressed by tumors have several sources;[71] some are derived from oncogenic viruses like human papillomavirus, which causes cervical cancer,[72] while others are the organism's own proteins that occur at low levels in normal cells but reach high levels in tumor cells. One example is an enzyme called tyrosinase that, when expressed at high levels, transforms certain skin cells (e.g. melanocytes) into tumors called melanomas.[73][74] A third possible source of tumor antigens are proteins normally important for regulating cell growth and survival, that commonly mutate into cancer inducing molecules called oncogenes.[71][75][76]
The main response of the immune system to tumors is to destroy the abnormal cells using killer T cells, sometimes with the assistance of helper T cells.[74][77] Tumor antigens are presented on MHC class I molecules in a similar way to viral antigens. This allows killer T cells to recognize the tumor cell as abnormal.[78] NK cells also kill tumorous cells in a similar way, especially if the tumor cells have fewer MHC class I molecules on their surface than normal; this is a common phenomenon with tumors.[79] Sometimes antibodies are generated against tumor cells allowing for their destruction by the complement system.[75]
Clearly, some tumors evade the immune system and go on to become cancers.[80] Tumor cells often have a reduced number of MHC class I molecules on their surface, thus avoiding detection by killer T cells.[78] Some tumor cells also release products that inhibit the immune response; for example by secreting the cytokine TGF-β, which suppresses the activity of macrophages and lymphocytes.[81] In addition, immunological tolerance may develop against tumor antigens, so the immune system no longer attacks the tumor cells.[80]
Paradoxically, macrophages can promote tumor growth [82] when tumor cells send out cytokines that attract macrophages which then generate cytokines and growth factors that nurture tumor development. In addition, a combination of hypoxia in the tumor and a cytokine produced by macrophages induces tumor cells to decrease production of a protein that blocks metastasis and thereby assists spread of cancer cells.
Physiological regulation
Hormones can act as immunomodulators, altering the sensitivity of the immune system. For example, female sex hormones are known immunostimulators of both adaptive[83] and innate immune responses.[84] Some autoimmune diseases such as lupus erythematosus strike women preferentially, and their onset often coincides with puberty. By contrast, male sex hormones such as testosterone seem to be immunosuppressive.[85] Other hormones appear to regulate the immune system as well, most notably prolactin, growth hormone and vitamin D.[86][87] It is conjectured that a progressive decline in hormone levels with age is partially responsible for weakened immune responses in aging individuals.[88] Conversely, some hormones are regulated by the immune system, notably thyroid hormone activity.[89]
The immune system is enhanced by sleep and rest,[90] and is impaired by stress.[91]
Diet may affect the immune system; for example, fresh fruits, vegetables, and foods rich in certain fatty acids may foster a healthy immune system.[92] Likewise, fetal undernourishment can cause a lifelong impairment of the immune system.[93] In traditional medicine, some herbs are believed to stimulate the immune system, such as echinacea, licorice, ginseng, astragalus, sage, garlic, elderberry, shitake and lingzhi mushrooms, and hyssop, as well as honey. Studies have suggested that such herbs can indeed stimulate the immune system,[94] although their mode of action is complex and difficult to characterize.
Manipulation in medicine
The immune response can be manipulated to suppress unwanted responses resulting from autoimmunity, allergy, and transplant rejection, and to stimulate protective responses against pathogens that largely elude the immune system (see immunization). Immunosuppressive drugs are used to control autoimmune disorders or inflammation when excessive tissue damage occurs, and to prevent transplant rejection after an organ transplant.[25][95]
Anti-inflammatory drugs are often used to control the effects of inflammation. The glucocorticoids are the most powerful of these drugs; however, these drugs can have many undesirable side effects (e.g., central obesity, hyperglycemia, osteoporosis) and their use must be tightly controlled.[96] Therefore, lower doses of anti-inflammatory drugs are often used in conjunction with cytotoxic or immunosuppressive drugs such as methotrexate or azathioprine. Cytotoxic drugs inhibit the immune response by killing dividing cells such as activated T cells. However, the killing is indiscriminate and other organs and cell types are affected, which causes toxic side effects.[95] Immunosuppressive drugs such as cyclosporin prevent T cells from responding to signals correctly by inhibiting signal transduction pathways.[97]
Larger drugs (>500 Da) can provoke a neutralizing immune response, particularly if the drugs are administered repeatedly, or in larger doses. This limits the effectiveness of drugs based on larger peptides and proteins (which are typically larger than 6000 Da). In some cases, the drug itself is not immunogenic, but may be co-administered with an immunogenic compound, as is sometimes the case for Taxol. Computational methods have been developed to predict the immunogenicity of peptides and proteins, which are particularly useful in designing therapeutic antibodies, assessing likely virulence of mutations in viral coat particles, and validation of proposed peptide-based drug treatments. Early techniques relied mainly on the observation that hydrophilic amino acids are overrepresented in epitope regions than hydrophobic amino acids;[98] however, more recent developments rely on machine learning techniques using databases of existing known epitopes, usually on well-studied virus proteins, as a training set.[99] A publicly accessible database has been established for the cataloguing of epitopes from pathogens known to be recognizable by B cells.[100] The emerging field of bioinformatics-based studies of immunogenicity is referred to as immunoinformatics.[101]
Manipulation by pathogens
The success of any pathogen is dependent on its ability to elude host immune responses. Therefore, pathogens have developed several methods that allow them to successfully infect a host, while evading immune-mediated destruction.[102] Bacteria often overcome physical barriers by secreting enzymes that digest the barrier — for example, by using a type II secretion system.[103] Alternatively, using a type III secretion system, they may insert a hollow tube into the host cell, which provides a direct conduit for proteins to move from the pathogen to the host; the proteins transported along the tube are often used to shut down host defenses.[104]
An evasion strategy used by several pathogens to circumvent the innate immune system is intracellular replication (also called intracellular pathogenesis). Here, a pathogen spends a majority of its life-cycle inside host cells, where it is shielded from direct contact with immune cells, antibodies and complement. Some examples of intracellular pathogens include viruses, the food poisoning bacterium Salmonella and the eukaryotic parasites that cause malaria (Plasmodium falciparum) and leishmaniasis (Leishmania spp.). Other bacteria, such as Mycobacterium tuberculosis, live inside a protective capsule that prevents lysis by complement.[105] Many pathogens secrete compounds that diminish or misdirect the host's immune response.[102] Some bacteria form biofilms to protect themselves from the cells and proteins of the immune system. Such biofilms are present in many successful infections, e.g., the chronic Pseudomonas aeruginosa and Burkholderia cenocepacia infections characteristic of cystic fibrosis.[106] Other bacteria generate surface proteins that bind to antibodies, rendering them ineffective; examples include Streptococcus (protein G), Staphylococcus aureus (protein A), and Peptostreptococcus magnus (protein L).[107]
The mechanisms used by viruses to evade the adaptive immune system are more complicated. The simplest approach is to rapidly change non-essential epitopes (amino acids and/or sugars) on the invader's surface, while keeping essential epitopes concealed. HIV, for example, regularly mutates the proteins on its viral envelope that are essential for entry into its host target cell. These frequent changes in antigens may explain the failures of vaccines directed at these proteins.[108] Masking antigens with host molecules is another common strategy for avoiding detection by the immune system. In HIV, the envelope that covers the viron is formed from the outermost membrane of the host cell; such "self-cloaked" viruses make it difficult for the immune system to identify them as "non-self".[109]
History of immunology

Immunology is a science that examines the structure and function of the immune system. It originates from medicine and early studies on the causes of immunity to disease. The earliest known mention of immunity was during the plague of Athens in 430 BC. Thucydides noted that people who had recovered from a previous bout of the disease could nurse the sick without contracting the illness a second time.[110] This observation of acquired immunity was later exploited by Louis Pasteur in his development of vaccination and his proposed germ theory of disease.[111] Pasteur's theory was in direct opposition to contemporary theories of disease, such as the miasma theory. It was not until Robert Koch's 1891 proofs, for which he was awarded a Nobel Prize in 1905, that microorganisms were confirmed as the cause of infectious disease.[112] Viruses were confirmed as human pathogens in 1901, with the discovery of the yellow fever virus by Walter Reed.[113]
Immunology made a great advance towards the end of the 19th century, through rapid developments, in the study of humoral immunity and cellular immunity.[114] Particularly important was the work of Paul Ehrlich, who proposed the side-chain theory to explain the specificity of the antigen-antibody reaction; his contributions to the understanding of humoral immunity were recognized by the award of a Nobel Prize in 1908, which was jointly awarded to the founder of cellular immunology, Elie Metchnikoff.[115]
See also
| Wikimedia Commons has media related to Immunology. |
- Clonal selection
- Epitope
- Hapten
- Human physiology
- Immunostimulator
- Monoclonal antibodies
- Original antigenic sin
- Polyclonal antibody
- Tumor antigens
References
- ↑ 1.0 1.1 1.2 Beck, Gregory (November 1996). "Immunity and the Invertebrates" (PDF). Scientific American: 60–66. Retrieved 2007-01-01. Unknown parameter
|coauthors=ignored (help) - ↑ 2.0 2.1 Litman G, Cannon J, Dishaw L (2005). "Reconstructing immune phylogeny: new perspectives". Nat Rev Immunol. 5 (11): 866–79. PMID 16261174.
- ↑ 3.0 3.1 3.2 Mayer, Gene (2006). "Immunology - Chapter One: Innate (non-specific) Immunity". Microbiology and Immunology On-Line Textbook. USC School of Medicine. Retrieved 2007-01-01.
- ↑ Smith A.D. (Ed) Oxford dictionary of biochemistry and molecular biology. (1997) Oxford University Press. ISBN 0-19-854768-4
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 5.8 Alberts, Bruce (2002). Molecular Biology of the Cell; Fourth Edition. New York and London: Garland Science. ISBN 0-8153-3218-1. Unknown parameter
|coauthors=ignored (help) - ↑ Boyton R, Openshaw P. "Pulmonary defences to acute respiratory infection". Br Med Bull. 61: 1–12. PMID 11997295.
- ↑ Agerberth B, Gudmundsson G. "Host antimicrobial defence peptides in human disease". Curr Top Microbiol Immunol. 306: 67–90. PMID 16909918.
- ↑ Moreau J, Girgis D, Hume E, Dajcs J, Austin M, O'Callaghan R (2001). "Phospholipase A(2) in rabbit tears: a host defense against Staphylococcus aureus". Invest Ophthalmol Vis Sci. 42 (10): 2347–54. PMID 11527949.
- ↑ Hankiewicz J, Swierczek E (1974). "Lysozyme in human body fluids". Clin Chim Acta. 57 (3): 205–9. PMID 4434640.
- ↑ Fair W, Couch J, Wehner N (1976). "Prostatic antibacterial factor. Identity and significance". Urology. 7 (2): 169–77. PMID 54972.
- ↑ Yenugu S, Hamil K, Birse C, Ruben S, French F, Hall S (2003). "Antibacterial properties of the sperm-binding proteins and peptides of human epididymis 2 (HE2) family; salt sensitivity, structural dependence and their interaction with outer and cytoplasmic membranes of Escherichia coli". Biochem J. 372 (Pt 2): 473–83. PMID 12628001.
- ↑ Gorbach S (1990). "Lactic acid bacteria and human health". Ann Med. 22 (1): 37–41. PMID 2109988.
- ↑ Hill L, Embil J (1986). "Vaginitis: current microbiologic and clinical concepts". CMAJ. 134 (4): 321–31. PMID 3510698.
- ↑ Reid G, Bruce A (2003). "Urogenital infections in women: can probiotics help?". Postgrad Med J. 79 (934): 428–32. PMID 12954951.
- ↑ Salminen S, Gueimonde M, Isolauri E (2005). "Probiotics that modify disease risk". J Nutr. 135 (5): 1294–8. PMID 15867327.
- ↑ Reid G, Jass J, Sebulsky M, McCormick J (2003). "Potential uses of probiotics in clinical practice". Clin Microbiol Rev. 16 (4): 658–72. PMID 14557292.
- ↑ Medzhitov R (2007). "Recognition of microorganisms and activation of the immune response". Nature. 449 (7164): 819–26. doi:10.1038/nature06246. PMID 17943118.
- ↑ Kawai T, Akira S (2006). "Innate immune recognition of viral infection". Nat Immunol. 7 (2): 131–7. PMID 16424890.
- ↑ Miller, SB (2006). "Prostaglandins in Health and Disease: An Overview". Seminars in Arthritis and Rheumatism. 36 (1): 37–49. PMID 16887467.
- ↑ Ogawa Y, Calhoun WJ. (2006). "The role of leukotrienes in airway inflammation". J Allergy Clin Immunol. 118 (4): 789–98. PMID 17030228.
- ↑ Le Y, Zhou Y, Iribarren P, Wang J (2004). "Chemokines and chemokine receptors: their manifold roles in homeostasis and disease" (PDF). Cell Mol Immunol. 1 (2): 95–104. PMID 16212895.
- ↑ Martin P, Leibovich S (2005). "Inflammatory cells during wound repair: the good, the bad and the ugly". Trends Cell Biol. 15 (11): 599–607. PMID 16202600.
- ↑ 23.0 23.1 Rus H, Cudrici C, Niculescu F (2005). "The role of the complement system in innate immunity". Immunol Res. 33 (2): 103–12. PMID 16234578.
- ↑ Mayer, Gene (2006). "Immunology - Chapter Two: Complement". Microbiology and Immunology On-Line Textbook. USC School of Medicine. Retrieved 2007-01-01.
- ↑ 25.0 25.1 25.2 25.3 25.4 25.5 Janeway CA, Jr.; et al. (2005). Immunobiology (6th ed. ed.). Garland Science. ISBN 0-443-07310-4.
- ↑ Liszewski M, Farries T, Lublin D, Rooney I, Atkinson J. "Control of the complement system". Adv Immunol. 61: 201–83. PMID 8834497.
- ↑ Sim R, Tsiftsoglou S (2004). "Proteases of the complement system" (PDF). Biochem Soc Trans. 32 (Pt 1): 21–7. PMID 14748705.
- ↑ Ryter A (1985). "Relationship between ultrastructure and specific functions of macrophages". Comp Immunol Microbiol Infect Dis. 8 (2): 119–33. PMID 3910340.
- ↑ Langermans J, Hazenbos W, van Furth R (1994). "Antimicrobial functions of mononuclear phagocytes". J Immunol Methods. 174 (1–2): 185–94. PMID 8083520.
- ↑ May R, Machesky L (2001). "Phagocytosis and the actin cytoskeleton". J Cell Sci. 114 (Pt 6): 1061–77. PMID 11228151.
- ↑ Salzet M, Tasiemski A, Cooper E (2006). "Innate immunity in lophotrochozoans: the annelids". Curr Pharm Des. 12 (24): 3043–50. PMID 16918433.
- ↑ Zen K, Parkos C (2003). "Leukocyte-epithelial interactions". Curr Opin Cell Biol. 15 (5): 557–64. PMID 14519390.
- ↑ 33.0 33.1 Stvrtinová, Viera (1995). Inflammation and Fever from Pathophysiology: Principles of Disease. Computing Centre, Slovak Academy of Sciences: Academic Electronic Press. Retrieved 2007-01-01. Unknown parameter
|coauthors=ignored (help) - ↑ Bowers, William (2006). "Immunology -Chapter Thirteen: Immunoregulation". Microbiology and Immunology On-Line Textbook. USC School of Medicine. Retrieved 2007-01-04.
- ↑ 35.0 35.1 Guermonprez P, Valladeau J, Zitvogel L, Théry C, Amigorena S. "Antigen presentation and T cell stimulation by dendritic cells". Annu Rev Immunol. 20: 621–67. PMID 11861614.
- ↑ Krishnaswamy G, Ajitawi O, Chi D. "The human mast cell: an overview". Methods Mol Biol. 315: 13–34. PMID 16110146.
- ↑ Kariyawasam H, Robinson D (2006). "The eosinophil: the cell and its weapons, the cytokines, its locations". Semin Respir Crit Care Med. 27 (2): 117–27. PMID 16612762.
- ↑ Middleton D, Curran M, Maxwell L (2002). "Natural killer cells and their receptors". Transpl Immunol. 10 (2–3): 147–64. PMID 12216946.
- ↑ Pancer Z, Cooper M. "The evolution of adaptive immunity". Annu Rev Immunol. 24: 497–518. PMID 16551257.
- ↑ Holtmeier W, Kabelitz D. "gammadelta T cells link innate and adaptive immune responses". Chem Immunol Allergy. 86: 151–83. PMID 15976493.
- ↑ 41.0 41.1 "Understanding the Immune System: How it Works" (PDF). National Institute of Allergy and Infectious Diseases (NIAID). Retrieved 2007-01-01.
- ↑ Harty J, Tvinnereim A, White D. "CD8+ T cell effector mechanisms in resistance to infection". Annu Rev Immunol. 18: 275–308. PMID 10837060.
- ↑ 43.0 43.1 Radoja S, Frey A, Vukmanovic S (2006). "T-cell receptor signaling events triggering granule exocytosis". Crit Rev Immunol. 26 (3): 265–90. PMID 16928189.
- ↑ Abbas A, Murphy K, Sher A (1996). "Functional diversity of helper T lymphocytes". Nature. 383 (6603): 787–93. PMID 8893001.
- ↑ McHeyzer-Williams L, Malherbe L, McHeyzer-Williams M. "Helper T cell-regulated B cell immunity". Curr Top Microbiol Immunol. 311: 59–83. PMID 17048705.
- ↑ Kovacs B, Maus M, Riley J, Derimanov G, Koretzky G, June C, Finkel T (2002). "Human CD8+ T cells do not require the polarization of lipid rafts for activation and proliferation". Proc Natl Acad Sci U S A. 99 (23): 15006–11. PMID 12419850.
- ↑ Grewal I, Flavell R. "CD40 and CD154 in cell-mediated immunity". Annu Rev Immunol. 16: 111–35. PMID 9597126.
- ↑ Girardi M (2006). "Immunosurveillance and immunoregulation by γδ T cells". J Invest Dermatol. 126 (1): 25–31. PMID 16417214.
- ↑ Holtmeier W, Kabelitz D (2005). "γδ T cells link innate and adaptive immune responses". Chem Immunol Allergy. 86: 151–183. PMID 15976493.
- ↑ 50.0 50.1 Sproul T, Cheng P, Dykstra M, Pierce S (2000). "A role for MHC class II antigen processing in B cell development". Int Rev Immunol. 19 (2–3): 139–55. PMID 10763706.
- ↑ Kehry M, Hodgkin P (1994). "B-cell activation by helper T-cell membranes". Crit Rev Immunol. 14 (3–4): 221–38. PMID 7538767.
- ↑ Bowers, William (2006). "Immunology - Chapter nine: Cells involved in immune responses". Microbiology and Immunology On-Line Textbook. USC School of Medicine. Retrieved 2007-01-04.
- ↑ M.N. Alder, I.B. Rogozin, L.M. Iyer, G.V. Glazko, M.D. Cooper, Z. Pancer (2005). "Diversity and Function of Adaptive Immune Receptors in a Jawless Vertebrate". Science. 310 (5756): 1970–1973. PMID 16373579.
- ↑ Saji F, Samejima Y, Kamiura S, Koyama M (1999). "Dynamics of immunoglobulins at the feto-maternal interface" (PDF). Rev Reprod. 4 (2): 81–9. PMID 10357095.
- ↑ Van de Perre P (2003). "Transfer of antibody via mother's milk". Vaccine. 21 (24): 3374–6. PMID 12850343.
- ↑ Keller, Margaret A. and E. Richard Stiehm (2000). "Passive Immunity in Prevention and Treatment of Infectious Diseases". Clinical Microbiology Reviews. 13 (4): 602–614. PMID 11023960.
- ↑ Death and DALY estimates for 2002 by cause for WHO Member States. World Health Organization. Retrieved on 2007-01-01.
- ↑ Singh M, O'Hagan D (1999). "Advances in vaccine adjuvants". Nat Biotechnol. 17 (11): 1075–81. PMID 10545912.
- ↑ Aw D, Silva A, Palmer D (2007). "Immunosenescence: emerging challenges for an ageing population". Immunology. 120 (4): 435–446. PMID 17313487.
- ↑ 60.0 60.1 60.2 60.3 Chandra, RK (1997). "Nutrition and the immune system: an introduction". American Journal of Clinical Nutrition. Vol 66: 460S–463S. PMID 9250133. Free full-text pdf available
- ↑ Miller JF (2002). "The discovery of thymus function and of thymus-derived lymphocytes". Immunol. Rev. 185: 7–14. PMID 12190917.
- ↑ Joos L, Tamm M (2005). "Breakdown of pulmonary host defense in the immunocompromised host: cancer chemotherapy". Proc Am Thorac Soc. 2 (5): 445–8. PMID 16322598.
- ↑ Copeland K, Heeney J (1996). "T helper cell activation and human retroviral pathogenesis". Microbiol Rev. 60 (4): 722–42. PMID 8987361.
- ↑ Miller J (1993). "Self-nonself discrimination and tolerance in T and B lymphocytes". Immunol Res. 12 (2): 115–30. PMID 8254222.
- ↑ 65.0 65.1 65.2 65.3 Ghaffar, Abdul (2006). "Immunology - Chapter Seventeen: Hypersensitivity Reactions". Microbiology and Immunology On-Line Textbook. USC School of Medicine. Retrieved 2007-01-01.
- ↑ Bickle T, Krüger D (1993). "Biology of DNA restriction". Microbiol Rev. 57 (2): 434–50. PMID 8336674.
- ↑ Stram Y, Kuzntzova L. (2006). "Inhibition of viruses by RNA interference". Virus Genes. 32 (3): 299–306. PMID 16732482.
- ↑ 68.0 68.1 Schneider, David (Spring 2005). "Innate Immunity - Lecture 4: Plant immune responses". Stanford University Department of Microbiology and Immunology. Retrieved 2007-01-01.
- ↑ Baulcombe D (2004). "RNA silencing in plants". Nature. 431 (7006): 356–63. PMID 15372043.
- ↑ Morgan R; et al. (2006). "Cancer regression in patients after transfer of genetically engineered lymphocytes". Science. 314: 126–129. PMID 16946036.
- ↑ 71.0 71.1 Andersen MH, Schrama D, Thor Straten P, Becker JC (2006). "Cytotoxic T cells". J Invest Dermatol. 126 (1): 32–41. PMID 16417215.
- ↑ Boon T, van der Bruggen P (1996). "Human tumor antigens recognized by T lymphocytes". J Exp Med. 183: 725–29. PMID 8642276.
- ↑ Castelli C, Rivoltini L, Andreola G, Carrabba M, Renkvist N, Parmiani G (2000). "T cell recognition of melanoma-associated antigens". J Cell Physiol. 182: 323–31. PMID 10653598.
- ↑ 74.0 74.1 Romero P, Cerottini JC, Speiser DE (2006). "The human T cell response to melanoma antigens". Adv Immunol. 92: 187–224. PMID 17145305.
- ↑ 75.0 75.1 Guevara-Patino JA, Turk MJ, Wolchok JD, Houghton AN (2003). "Immunity to cancer through immune recognition of altered self: studies with melanoma". Adv Cancer Res. 90: 157–77. PMID 14710950.
- ↑ Renkvist N, Castelli C, Robbins PF, Parmiani G (2001). "A listing of human tumor antigens recognized by T cells". Cancer Immunol Immunother. 50: 3–15. PMID 11315507.
- ↑ Gerloni M, Zanetti M. (2005). "CD4 T cells in tumor immunity". . Springer Semin Immunopathol. 27 (1): 37–48. PMID 15965712.
- ↑ 78.0 78.1 Seliger B, Ritz U, Ferrone S (2006). "Molecular mechanisms of HLA class I antigen abnormalities following viral infection and transformation". Int J Cancer. 118 (1): 129–38. PMID 16003759.
- ↑ Hayakawa Y, Smyth MJ. (2006). "Innate immune recognition and suppression of tumors". Adv Cancer Res. 95: 293–322. PMID 16860661.
- ↑ 80.0 80.1 Seliger B (2005). "Strategies of tumor immune evasion". BioDrugs. 19 (6): 347–54. PMID 16392887.
- ↑ Frumento G, Piazza T, Di Carlo E, Ferrini S (2006). "Targeting tumor-related immunosuppression for cancer immunotherapy". Endocr Metab Immune Disord Drug Targets. 6 (3): 233–7. PMID 17017974.
- ↑ Stix, Gary (July 2007). "A Malignant Flame" (PDF). Scientific American: 60–67. Retrieved 2007-01-01.
- ↑ Wira, CR (2004). "Endocrine regulation of the mucosal immune system in the female reproductive tract". In In: Ogra PL, Mestecky J, Lamm ME, Strober W, McGhee JR, Bienenstock J (eds.). Mucosal Immunology. San Francisco: Elsevier. ISBN 0124915434. Unknown parameter
|coauthors=ignored (help) - ↑ Lang, TJ (2004). "Estrogen as an immunomodulator". Clin Immunol. 113: 224&ndash, 230. PMID 15507385.
Moriyama, A (1999). "Secretory leukocyte protease inhibitor (SLPI) concentrations in cervical mucus of women with normal menstrual cycle". Molecular Human Reproduction. 5: 656&ndash, 661. PMID 10381821. Unknown parameter|coauthors=ignored (help)
Cutolo, M (2004). "Sex hormones influence on the immune system: basic and clinical aspects in autoimmunity". Lupus. 13: 635&ndash, 638. PMID 15485092. Unknown parameter|coauthors=ignored (help)
King, AE (2000). "Presence of secretory leukocyte protease inhibitor in human endometrium and first trimester decidua suggests an antibacterial role". Molecular Human Reproduction. 6: 191&ndash, 196. PMID 10655462. Unknown parameter|coauthors=ignored (help) - ↑ Fimmel (2005). "Influence of physiological androgen levels on wound healing and immune status in men". Aging Male. 8: 166&ndash, 174. PMID 16390741. Unknown parameter
|coauthors=ignored (help); Unknown parameter|fist=ignored (help) - ↑ Dorshkind, K (2000). "The Roles of Prolactin, Growth Hormone, Insulin-Like Growth Factor-I, and Thyroid Hormones in Lymphocyte Development and Function: Insights from Genetic Models of Hormones and Hormone Receptor Deficiency". Endocrine Reviews. 21: 292&ndash, 312. PMID 10857555. Unknown parameter
|coauthors=ignored (help) - ↑ Nagpal, Sunil (2005). "Noncalcemic Actions of Vitamin D Receptor Ligands". Endocrine Reviews. 26 (5): 662&ndash, 687. PMID 15798098. Unknown parameter
|coauthors=ignored (help). - ↑ Hertoghe, T (2005). "The "multiple hormone deficiency" theory of aging: Is human senescence caused mainly by multiple hormone deficiencies?". Annals of the New York Academy of Science. 1051: 448&ndash, 465. PMID 16399912.
- ↑ Klein, JR (2006). "The immune system as a regulator of thyroid hormone activity". Exp Biol Med. 231: 229&ndash, 236. PMID 16514168.
- ↑ Lange, T (2003). "Sleep Enhances the Human Antibody response to Hepatitis A Vaccination". Psychosomatic Medicine. 65: 831&ndash, 835. PMID 14508028. Unknown parameter
|coauthors=ignored (help) - ↑ Khansari, DN (1990). "Effects of stress on the immune system". Immunology Today. 11: 170&ndash, 175. PMID 2186751. Unknown parameter
|coauthors=ignored (help) - ↑ Pond, CM (2005). "Adipose tissue and the immune system". Prostaglandins, Leukotrienes, and Essential Fatty Acids. 73: 17–30. PMID 15946832.
- ↑ Langley-Evans, SC (2006). "Diet and the developing immune system". Lupus. 15: 746&ndash, 752. PMID 17153845. Unknown parameter
|coauthors=ignored (help) - ↑ Spelman, K (2006). "Modulation of cytokine expression by traditional medicines: a review of herbal immunomodulators". Alternative Medicine reviews: 128&ndash, 150. PMID 16813462. Text " volume 11 " ignored (help); Unknown parameter
|coauthors=ignored (help)
Brush, J (2006). "The effect of Echinacea purpurea, Astragalus membranaceus and Glycyrrhiza glabra on CD69 expression and immune cell activation in humans". Phytotherapy Research. 20: 687&ndash, 695. PMID 16807880. Unknown parameter|coauthors=ignored (help) - ↑ 95.0 95.1 Taylor A, Watson C, Bradley J (2005). "Immunosuppressive agents in solid organ transplantation: Mechanisms of action and therapeutic efficacy". Crit Rev Oncol Hematol. 56 (1): 23–46. PMID 16039869.
- ↑ Barnes P (2006). "Corticosteroids: the drugs to beat". Eur J Pharmacol. 533 (1–3): 2–14. PMID 16436275.
- ↑ Masri M (2003). "The mosaic of immunosuppressive drugs". Mol Immunol. 39 (17–18): 1073–7. PMID 12835079.
- ↑ Welling GW, Wiejer WJ, van der Zee R, Welling-Werster S. (1985). "Prediction of sequential antigenic regions in proteins". J Mol Recognit. 88 (2): 215–8. PMID 2411595.
- ↑ Sollner J, Mayer B. (2006). "Machine learning approaches for prediction of linear B-cell epitopes on proteins". 19 (3): 200–8. PMID 16598694.
- ↑ Saha S, Bhasin M, Raghava GP. (2005). "Bcipep: a database of B-cell epitopes". BMC Bioinformatics. 6 (1): 79. PMID 15921533.
- ↑ Flower DR, Doytchinova IA. (2002). "Immunoinformatics and the prediction of immunogenicity". Appl Bioinformatics. 1 (4): 167–76. PMID 15130835.
- ↑ 102.0 102.1 Finlay B, McFadden G (2006). "Anti-immunology: evasion of the host immune system by bacterial and viral pathogens". Cell. 124 (4): 767–82. PMID 16497587.
- ↑ Cianciotto NP. (2005). "Type II secretion: a protein secretion system for all seasons". Trends Microbiol. 13 (12): 581–8. PMID 16216510.
- ↑ Winstanley C, Hart CA (2001). "Type III secretion systems and pathogenicity islands". J Med Microbiol. 50 (2): 116–26. PMID 11211218.
- ↑ Finlay B, Falkow S (1997). "Common themes in microbial pathogenicity revisited" (PDF). Microbiol Mol Biol Rev. 61 (2): 136–69. PMID 9184008.
- ↑ Kobayashi H (2005). "Airway biofilms: implications for pathogenesis and therapy of respiratory tract infections". Treat Respir Med. 4 (4): 241–53. PMID 16086598.
- ↑ Housden N, Harrison S, Roberts S, Beckingham J, Graille M, Stura E, Gore M (2003). "Immunoglobulin-binding domains: Protein L from Peptostreptococcus magnus" (PDF). Biochem Soc Trans. 31 (Pt 3): 716–8. PMID 12773190.
- ↑ Burton, Dennis R. (2005). "Antibody vs. HIV in a clash of evolutionary titans". Proc Natl Acad Sci U S A. 102 (42): 14943–8. PMID 16219699. Unknown parameter
|coauthors=ignored (help) - ↑ Cantin R, Methot S, Tremblay MJ. (2005). "Plunder and stowaways: incorporation of cellular proteins by enveloped viruses". J Virol. 79 (11): 6577–87. PMID 15890896.
- ↑ Retief F, Cilliers L (1998). "The epidemic of Athens, 430-426 BC". S Afr Med J. 88 (1): 50–3. PMID 9539938.
- ↑ Plotkin S (2005). "Vaccines: past, present and future". Nat Med. 11 (4 Suppl): S5–11. PMID 15812490.
- ↑ The Nobel Prize in Physiology or Medicine 1905 Nobelprize.org Accessed January 8 2007.
- ↑ Major Walter Reed, Medical Corps, U.S. Army Walter Reed Army Medical Center. Accessed January 8 2007.
- ↑
Metchnikoff, Elie (1905). Immunity in Infective Diseases (Full Text Version: Google Books). Cambridge University Press. ISBN 68025143. Unknown parameter
|coauthors=ignored (help) - ↑ The Nobel Prize in Physiology or Medicine 1908 Nobelprize.org Accessed January 8 2007
External links
- How Your Immune System Works - from HowStuffWorks
- Immune System - from the University of Hartford
- Immunobiology; Fifth Edition – Online version of the textbook by Charles Janeway (Advanced undergraduate/graduate level)
- Immunology - BioMed Central (free content) scientific journal
- The Inner Life of a Cell - Rendering of the inner functions of the human body
- The Microbial World - Animal defenses against microbes - Chapter in on-line microbiology textbook
- Microbiology and Immunology On-Line Textbook - from the University of South Carolina School of Medicine
- Plant Immunity - Institute of Biochemical Plant Pathology at the GSF-National Research Center for Environment and Health
Template:Lymphatic system Template:Organ systems
Template:Link FA ar:جهاز مناعي bg:Имунна система cs:Imunita (biologie) da:Immunforsvar de:Immunsystem et:Immuunsüsteem ko:면역 id:Imunitas it:Sistema immunitario he:מערכת החיסון mk:Имунолошки систем nl:Afweer no:Immunforsvar sk:Imunitný systém sl:Imnuski sistem sr:Имунски систем fi:Immuunijärjestelmä sv:Immunförsvar tl:Sistemang pananggalang yi:אימיון סיסטעם
- Pages with script errors
- Pages with citations using unsupported parameters
- CS1 maint: Multiple names: authors list
- CS1 maint: Explicit use of et al.
- CS1 maint: Extra text
- CS1 maint: Multiple names: editors list
- CS1 maint: Extra text: editors list
- Pages with citations using unnamed parameters
- Commons category link is locally defined
- Commons category link is on Wikidata using P373
- Immune system
- Immunology