Xanthine oxidase: Difference between revisions
m (Robot: Automated text replacement (-{{SIB}} +, -{{EH}} +, -{{EJ}} +, -{{Editor Help}} +, -{{Editor Join}} +)) |
No edit summary |
||
| Line 1: | Line 1: | ||
{{enzyme | |||
| Name = xanthine oxidase/dehydrogenase | |||
| EC_number = 1.17.3.2 | |||
| IUBMB_EC_number = 1/17/3/2 | |||
| CAS_number= 9002-17-9 | |||
| GO_code = 0004855 | |||
| image = XanthineOxidase-1FIQ.png | |||
| width = | |||
| caption = Crystallographic structure (monomer) of bovine xanthine oxidase.<ref name="pmid11005854">{{PDB|1FIQ}}; {{cite journal | vauthors = Enroth C, Eger BT, Okamoto K, Nishino T, Nishino T, Pai EF | title = Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 97 | issue = 20 | pages = 10723–8 | date = September 2000 | pmid = 11005854 | pmc = 27090 | doi = 10.1073/pnas.97.20.10723 | url = http://www.pnas.org/content/97/20/10723.abstract }}</ref><br>The bounded FAD (red), FeS-cluster (orange), the molybdopterin cofactor with molybdenum (yellow) and salicylate (blue) are indicated. | |||
}} | |||
{{Infobox protein | |||
| Name = xanthine oxidase/dehydrogenase | |||
| caption = | |||
| image = | |||
| width = | |||
| HGNCid = 12805 | |||
| Symbol = [[Xanthine dehydrogenase|XDH]] | |||
| AltSymbols = | |||
| EntrezGene = 7498 | |||
| OMIM = 607633 | |||
| RefSeq = NM_000379 | |||
| UniProt = P47989 | |||
| PDB = 1FIQ | |||
| ECnumber = 1.17.3.2 | |||
| Chromosome = 2 | |||
| Arm = p | |||
| Band = 23.1 | |||
| LocusSupplementaryData = | |||
}} | |||
'''Xanthine oxidase''' ('''XO''', sometimes {{'}}'''XAO'''{{'}}) is a form of [[xanthine oxidoreductase]], a type of [[enzyme]] that generates [[reactive oxygen species]].<ref>{{cite journal | vauthors = Ardan T, Kovaceva J, Cejková J | title = Comparative histochemical and immunohistochemical study on xanthine oxidoreductase/xanthine oxidase in mammalian corneal epithelium | journal = Acta Histochemica | volume = 106 | issue = 1 | pages = 69–75 | date = February 2004 | pmid = 15032331 | doi = 10.1016/j.acthis.2003.08.001 }}</ref> These enzymes catalyze the [[oxidation]] of [[hypoxanthine]] to [[xanthine]] and can further catalyze the oxidation of xanthine to [[uric acid]]. These enzymes play an important role in the catabolism of [[purines]] in some species, including humans.<ref>{{cite journal | vauthors = Hille R | title = Molybdenum-containing hydroxylases | journal = Archives of Biochemistry and Biophysics | volume = 433 | issue = 1 | pages = 107–16 | date = Jan 2005 | pmid = 15581570 | doi = 10.1016/j.abb.2004.08.012 }}</ref><ref>{{cite journal | vauthors = Harrison R | title = Structure and function of xanthine oxidoreductase: where are we now? | journal = Free Radical Biology & Medicine | volume = 33 | issue = 6 | pages = 774–97 | date = September 2002 | pmid = 12208366 | doi = 10.1016/S0891-5849(02)00956-5 }}</ref> | |||
Xanthine oxidase is defined as an ''enzyme activity'' (EC 1.17.3.2).<ref>{{cite web|url=http://www.genome.jp/dbget-bin/www_bget?ec:1.17.3.2|title=KEGG record for EC 1.17.3.2}}</ref> The same protein, which in humans has the [[HGNC]] approved gene symbol ''XDH'', can also have [[xanthine dehydrogenase]] activity (EC 1.17.1.4).<ref name="KEGG record for EC 1.17.1.4">{{cite web|url=http://www.genome.jp/dbget-bin/www_bget?ec:1.17.1.4|title=KEGG record for EC 1.17.1.4}}</ref> Most of the protein in the liver exists in a form with xanthine dehydrogenase activity, but it can be converted to xanthine oxidase by reversible sulfhydryl oxidation or by irreversible proteolytic modification.<ref name="entrez">{{cite web | title = Entrez Gene: XDH xanthine dehydrogenase| url =https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=7498| accessdate = }}</ref><ref>{{cite web|url=http://www.omim.org/entry/607633?search=xanthine%20dehydrogenase&highlight=xanthine%20dehydrogenase |title=*607633 XANTHINE DEHYDROGENASE; XDH |deadurl=yes }}</ref> | |||
The [[ | |||
==Reaction== | ==Reaction== | ||
* hypoxanthine + | The following chemical reactions are catalyzed by xanthine oxidase: | ||
* xanthine + O<sub>2</sub> + H<sub>2</sub>O | * hypoxanthine + H<sub>2</sub>O + O<sub>2</sub> <math>\rightleftharpoons</math> xanthine + H<sub>2</sub>O<sub>2</sub> | ||
* xanthine + H<sub>2</sub>O + O<sub>2</sub> <math>\rightleftharpoons</math> uric acid + H<sub>2</sub>O<sub>2</sub> | |||
* Xanthine oxidase can also act on certain other purines, pterins, and aldehydes. For example, it efficiently converts 1-methylxanthine (a metabolite of [[caffeine]]) to 1-methyluric acid, but has little activity on 3-methylxanthine.<ref>{{cite journal | vauthors = Birkett DJ, Miners JO, Valente L, Lillywhite KJ, Day RO | title = 1-Methylxanthine derived from caffeine as a pharmacodynamic probe of oxypurinol effect | journal = British Journal of Clinical Pharmacology | volume = 43 | issue = 2 | pages = 197–200 | date = February 1997 | pmid = 9131954 | pmc = 2042732 | doi = 10.1046/j.1365-2125.1997.53711.x | url = https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2042732/pdf/bcp_0537.pdf }}</ref> | |||
* Under some circumstances it can produce [[superoxide]] ion RH + H<sub>2</sub>O + 2 O<sub>2</sub> <math>\rightleftharpoons</math> ROH + 2 O<sub>2</sub><sup>−</sup> + 2 H<sup>+</sup>.<ref name="KEGG record for EC 1.17.1.4"/> | |||
<gallery> | <gallery> | ||
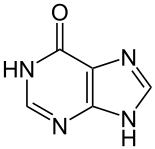
Image:Hypoxanthin - Hypoxanthine.svg|[[hypoxanthine]] ( | Image:Hypoxanthin - Hypoxanthine.svg|[[hypoxanthine]] (one oxygen atom) | ||
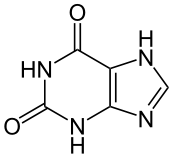
Image:Xanthin - Xanthine.svg|[[xanthine]] ( | Image:Xanthin - Xanthine.svg|[[xanthine]] (two oxygens) | ||
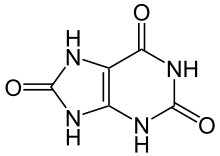
Image:Harnsäure Ketoform.svg|[[uric acid]] ( | Image:Harnsäure Ketoform.svg|[[uric acid]] (three oxygens) | ||
</gallery> | </gallery> | ||
=== Other reactions === | |||
Because XO is a superoxide-producing enzyme, with general low specificity,<ref name="MG Bonini et al">{{cite journal | vauthors = Bonini MG, Miyamoto S, Di Mascio P, Augusto O | title = Production of the carbonate radical anion during xanthine oxidase turnover in the presence of bicarbonate | journal = The Journal of Biological Chemistry | volume = 279 | issue = 50 | pages = 51836–43 | date = December 2004 | pmid = 15448145 | doi = 10.1074/jbc.M406929200 | url = http://www.jbc.org/content/279/50/51836.abstract }}</ref> it can be combined with other compounds and enzymes and create reactive oxidants, as well as oxidize other substrates. | |||
Bovine xanthine oxidase (from milk) was originally thought to have a binding site to reduce [[cytochrome c]] with, but it has been found that the mechanism to reduce this protein is through XO's superoxide anion byproduct, with competitive inhibition by [[carbonic anhydrase]].<ref name="JM McCord & I. Fridovich">{{cite journal | vauthors = McCord JM, Fridovich I | title = The reduction of cytochrome c by milk xanthine oxidase | journal = The Journal of Biological Chemistry | volume = 243 | issue = 21 | pages = 5753–60 | date = November 1968 | pmid = 4972775 | url = http://www.jbc.org/content/243/21/5753.short }}</ref> | |||
Another reaction catalyzed by xanthine oxidase is the decomposition of S-Nitrosothiols (RSNO), a reactive nitrogen species, to nitric oxide (NO), which reacts with a superoxide anion to form peroxynitrite under aerobic conditions.<ref name="M Trujillo">{{cite journal|last=Trujillo|first=M.|title=Xanthine Oxidase-mediated Decomposition of S-Nitrosothiols|journal=Journal of Biological Chemistry|date=3 April 1998|volume=273|issue=14|pages=7828–7834|doi=10.1074/jbc.273.14.7828|url=http://www.jbc.org/content/273/14/7828.abstract}}</ref> | |||
XO has also been found to produce the strong one-electron oxidant carbonate radical anion from oxidation with acetaldehyde in the presence of catalase and bicarbonate. It was suggested that the carbonate radical was likely produced in one of the enzyme's redox centers with a peroxymonocarbonate intermediate.<ref name="MG Bonini et al" /> | |||
Here is a diagram highlighting the pathways catalyzed by xanthine oxidase. | |||
[[File:Xanthine oxidase pathways.jpg|frameless|A diagram illustrating many of the pathways catalyzed by xanthine oxidase.]] | |||
==Protein structure== | ==Protein structure== | ||
The protein is large, having a [[molecular weight]] of 270 | The protein is large, having a [[molecular weight]] of 270 kDa, and has 2 [[Flavin group|flavin]] molecules (bound as FAD), 2 [[molybdenum]] atoms, and 8 [[iron]] atoms bound per enzymatic unit. The molybdenum atoms are contained as [[molybdopterin]] cofactors and are the active sites of the enzyme. The iron atoms are part of [2Fe-2S] [[ferredoxin]] [[iron-sulfur cluster]]s and participate in electron transfer reactions. | ||
== | ==Catalytic mechanism== | ||
The active site of XO is composed of a molybdopterin unit with the molybdenum atom also coordinated by terminal oxygen ([[oxo]]), sulfur atoms and a terminal [[hydroxide]]. In the reaction with xanthine to form uric acid, an oxygen atom is transferred from molybdenum to xanthine. The reformation of the active molybdenum center occurs by the addition of water. Like other known molybdenum-containing oxidoreductases, the oxygen atom introduced to the [[Substrate (biochemistry)|substrate]] by XO originates from water rather than from [[dioxygen]] (O<sub>2</sub>). | The active site of XO is composed of a molybdopterin unit with the molybdenum atom also coordinated by terminal oxygen ([[oxo ligand|oxo]]), sulfur atoms and a terminal [[hydroxide]].<ref>{{cite journal | vauthors = Hille R | title = Structure and Function of Xanthine Oxidoreductase | journal = [[European Journal of Inorganic Chemistry]] | year = 2006 | volume = 2006= | issue = 10 | pages = 1905–2095 | doi = 10.1002/ejic.200600087 }}</ref> In the reaction with xanthine to form uric acid, an oxygen atom is transferred from molybdenum to xanthine, whereby several intermediates are assumed to be involved.<ref>{{cite journal | vauthors = Metz S, Thiel W | title = A combined QM/MM study on the reductive half-reaction of xanthine oxidase: substrate orientation and mechanism | journal = Journal of the American Chemical Society | volume = 131 | issue = 41 | pages = 14885–902 | date = October 2009 | pmid = 19788181 | doi = 10.1021/ja9045394 }}</ref> The reformation of the active molybdenum center occurs by the addition of water. Like other known molybdenum-containing oxidoreductases, the oxygen atom introduced to the [[Substrate (biochemistry)|substrate]] by XO originates from water rather than from [[dioxygen]] (O<sub>2</sub>). | ||
== | ==Clinical significance== | ||
Xanthine oxidase is a [[superoxide]]-producing enzyme found normally in [[Serum (blood)|serum]] and the lungs, and its activity is increased during [[Influenza A virus|influenza A]] infection.<ref>http://www.colorado.edu/intphys/iphy3700/vitCHemila92.pdf</ref> | |||
During severe liver damage, xanthine oxidase is released into the blood, so a blood assay for XO is a way to determine if [[liver]] damage has happened.<ref>{{cite journal|last1=Battelli|first1=Maria Giulia|last2=Musiani|first2=Silvia|last3=Valgimigli|first3=Marco|last4=Gramantieri|first4=Laura|last5=Tomassoni|first5=Federica|last6=Bolondi|first6=Luigi|last7=Stirpe|first7=Fiorenzo|title=Serum xanthine oxidase in human liver disease|journal=The American Journal of Gastroenterology|date=April 2001|volume=96|issue=4|pages=1194–1199|doi=10.1111/j.1572-0241.2001.03700.x}}</ref> | |||
Because xanthine oxidase is a [[metabolic pathway]] for [[uric acid]] formation, the xanthine oxidase inhibitor [[allopurinol]] is used in the treatment of [[gout]]. Since xanthine oxidase is involved in the metabolism of [[6-mercaptopurine]], caution should be taken before administering allopurinol and 6-mercaptopurine, or its prodrug [[azathioprine]], in conjunction. | |||
[[Xanthinuria]] is a rare [[genetic disorder]] where the lack of xanthine oxidase leads to high concentration of xanthine in blood and can cause health problems such as [[renal failure]]. There is no specific treatment, sufferers are advised by doctors to avoid foods high in [[purine]] and to maintain a high fluid intake. Type I xanthinuria has been traced directly to mutations of the ''XDH'' gene which mediates xanthine oxidase activity. Type II xanthinuria may result from a failure of the mechanism which inserts sulfur into the active sites of xanthine oxidase and [[aldehyde oxidase]], a related enzyme with some overlapping activities (such as conversion of [[allopurinol]] to [[oxypurinol]]).<ref>{{cite web|url=http://www.omim.org/entry/603592?search=type%20II%20xanthinuria&highlight=ii%20xanthinuria%20type |title=OMIM: Xanthinuria type II |deadurl=yes }}</ref> | |||
== | Inhibition of xanthine oxidase has been proposed as a mechanism for improving cardiovascular health.<ref name="pmid21894646">{{cite journal | vauthors = Dawson J, Walters M | title = Uric acid and xanthine oxidase: future therapeutic targets in the prevention of cardiovascular disease? | journal = British Journal of Clinical Pharmacology | volume = 62 | issue = 6 | pages = 633–44 | date = December 2006 | pmid = 21894646 | pmc = 1885190 | doi = 10.1111/j.1365-2125.2006.02785.x }}</ref> A study found that patients with chronic obstructive pulmonary disease ([[COPD]]) had a decrease in oxidative stress, including glutathione oxidation and lipid peroxidation, when xanthine oxidase was inhibited using allopurinol.<ref name="LM Heunks et al.">{{cite journal | vauthors = Heunks LM, Viña J, van Herwaarden CL, Folgering HT, Gimeno A, Dekhuijzen PN | title = Xanthine oxidase is involved in exercise-induced oxidative stress in chronic obstructive pulmonary disease | journal = The American Journal of Physiology | volume = 277 | issue = 6 Pt 2 | pages = R1697–704 | date = December 1999 | pmid = 10600916 }}</ref> Oxidative stress can be caused by hydroxyl free radicals and hydrogen peroxide, both of which are byproducts of XO activity.<ref name="P. Higgins et al.">{{cite journal | vauthors = Higgins P, Dawson J, Walters M | title = The potential for xanthine oxidase inhibition in the prevention and treatment of cardiovascular and cerebrovascular disease | journal = Cardiovascular Psychiatry and Neurology | volume = 2009 | pages = 282059 | year = 2009 | pmid = 20029618 | doi = 10.1155/2009/282059 | pmc=2790135}}</ref> | ||
Increased concentration of serum uric acid has been under research as an indicator for cardiovascular health factors, and has been used to strongly predict mortality, heart transplant, and more in patients.<ref name=pmid21894646 /> But it is not clear whether this could be a direct or casual association or link between serum uric acid concentration (and by proxy, xanthine oxidase activity) and cardiovascular health.<ref name="J. Dawson et al">{{cite journal | vauthors = Dawson J, Quinn T, Walters M | title = Uric acid reduction: a new paradigm in the management of cardiovascular risk? | journal = Current Medicinal Chemistry | volume = 14 | issue = 17 | pages = 1879–86 | year = 2007 | pmid = 17627523 | doi = 10.2174/092986707781058797 }}</ref> States of high cell turnover and alcohol ingestion are some of the most prominent cases of high serum uric acid concentrations.<ref name="P. Higgins et al." /> | |||
Reactive nitrogen species, such as peroxynitrite that xanthine oxidase can form, have been found to react with DNA, proteins, and cells, causing cellular damage or even toxicity. Reactive nitrogen signaling, coupled with reactive oxygen species, have been found to be a central part of myocardial and vascular function, explaining why xanthine oxidase is being researched for links to cardiovascular health.<ref name="JM Zimmet & JM Hare">{{cite journal | vauthors = Zimmet JM, Hare JM | title = Nitroso-redox interactions in the cardiovascular system | journal = Circulation | volume = 114 | issue = 14 | pages = 1531–44 | date = October 2006 | pmid = 17015805 | doi = 10.1161/CIRCULATIONAHA.105.605519 }}</ref> | |||
Both xanthine oxidase and xanthine [[oxidoreductase]] are also present in [[cornea]]l epithelium and endothelium and may be involved in oxidative eye injury.<ref name="pmid12168784">{{cite journal | vauthors = Cejková J, Ardan T, Filipec M, Midelfart A | title = Xanthine oxidoreductase and xanthine oxidase in human cornea | journal = Histology and Histopathology | volume = 17 | issue = 3 | pages = 755–60 | year = 2002 | pmid = 12168784 | doi = | url = http://www.hh.um.es/Abstracts/Vol_17/17_3/17_3_755.htm }}</ref> | |||
| | |||
== | ==Inhibitors== | ||
{{main|Xanthine oxidase inhibitor}} | |||
Inhibitors of XO include [[allopurinol]],<ref name="pmid16507884">{{cite journal | vauthors = Pacher P, Nivorozhkin A, Szabó C | title = Therapeutic effects of xanthine oxidase inhibitors: renaissance half a century after the discovery of allopurinol | journal = Pharmacological Reviews | volume = 58 | issue = 1 | pages = 87–114 | date = March 2006 | pmid = 16507884 | pmc = 2233605 | doi = 10.1124/pr.58.1.6 }}</ref> [[oxypurinol]],<ref name="pmid2829916">{{cite journal | vauthors = Spector T | title = Oxypurinol as an inhibitor of xanthine oxidase-catalyzed production of superoxide radical | journal = Biochemical Pharmacology | volume = 37 | issue = 2 | pages = 349–52 | date = Jan 1988 | pmid = 2829916 | doi = 10.1016/0006-2952(88)90739-3 }}</ref> and [[phytic acid]].<ref name="pmid14738912">{{cite journal | vauthors = Muraoka S, Miura T | title = Inhibition of xanthine oxidase by phytic acid and its antioxidative action | journal = Life Sciences | volume = 74 | issue = 13 | pages = 1691–700 | date = February 2004 | pmid = 14738912 | doi = 10.1016/j.lfs.2003.09.040 }}</ref> It has also been found to be inhibited by [[flavonoids]],<ref name="Cos et al.">{{cite journal | vauthors = Cos P, Ying L, Calomme M, Hu JP, Cimanga K, Van Poel B, Pieters L, Vlietinck AJ, Vanden Berghe D | title = Structure-activity relationship and classification of flavonoids as inhibitors of xanthine oxidase and superoxide scavengers | journal = Journal of Natural Products | volume = 61 | issue = 1 | pages = 71–6 | date = Jan 1998 | pmid = 9461655 | doi = 10.1021/np970237h }}</ref> including those found in Bougainvillea spectabilis Willd ([[Nyctaginaceae]]) leaves (with an [[IC50]] of 7.23 μM), typically used in [[folk medicine]].<ref name="Chang, WS">{{cite journal | vauthors = Chang WS, Lee YJ, Lu FJ, Chiang HC | title = Inhibitory effects of flavonoids on xanthine oxidase | journal = Anticancer Research | volume = 13 | issue = 6A | pages = 2165–70 | date = Nov–Dec 1993 | pmid = 8297130 }}</ref> | |||
[[ | == See also == | ||
* [[xanthine dehydrogenase]] | |||
* [[sodium molybdate]] | |||
== References == | |||
{{reflist|33em}} | |||
== External links == | |||
{{ | * {{MeshName|Xanthine+Oxidase}} | ||
{{Nucleotide metabolism}} | |||
{{Other oxidoreductases}} | {{Other oxidoreductases}} | ||
{{ | {{Enzymes}} | ||
[[ | {{Portal bar|Molecular and Cellular Biology|border=no}} | ||
[[ | {{Use dmy dates|date=April 2017}} | ||
[[ | |||
[[Category:EC 1.17.3]] | |||
[[Category:Metalloproteins]] | |||
[[Category:Molybdenum enzymes]] | |||
[[Category:Iron-sulfur enzymes]] | |||
[[Category:Superoxide generating enzymes]] | |||
Revision as of 15:11, 6 December 2017
| xanthine oxidase/dehydrogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
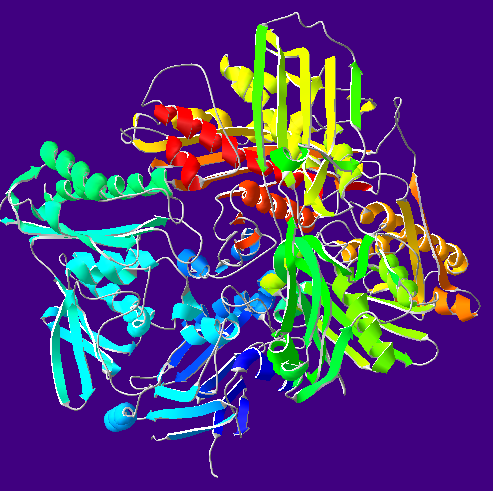 Crystallographic structure (monomer) of bovine xanthine oxidase.[1] The bounded FAD (red), FeS-cluster (orange), the molybdopterin cofactor with molybdenum (yellow) and salicylate (blue) are indicated. | |||||||||
| Identifiers | |||||||||
| EC number | 1.17.3.2 | ||||||||
| CAS number | 9002-17-9 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| xanthine oxidase/dehydrogenase | |
|---|---|
| Identifiers | |
| Symbol | XDH |
| Entrez | 7498 |
| HUGO | 12805 |
| OMIM | 607633 |
| PDB | 1FIQ |
| RefSeq | NM_000379 |
| UniProt | P47989 |
| Other data | |
| EC number | 1.17.3.2 |
| Locus | Chr. 2 p23.1 |
Xanthine oxidase (XO, sometimes 'XAO') is a form of xanthine oxidoreductase, a type of enzyme that generates reactive oxygen species.[2] These enzymes catalyze the oxidation of hypoxanthine to xanthine and can further catalyze the oxidation of xanthine to uric acid. These enzymes play an important role in the catabolism of purines in some species, including humans.[3][4]
Xanthine oxidase is defined as an enzyme activity (EC 1.17.3.2).[5] The same protein, which in humans has the HGNC approved gene symbol XDH, can also have xanthine dehydrogenase activity (EC 1.17.1.4).[6] Most of the protein in the liver exists in a form with xanthine dehydrogenase activity, but it can be converted to xanthine oxidase by reversible sulfhydryl oxidation or by irreversible proteolytic modification.[7][8]
Reaction
The following chemical reactions are catalyzed by xanthine oxidase:
- hypoxanthine + H2O + O2 <math>\rightleftharpoons</math> xanthine + H2O2
- xanthine + H2O + O2 <math>\rightleftharpoons</math> uric acid + H2O2
- Xanthine oxidase can also act on certain other purines, pterins, and aldehydes. For example, it efficiently converts 1-methylxanthine (a metabolite of caffeine) to 1-methyluric acid, but has little activity on 3-methylxanthine.[9]
- Under some circumstances it can produce superoxide ion RH + H2O + 2 O2 <math>\rightleftharpoons</math> ROH + 2 O2− + 2 H+.[6]
-
hypoxanthine (one oxygen atom)
-
xanthine (two oxygens)
-
uric acid (three oxygens)
Other reactions
Because XO is a superoxide-producing enzyme, with general low specificity,[10] it can be combined with other compounds and enzymes and create reactive oxidants, as well as oxidize other substrates.
Bovine xanthine oxidase (from milk) was originally thought to have a binding site to reduce cytochrome c with, but it has been found that the mechanism to reduce this protein is through XO's superoxide anion byproduct, with competitive inhibition by carbonic anhydrase.[11]
Another reaction catalyzed by xanthine oxidase is the decomposition of S-Nitrosothiols (RSNO), a reactive nitrogen species, to nitric oxide (NO), which reacts with a superoxide anion to form peroxynitrite under aerobic conditions.[12]
XO has also been found to produce the strong one-electron oxidant carbonate radical anion from oxidation with acetaldehyde in the presence of catalase and bicarbonate. It was suggested that the carbonate radical was likely produced in one of the enzyme's redox centers with a peroxymonocarbonate intermediate.[10]
Here is a diagram highlighting the pathways catalyzed by xanthine oxidase.
A diagram illustrating many of the pathways catalyzed by xanthine oxidase.
Protein structure
The protein is large, having a molecular weight of 270 kDa, and has 2 flavin molecules (bound as FAD), 2 molybdenum atoms, and 8 iron atoms bound per enzymatic unit. The molybdenum atoms are contained as molybdopterin cofactors and are the active sites of the enzyme. The iron atoms are part of [2Fe-2S] ferredoxin iron-sulfur clusters and participate in electron transfer reactions.
Catalytic mechanism
The active site of XO is composed of a molybdopterin unit with the molybdenum atom also coordinated by terminal oxygen (oxo), sulfur atoms and a terminal hydroxide.[13] In the reaction with xanthine to form uric acid, an oxygen atom is transferred from molybdenum to xanthine, whereby several intermediates are assumed to be involved.[14] The reformation of the active molybdenum center occurs by the addition of water. Like other known molybdenum-containing oxidoreductases, the oxygen atom introduced to the substrate by XO originates from water rather than from dioxygen (O2).
Clinical significance
Xanthine oxidase is a superoxide-producing enzyme found normally in serum and the lungs, and its activity is increased during influenza A infection.[15]
During severe liver damage, xanthine oxidase is released into the blood, so a blood assay for XO is a way to determine if liver damage has happened.[16]
Because xanthine oxidase is a metabolic pathway for uric acid formation, the xanthine oxidase inhibitor allopurinol is used in the treatment of gout. Since xanthine oxidase is involved in the metabolism of 6-mercaptopurine, caution should be taken before administering allopurinol and 6-mercaptopurine, or its prodrug azathioprine, in conjunction.
Xanthinuria is a rare genetic disorder where the lack of xanthine oxidase leads to high concentration of xanthine in blood and can cause health problems such as renal failure. There is no specific treatment, sufferers are advised by doctors to avoid foods high in purine and to maintain a high fluid intake. Type I xanthinuria has been traced directly to mutations of the XDH gene which mediates xanthine oxidase activity. Type II xanthinuria may result from a failure of the mechanism which inserts sulfur into the active sites of xanthine oxidase and aldehyde oxidase, a related enzyme with some overlapping activities (such as conversion of allopurinol to oxypurinol).[17]
Inhibition of xanthine oxidase has been proposed as a mechanism for improving cardiovascular health.[18] A study found that patients with chronic obstructive pulmonary disease (COPD) had a decrease in oxidative stress, including glutathione oxidation and lipid peroxidation, when xanthine oxidase was inhibited using allopurinol.[19] Oxidative stress can be caused by hydroxyl free radicals and hydrogen peroxide, both of which are byproducts of XO activity.[20]
Increased concentration of serum uric acid has been under research as an indicator for cardiovascular health factors, and has been used to strongly predict mortality, heart transplant, and more in patients.[18] But it is not clear whether this could be a direct or casual association or link between serum uric acid concentration (and by proxy, xanthine oxidase activity) and cardiovascular health.[21] States of high cell turnover and alcohol ingestion are some of the most prominent cases of high serum uric acid concentrations.[20]
Reactive nitrogen species, such as peroxynitrite that xanthine oxidase can form, have been found to react with DNA, proteins, and cells, causing cellular damage or even toxicity. Reactive nitrogen signaling, coupled with reactive oxygen species, have been found to be a central part of myocardial and vascular function, explaining why xanthine oxidase is being researched for links to cardiovascular health.[22]
Both xanthine oxidase and xanthine oxidoreductase are also present in corneal epithelium and endothelium and may be involved in oxidative eye injury.[23]
Inhibitors
Inhibitors of XO include allopurinol,[24] oxypurinol,[25] and phytic acid.[26] It has also been found to be inhibited by flavonoids,[27] including those found in Bougainvillea spectabilis Willd (Nyctaginaceae) leaves (with an IC50 of 7.23 μM), typically used in folk medicine.[28]
See also
References
- ↑ PDB: 1FIQ; Enroth C, Eger BT, Okamoto K, Nishino T, Nishino T, Pai EF (September 2000). "Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion". Proceedings of the National Academy of Sciences of the United States of America. 97 (20): 10723–8. doi:10.1073/pnas.97.20.10723. PMC 27090. PMID 11005854.
- ↑ Ardan T, Kovaceva J, Cejková J (February 2004). "Comparative histochemical and immunohistochemical study on xanthine oxidoreductase/xanthine oxidase in mammalian corneal epithelium". Acta Histochemica. 106 (1): 69–75. doi:10.1016/j.acthis.2003.08.001. PMID 15032331.
- ↑ Hille R (Jan 2005). "Molybdenum-containing hydroxylases". Archives of Biochemistry and Biophysics. 433 (1): 107–16. doi:10.1016/j.abb.2004.08.012. PMID 15581570.
- ↑ Harrison R (September 2002). "Structure and function of xanthine oxidoreductase: where are we now?". Free Radical Biology & Medicine. 33 (6): 774–97. doi:10.1016/S0891-5849(02)00956-5. PMID 12208366.
- ↑ "KEGG record for EC 1.17.3.2".
- ↑ 6.0 6.1 "KEGG record for EC 1.17.1.4".
- ↑ "Entrez Gene: XDH xanthine dehydrogenase".
- ↑ "*607633 XANTHINE DEHYDROGENASE; XDH".
- ↑ Birkett DJ, Miners JO, Valente L, Lillywhite KJ, Day RO (February 1997). "1-Methylxanthine derived from caffeine as a pharmacodynamic probe of oxypurinol effect" (PDF). British Journal of Clinical Pharmacology. 43 (2): 197–200. doi:10.1046/j.1365-2125.1997.53711.x. PMC 2042732. PMID 9131954.
- ↑ 10.0 10.1 Bonini MG, Miyamoto S, Di Mascio P, Augusto O (December 2004). "Production of the carbonate radical anion during xanthine oxidase turnover in the presence of bicarbonate". The Journal of Biological Chemistry. 279 (50): 51836–43. doi:10.1074/jbc.M406929200. PMID 15448145.
- ↑ McCord JM, Fridovich I (November 1968). "The reduction of cytochrome c by milk xanthine oxidase". The Journal of Biological Chemistry. 243 (21): 5753–60. PMID 4972775.
- ↑ Trujillo, M. (3 April 1998). "Xanthine Oxidase-mediated Decomposition of S-Nitrosothiols". Journal of Biological Chemistry. 273 (14): 7828–7834. doi:10.1074/jbc.273.14.7828.
- ↑ Hille R (2006). "Structure and Function of Xanthine Oxidoreductase". European Journal of Inorganic Chemistry. 2006= (10): 1905–2095. doi:10.1002/ejic.200600087.
- ↑ Metz S, Thiel W (October 2009). "A combined QM/MM study on the reductive half-reaction of xanthine oxidase: substrate orientation and mechanism". Journal of the American Chemical Society. 131 (41): 14885–902. doi:10.1021/ja9045394. PMID 19788181.
- ↑ http://www.colorado.edu/intphys/iphy3700/vitCHemila92.pdf
- ↑ Battelli, Maria Giulia; Musiani, Silvia; Valgimigli, Marco; Gramantieri, Laura; Tomassoni, Federica; Bolondi, Luigi; Stirpe, Fiorenzo (April 2001). "Serum xanthine oxidase in human liver disease". The American Journal of Gastroenterology. 96 (4): 1194–1199. doi:10.1111/j.1572-0241.2001.03700.x.
- ↑ "OMIM: Xanthinuria type II".
- ↑ 18.0 18.1 Dawson J, Walters M (December 2006). "Uric acid and xanthine oxidase: future therapeutic targets in the prevention of cardiovascular disease?". British Journal of Clinical Pharmacology. 62 (6): 633–44. doi:10.1111/j.1365-2125.2006.02785.x. PMC 1885190. PMID 21894646.
- ↑ Heunks LM, Viña J, van Herwaarden CL, Folgering HT, Gimeno A, Dekhuijzen PN (December 1999). "Xanthine oxidase is involved in exercise-induced oxidative stress in chronic obstructive pulmonary disease". The American Journal of Physiology. 277 (6 Pt 2): R1697–704. PMID 10600916.
- ↑ 20.0 20.1 Higgins P, Dawson J, Walters M (2009). "The potential for xanthine oxidase inhibition in the prevention and treatment of cardiovascular and cerebrovascular disease". Cardiovascular Psychiatry and Neurology. 2009: 282059. doi:10.1155/2009/282059. PMC 2790135. PMID 20029618.
- ↑ Dawson J, Quinn T, Walters M (2007). "Uric acid reduction: a new paradigm in the management of cardiovascular risk?". Current Medicinal Chemistry. 14 (17): 1879–86. doi:10.2174/092986707781058797. PMID 17627523.
- ↑ Zimmet JM, Hare JM (October 2006). "Nitroso-redox interactions in the cardiovascular system". Circulation. 114 (14): 1531–44. doi:10.1161/CIRCULATIONAHA.105.605519. PMID 17015805.
- ↑ Cejková J, Ardan T, Filipec M, Midelfart A (2002). "Xanthine oxidoreductase and xanthine oxidase in human cornea". Histology and Histopathology. 17 (3): 755–60. PMID 12168784.
- ↑ Pacher P, Nivorozhkin A, Szabó C (March 2006). "Therapeutic effects of xanthine oxidase inhibitors: renaissance half a century after the discovery of allopurinol". Pharmacological Reviews. 58 (1): 87–114. doi:10.1124/pr.58.1.6. PMC 2233605. PMID 16507884.
- ↑ Spector T (Jan 1988). "Oxypurinol as an inhibitor of xanthine oxidase-catalyzed production of superoxide radical". Biochemical Pharmacology. 37 (2): 349–52. doi:10.1016/0006-2952(88)90739-3. PMID 2829916.
- ↑ Muraoka S, Miura T (February 2004). "Inhibition of xanthine oxidase by phytic acid and its antioxidative action". Life Sciences. 74 (13): 1691–700. doi:10.1016/j.lfs.2003.09.040. PMID 14738912.
- ↑ Cos P, Ying L, Calomme M, Hu JP, Cimanga K, Van Poel B, Pieters L, Vlietinck AJ, Vanden Berghe D (Jan 1998). "Structure-activity relationship and classification of flavonoids as inhibitors of xanthine oxidase and superoxide scavengers". Journal of Natural Products. 61 (1): 71–6. doi:10.1021/np970237h. PMID 9461655.
- ↑ Chang WS, Lee YJ, Lu FJ, Chiang HC (Nov–Dec 1993). "Inhibitory effects of flavonoids on xanthine oxidase". Anticancer Research. 13 (6A): 2165–70. PMID 8297130.
External links
- Xanthine+Oxidase at the US National Library of Medicine Medical Subject Headings (MeSH)