HLA-DQ: Difference between revisions
m (Robot: Automated text replacement (-{{SIB}} + & -{{EH}} + & -{{EJ}} + & -{{Editor Help}} + & -{{Editor Join}} +)) |
(→Effects of heterogeneity of isoform pairing: deminished typo-itude) |
||
| Line 1: | Line 1: | ||
{| | {{heteropolypeptide | ||
| | | heteropolymer = [[MHC class II]], DQ | ||
| | | polymer_type = heterodimer | ||
| protein_type = [[Transmembrane receptor|cell surface receptor]] | |||
| | | function = Immune recognition and<br> antigen presentation | ||
| | | image = DQ_Illustration.PNG | ||
| image_source = DQ1 binding pocket with ligand | |||
| SubunitCount = 2 | |||
| | | subunit1 = α | ||
| gene1 = [[HLA-DQA1]] | |||
| locus1 = [[Chromosome 6]]p21.31 | |||
| | | subunit2 = β | ||
| | | gene2 = [[HLA-DQB1]] | ||
| | | locus2 = [[Chromosome 6]]p21.31 | ||
}} | |||
'''HLA-DQ''' ('''DQ''') is a cell surface receptor [[protein]] found on [[antigen presenting cell]]s. It is an αβ [[protein dimer|heterodimer]] of type [[MHC class II]]. The α and β [[peptide|chains]] are encoded by two [[locus (genetics)|loci]], [[HLA-DQA1]] and [[HLA-DQB1]], that are adjacent to each other on [[chromosome 6 (human)|chromosome band 6p21.3]]. Both α-chain and β-chain vary greatly. A person often produces two α-chain and two β-chain [[wikt:variant|variants]] and thus 4 [[wikt:isoform|isoforms]] of DQ. The DQ loci are in close [[genetic linkage]] to [[HLA-DR]], and less closely linked to [[HLA-DP]], [[HLA-A]], [[HLA-B]] and [[HLA-C]]. | |||
| | |||
| | |||
| | |||
| | |||
Different isoforms of DQ can bind to and present different antigens to [[T-cell]]s. In this process T-cells are stimulated to grow and can signal [[B-cell]]s to produce [[antibodies]]. DQ functions in recognizing and presenting foreign antigens (proteins derived from potential [[wikt:pathogen|pathogens]]). But DQ is also involved in recognizing common [[autoantigen|self-antigen]]s and presenting those antigens to the immune system in order to develop [[immune tolerance|tolerance]] from a very young age. | |||
When tolerance to self proteins is lost, DQ may become involved in [[autoimmune disease]]. Two autoimmune diseases in which HLA-DQ is involved are [[coeliac disease]] and [[diabetes mellitus type 1]]. DQ is one of several antigens involved in [[transplant rejection|rejection of organ transplants]]. As a variable cell surface receptor on [[immune cell]]s, these D antigens, originally [[Human leukocyte antigen#Lymphocyte bearing antigens recognized|HL-A4]] antigens, are involved in [[graft versus host disease]] when [[lymphoid tissue]]s are transplanted between people. [[Serology|Serological studies]] of DQ recognized that antibodies to DQ bind primarily to the β-chain. The currently used serotypes are [[HLA-DQ2]], -[[HLA-DQ3|DQ3]], -[[HLA-DQ4|DQ4]], -[[HLA-DQ5|DQ5]], -[[HLA-DQ6|DQ6]], -[[HLA-DQ7|DQ7]], -[[HLA-DQ8|DQ8]], -[[HLA-DQ9|DQ9]]. [[HLA-DQ1]] is a weak reaction to the α-chain and was replaced by DQ5 and DQ6 serology. Serotyping is capable of identifying most aspects of DQ isoform structure and function, however sequence specific [[PCR]] is now the preferred method of determining [[HLA-DQA1#Alleles|HLA-DQA1]] and [[HLA-DQB1#Alleles|HLA-DQB1 alleles]], as serotyping cannot resolve, often, the critical contribution of the DQ α-chain. This can be compensated for by examining DR serotypes as well as DQ serotypes. | |||
HLA-DQ | |||
==Structure, Functions, Genetics== | |||
[[File:TCELL-APC DQ.png|thumb|left|HLA DQ Receptor with bound peptide and TCR]] | |||
===Function=== | ===Function=== | ||
The name 'HLA DQ' originally describes a transplantation antigen of MHC class II category of the [[major histocompatibility complex]] of humans; however, this status is an artifact of the early era of organ transplantation. | The name 'HLA DQ' originally describes a transplantation antigen of MHC class II category of the [[major histocompatibility complex]] of humans; however, this status is an artifact of the early era of organ transplantation. | ||
HLA DQ functions as a cell surface receptor for foreign or self antigens. The immune system surveys antigens for foreign pathogens when presented by MHC receptors (like HLA DQ). The MHC Class II antigens are found on [[antigen presenting cells]] (APC) (macrophages, dendritic cells, and B-lymphocytes). Normally, these APC 'present' class II receptor/antigens to a great many T-cells, each with unique T-cell receptor (TCR) variants. A few TCR variants that recognize these DQ/antigen complexes are on CD4 positive (CD4+) T-cells. These T-cells, called T-helper cells, can promote the amplification of B-cells which, in turn recognize a different portion of the same antigen. Alternatively, macrophages and other megalocytes consume cells by apoptotic signaling and present self-antigens. Self antigens, in the right context, form a suppressor T-cell population that protects self tissues from immune attack or autoimmunity. | HLA DQ functions as a cell surface receptor for foreign or self antigens. The immune system surveys antigens for foreign pathogens when presented by MHC receptors (like HLA DQ). The MHC Class II antigens are found on [[antigen presenting cells]] (APC) (macrophages, dendritic cells, and B-lymphocytes). Normally, these APC 'present' class II receptor/antigens to a great many T-cells, each with unique T-cell receptor (TCR) variants. A few TCR variants that recognize these DQ/antigen complexes are on CD4 positive (CD4+) T-cells. These T-cells, called T-helper cells, can promote the amplification of B-cells which, in turn recognize a different portion of the same antigen. Alternatively, macrophages and other megalocytes consume cells by apoptotic signaling and present self-antigens. Self antigens, in the right context, form a suppressor T-cell population that protects self tissues from immune attack or autoimmunity. | ||
===Genetics=== | ===Genetics=== | ||
[[ | [[File:HLA.svg|right|thumb|150px|HLA region of Chromosome 6]] | ||
HLA-DQ (DQ) is encoded on the [[Human leukocyte antigen|HLA]] region of chromosome 6, in what was classically known as the "D" antigen region. This region encoded the subunits for DP,-Q and -R which are the major [[MHC class II]] antigens in humans. Each of these proteins have slightly different functions and are regulated in slightly different ways. | HLA-DQ (DQ) is encoded on the [[Human leukocyte antigen|HLA]] region of [[chromosome 6]]p21.3, in what was classically known as the "D" antigen region. This region encoded the subunits for DP,-Q and -R which are the major [[MHC class II]] antigens in humans. Each of these proteins have slightly different functions and are regulated in slightly different ways. | ||
DQ is made up of two different subunits to form an αβ-heterodimer. | DQ is made up of two different subunits to form an αβ-heterodimer. Each subunit is encoded by its own "gene" (a coding locus). The | ||
DQ α subunit is encoded by the [[HLA-DQA1]] gene and the DQ β subunit is encoded by the [[HLA-DQB1]] gene. Both loci are variable in the human population (see [[#Regional evolution|regional evolution]]). | |||
====Detecting DQ isoforms==== | ====Detecting DQ isoforms==== | ||
In the human population DQ is highly variable, the β subunit | In the human population DQ is highly variable, the β subunit more so than the alpha chain. The variants are encoded by the HLA DQ genes | ||
and are the result of [[single nucleotide polymorphism]]s (SNP). Some | and are the result of [[single nucleotide polymorphism]]s (SNP). Some | ||
SNP result in no change in amino-acid sequence. Others result in changes in regions that are removed when the proteins is processed to the cell surface, still others result in change in the non-functional regions of the protein, and some changes result in a change of function of the DQ isoform that is produced. The isoforms generally | SNP result in no change in amino-acid sequence. Others result in changes in regions that are removed when the proteins is processed to the cell surface, still others result in change in the non-functional regions of the protein, and some changes result in a change of function of the DQ isoform that is produced. The isoforms generally | ||
change in the peptides they bind and present to T-cells. Much of | change in the peptides they bind and present to T-cells. Much of | ||
the isoform variation in DQ is within these 'functional' regions. | the isoform variation in DQ is within these 'functional' regions. | ||
''' | '''Serotyping'''. [[Antibodies]] raised against DQ tend to recognize these functional regions, in most cases the β-subunit. As a result, these antibodies can discriminate different classes of DQ based on the recognition similar DQβ proteins known as [[serotype]]s. | ||
An example of a serotype is '''DQ2'''. | An example of a serotype is '''DQ2'''. | ||
*Recognize HLA-DQB1*02 gene products which include gene products of the following alleles: | *Recognize HLA-DQB1*02 gene products which include gene products of the following alleles: | ||
** HLA-DQB1* | ** HLA-DQB1*02:01 | ||
** HLA-DQB1* | ** HLA-DQB1*02:02 | ||
** HLA-DQB1* | ** HLA-DQB1*02:03 | ||
Sometimes '''DQ2''' antibodies recognize other gene products, such as DQB1* | Sometimes '''DQ2''' antibodies recognize other gene products, such as DQB1*03:03, resulting in serotyping errors. Because of this mistyping serotyping is not as reliable as gene sequencing | ||
serotyping errors. Because of this mistyping serotyping is not as reliable as gene sequencing | or SSP-PCR. | ||
or SSP-PCR. | |||
While the DQ2 isoforms are recognized by the same antibodies, and all | While the DQ2 isoforms are recognized by the same antibodies, and all | ||
DQB1*02 are functionally similar, they can bind different α subunit and these αβ isoform variants can bind different sets of peptides. This difference in binding is an important feature that helps to understand autoimmune disease. | DQB1*02 are functionally similar, they can bind different α subunit and these αβ isoform variants can bind different sets of peptides. This difference in binding is an important feature that helps to understand autoimmune disease. | ||
The first identified DQ were DQw1 to DQw3. DQw1 (DQ1) recognized the alpha chain of DQA1*01 alleles. This group was later split by beta chain recognition to DQ5 and DQ6. DQ3 is known as broad antigen serotypes, because they recognize a broad group of antigens. However, because of this broad antigen recognition their specificity and usefulness is somewhat less than desirable. | |||
For most modern typing the DQ2, DQ4 - DQ9 set is used. | |||
{| border="0" cellspacing="0" cellpadding="0" align=" | {| border="0" cellspacing="0" cellpadding="0" align="left" style="text-align:center; background:#ffffff; margin-right: 2em; border:2px #e0e0ff solid;" | ||
|+ | |+ DQA1-B1 haplotypes in Caucasian Americans | ||
|- style = "background:#f0f0ff" | |- style = "background:#f0f0ff" | ||
! style = "background:#e8e8f8" |< | ! style = "background:#e8e8f8" |<span style="color:purple;">DQ</span> | ||
! < | ! <span style="color:purple;">DQ</span> | ||
! colspan = "3" style = "background:#f7e7ff" |<span style="color:purple;">DQ</span> | |||
! colspan = " | |||
! colspan = "2" | Freq | ! colspan = "2" | Freq | ||
| width = "15" | || width = "5" | | | width = "15" | || width = "5" | | ||
|- style = "background:#f0f0ff" | |- style = "background:#f0f0ff" | ||
! style = "background:#e8e8f8; font-size:80%" | Serotype | ! style = "background:#e8e8f8; font-size:80%" | Serotype | ||
| width = "80" | | | width = "80" | cis-isoform | ||
| width = "30" style = "background:#f7e7ff" | <span style="color:green;">Subtype</span> | |||
! width = "45" style = "background:#f7e7ff" | < | ! width = "45" style = "background:#f7e7ff" | <span style="color:#bb00bb;">A1</span> | ||
! width = "45" style = "background:#f7e7ff" | < | ! width = "45" style = "background:#f7e7ff" | <span style="color:blue;">B1</span> | ||
| colspan = "2" | '''%'''<ref name=" | | colspan = "2" | '''%'''<ref name="pmid12974796">{{cite journal |vauthors=Klitz W, Maiers M, Spellman S, etal |title=New HLA haplotype frequency reference standards: high-resolution and large sample typing of HLA DR-DQ haplotypes in a sample of European Americans |journal=Tissue Antigens |volume=62 |issue=4 |pages=296–307 |date=October 2003 |pmid=12974796 |doi= 10.1034/j.1399-0039.2003.00103.x|url=http://www.blackwell-synergy.com/openurl?genre=article&sid=nlm:pubmed&issn=0001-2815&date=2003&volume=62&issue=4&spage=296}}</ref> | ||
| colspan ="2" | <sup>rank</sup> | | colspan ="2" | <sup>rank</sup> | ||
|- style = "background:#ffffdf" | |- style = "background:#ffffdf" | ||
| style = "background:#f7f7d7" rowspan = " | | style = "background:#f7f7d7" rowspan = "3" | [[HLA-DQ2|DQ2]] | ||
| < | | <span style="color:#bb00bb;">α<sup>5</sup>-<span style="color:blue;">β<sup>2</sup></span> || <span style="color:green;">2.5</span> | ||
! style="background:#fbfbfb" align=right | | <span style="color:#bb00bb;">05:01</span><sup>♣</sup> || <span style="color:blue;">02:01</span> | ||
! style = "background:#fbfbfb" align = left width = "15" | | ! style="background:#fbfbfb" align=right width = "30"| 13. | ||
| style = "background:#fbfbfb" align = right | | ! style = "background:#fbfbfb" align = left width = "15" | 16 | ||
| style = "background:#fbfbfb" align = right |<sup>2nd</sup> | |||
|- style = "background:#ffffdf" | |- style = "background:#ffffdf" | ||
| < | | <span style="color:#bb00bb;">α<sup>2</sup>-<span style="color:blue;">β<sup>2</sup></span> || <span style="color:green;">2.2</span> | ||
|<span style="color:#bb00bb;">02:01 </span> || <span style="color:blue;">02:02</span> | |||
! style = "background:#fbfbfb" align = right | 11. | ! style = "background:#fbfbfb" align = right | 11. | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = left | 08 | ||
| style = "background:#fbfbfb" align = right | | | style = "background:#fbfbfb" align = right |<sup>3rd</sup> | ||
|- style = "background:#ffffdf" | |||
| <span style="color:#bb00bb;">α<sup>3</sup>-<span style="color:blue;">β<sup>2</sup></span> || <span style="color:green;">2.3</span> | |||
| <span style="color:#bb00bb;">03:02</span><sup>♠</sup> || <span style="color:blue;">02:02</span> | |||
! style = "background:#fbfbfb" align = right | 0. | |||
! style = "background:#fbfbfb" align = left | 08 | |||
| style = "background:#fbfbfb" align = right | | |||
|- style = "background:#f7ffe7" | |||
| style = "background:#eff8df" rowspan = 3 | [[HLA-DQ4|DQ4]] | |||
| rowspan = 2 | <span style="color:#bb00bb;">α<sup>3</sup>-<span style="color:blue;">β<sup>4</sup></span> || rowspan = 2 |<span style="color:green;">4.3</span> | |||
| <span style="color:#bb00bb;">03:01 </span> || <span style="color:blue;">04:02</span> | |||
! style = "background:#fbfbfb" align = right | 0. | |||
! style = "background:#fbfbfb" align = left | 03 | |||
| style = "background:#fbfbfb" align = right | | |||
|- style = "background:#f7ffe7" | |||
| <span style="color:#bb00bb;">03:02</span><sup>♠</sup> || <span style="color:blue;">04:02</span> | |||
! style = "background:#fbfbfb" align = right | 0. | |||
! style = "background:#fbfbfb" align = left | 11 | |||
| style = "background:#fbfbfb" align = right | | |||
|- style = "background:#f7ffe7" | |- style = "background:#f7ffe7" | ||
| style = " | | <span style="color:#bb00bb;">α<sup>4</sup>-<span style="color:blue;">β<sup>4</sup></span> || <span style="color:green;">4.4</span> | ||
|<span style="color:#bb00bb;">04:01 </span> || <span style="color:blue;">04:02</span> | |||
! style = "background:#fbfbfb" align = right | 2. | ! style = "background:#fbfbfb" align = right | 2. | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = left | 26 | ||
| style = "background:#fbfbfb" align = right | | | style = "background:#fbfbfb" align = right | | ||
|- style = "background:#eef7ef" | |- style = "background:#eef7ef" | ||
| style = "background:#e8efe8" rowspan = " | | style = "background:#e8efe8" rowspan = "8" | [[HLA-DQ5|DQ5]] | ||
| rowspan = " | | rowspan = "4" | <span style="color:#bb00bb;">α<sup>1</sup>-<span style="color:blue;">β<sup>5.1</sup></span> | ||
| < | | rowspan = 4 | <span style="color:green;">5.1</span> | ||
! style = "background:#fbfbfb" align = right | | | <span style="color:#bb00bb;">01:01</span> || <span style="color:blue;">05:01</span> | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = right | 10. | ||
| style = "background:#fbfbfb" align = right | <sup> | ! style = "background:#fbfbfb" align = left | 85 | ||
| style = "background:#fbfbfb" align = right | <sup>5th</sup> | |||
|- style = "background:#eef8ee" | |- style = "background:#eef8ee" | ||
| < | | <span style="color:#bb00bb;">01:02</span> || <span style="color:blue;">05:01</span> | ||
! style = "background:#fbfbfb" align = right | | ! style = "background:#fbfbfb" align = right | 0. | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = left | 03 | ||
| style = "background:#fbfbfb" align = right | | | style = "background:#fbfbfb" align = right | | ||
|- style = "background:#eef8ee" | |- style = "background:#eef8ee" | ||
| < | | <span style="color:#bb00bb;">01:03</span> || <span style="color:blue;">05:01</span> | ||
! style = "background:#fbfbfb" align = right | 0. | ! style = "background:#fbfbfb" align = right | 0. | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = left | 03 | ||
| style = "background:#fbfbfb" align = right | | | style = "background:#fbfbfb" align = right | | ||
|- style = "background:#eef8ee" | |- style = "background:#eef8ee" | ||
| < | | <span style="color:#bb00bb;">01:04</span> || <span style="color:blue;">05:01</span> | ||
! style = "background:#fbfbfb" align = right | 0. | ! style = "background:#fbfbfb" align = right | 0. | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = left | 71 | ||
| style = "background:#fbfbfb" align = right | | | style = "background:#fbfbfb" align = right | | ||
|- style = "background:#eef7ee" | |- style = "background:#eef7ee" | ||
|< | |rowspan = 2 |<span style="color:#bb00bb;">α<sup>1</sup>-<span style="color:blue;">β<sup>5.2</sup></span> | ||
| rowspan = 2 |<span style="color:green;">5.2</span> | |||
| <span style="color:#bb00bb;">01:02</span> || <span style="color:blue;">05:02</span> | |||
! style = "background:#fbfbfb" align = right | 1. | ! style = "background:#fbfbfb" align = right | 1. | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = left | 20 | ||
| style = "background:#fbfbfb" align = right | < | | style = "background:#fbfbfb" align = right | | ||
|- style = "background:#eef8ee" | |||
| <span style="color:#bb00bb;">01:03</span> || <span style="color:blue;">05:02</span> | |||
! style = "background:#fbfbfb" align = right | 0. | |||
! style = "background:#fbfbfb" align = left | 05 | |||
| style = "background:#fbfbfb" align = right | | |||
|- style = "background:#eef8ee" | |- style = "background:#eef8ee" | ||
|< | | <span style="color:#bb00bb;">α<sup>1</sup>-<span style="color:blue;">β<sup>5.3</sup></span> | ||
| <span style="color:green;">5.3</span> | |||
|<span style="color:#bb00bb;">01:04</span> || <span style="color:blue;">05:03</span> | |||
! style = "background:#fbfbfb" align = right | 2. | ! style = "background:#fbfbfb" align = right | 2. | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = left | 03 | ||
| style = "background:#fbfbfb" align = right | <sup> | | style = "background:#fbfbfb" align = right | | ||
|- style = "background:#eef8ee" | |||
|<span style="color:#bb00bb;">α<sup>1</sup>-<span style="color:blue;">β<sup>5.4</sup></span> | |||
| <span style="color:green;">5.4</span> | |||
|<span style="color:#bb00bb;">01:02</span> || <span style="color:blue;">05:04</span> | |||
! style = "background:#fbfbfb" align = right | 0. | |||
! style = "background:#fbfbfb" align = left | 08 | |||
| style = "background:#fbfbfb" align = right | | |||
|- style = "background:#efefff" | |- style = "background:#efefff" | ||
| style = "background:#e8e8f8" rowspan = " | | style = "background:#e8e8f8" rowspan = "8" | [[HLA-DQ6|DQ6]] | ||
| | |<span style="color:#bb00bb;">α<sup>1</sup>-<span style="color:blue;">β<sup>6.1</sup></span> || <span style="color:green;">6.1</span> | ||
| <span style="color:#bb00bb;">01:03</span> || <span style="color:blue;">06:01</span> | |||
! style = "background:#fbfbfb" align = right | 0. | ! style = "background:#fbfbfb" align = right | 0. | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = left | 66 | ||
| style = "background:#fbfbfb" align = right | | | style = "background:#fbfbfb" align = right | | ||
|- style = "background:#eeeeff" | |- style = "background:#eeeeff" | ||
| < | | rowspan = 3 |<span style="color:#bb00bb;">α<sup>1</sup>-<span style="color:blue;">β<sup>6.2</sup></span> | ||
| rowspan = 3 | <span style="color:green;">6.2</span> | |||
| <span style="color:#bb00bb;">01:02</span> || <span style="color:blue;">06:02</span> | |||
! style = "background:#fbfbfb" align = right | 14. | ! style = "background:#fbfbfb" align = right | 14. | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = left | 27 | ||
| style = "background:#fbfbfb" align = right | <sup> | | style = "background:#fbfbfb" align = right | <sup>1st</sup> | ||
|- style = "background:#eeeeff" | |||
| <span style="color:#bb00bb;">01:03</span> || <span style="color:blue;">06:02</span> | |||
! style = "background:#fbfbfb" align = right | 0. | |||
! style = "background:#fbfbfb" align = left | 03 | |||
| style = "background:#fbfbfb" align = right | | |||
|- style = "background:#eeeeff" | |- style = "background:#eeeeff" | ||
| <span style="color:#bb00bb;">01:04</span> || <span style="color:blue;">06:02</span> | |||
! style = "background:#fbfbfb" align = right | 0. | |||
! style = "background:#fbfbfb" align = right | | ! style = "background:#fbfbfb" align = left | 03 | ||
! style = "background:#fbfbfb" align = left | | | style = "background:#fbfbfb" align = right | | ||
| style = "background:#fbfbfb" align = right | | |||
|- style = "background:#eeeeff" | |- style = "background:#eeeeff" | ||
| < | |rowspan = "2" |<span style="color:#bb00bb;">α<sup>1</sup>-<span style="color:blue;">β<sup>6.3</sup></span> | ||
| rowspan = 2 | <span style="color:green;">6.3</span> | |||
| <span style="color:#bb00bb;">01:02</span> || <span style="color:blue;">06:03</span> | |||
! style = "background:#fbfbfb" align = right | 0. | ! style = "background:#fbfbfb" align = right | 0. | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = left | 27 | ||
| style = "background:#fbfbfb" align = right | <sup> | | style = "background:#fbfbfb" align = right | | ||
|- style = "background:#eeeeff" | |||
| <span style="color:#bb00bb;">01:03</span> || <span style="color:blue;">06:03</span> | |||
! style = "background:#fbfbfb" align = right | 5. | |||
! style = "background:#fbfbfb" align = left | 66 | |||
| style = "background:#fbfbfb" align = right | <sup>8th</sup> | |||
|- style = "background:#eeeeff" | |- style = "background:#eeeeff" | ||
| < | | <span style="color:#bb00bb;">α<sup>1</sup>-<span style="color:blue;">β<sup>6.4</sup></span> || <span style="color:green;">6.4</span> | ||
| <span style="color:#bb00bb;">01:02</span> || <span style="color:blue;">06:04</span> | |||
! style = "background:#fbfbfb" align = right | 3. | ! style = "background:#fbfbfb" align = right | 3. | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = left | 40 | ||
| style = "background:#fbfbfb" align = right | <sup> | | style = "background:#fbfbfb" align = right | <sup>10th</sup> | ||
|- style = "background:# | |- style = "background:#eeeeff" | ||
| style = "background:#efdff8" rowspan = " | | <span style="color:#bb00bb;">α<sup>1</sup>-<span style="color:blue;">β<sup>6.9</sup></span> || <span style="color:green;">6.9</span> | ||
| | | <span style="color:#bb00bb;">01:02</span> || <span style="color:blue;">06:09</span> | ||
| < | ! style = "background:#fbfbfb" align = right | 0. | ||
! style = "background:#fbfbfb" align = right | | ! style = "background:#fbfbfb" align = left | 71 | ||
! style = "background:#fbfbfb" align = left | | | style = "background:#fbfbfb" align = right | | ||
| style = "background:#fbfbfb" align = right | <sup> | |- style = "background:#f8e8ff" | ||
| style = "background:#efdff8" rowspan = "8" | [[HLA-DQ7|DQ7]] | |||
| <span style="color:#bb00bb;">α<sup>2</sup>-<span style="color:blue;">β<sup>7</sup></span> | |||
| <span style="color:green;">7.2</span> | |||
| <span style="color:#bb00bb;">02:01</span> || <span style="color:blue;">03:01</span> | |||
! style = "background:#fbfbfb" align = right | 0. | |||
! style = "background:#fbfbfb" align = left | 05 | |||
| style = "background:#fbfbfb" align = right | | |||
|- style = "background:#f8e8ff" | |||
|Rowspan = 4 | <span style="color:#bb00bb;">α<sup>3</sup>-<span style="color:blue;">β<sup>7</sup></span> | |||
| rowspan = 4 |<span style="color:green;">7.3</span> | |||
| <span style="color:#bb00bb;">03:01</span> || <span style="color:blue;">03:01</span> | |||
! style = "background:#fbfbfb" align = right | 0. | |||
! style = "background:#fbfbfb" align = left | 16 | |||
| style = "background:#fbfbfb" align = right | | |||
|- style = "background:#f8e8ff" | |||
| <span style="color:#bb00bb;">03:03</span><sup>♠</sup> || <span style="color:blue;">03:01</span> | |||
! style = "background:#fbfbfb" align = right | 6. | |||
! style = "background:#fbfbfb" align = left | 45 | |||
| style = "background:#fbfbfb" align = right | <sup>7th</sup> | |||
|- style = "background:#f8e8ff" | |- style = "background:#f8e8ff" | ||
| < | | <span style="color:#bb00bb;">03:01</span> || <span style="color:blue;">03:04</span> | ||
! style = "background:#fbfbfb" align = right | | ! style = "background:#fbfbfb" align = right | 0. | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = left | 09 | ||
| style = "background:#fbfbfb" align = right | | | style = "background:#fbfbfb" align = right | | ||
|- style = "background:#f8e8ff" | |- style = "background:#f8e8ff" | ||
|< | | <span style="color:#bb00bb;">03:02</span><sup>♠</sup> || <span style="color:blue;">03:04</span> | ||
! style = "background:#fbfbfb" align = right | | ! style = "background:#fbfbfb" align = right | 0. | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = left | 09 | ||
| style = "background:#fbfbfb" align = right | | | style = "background:#fbfbfb" align = right | | ||
|- style = "background:#f8e8ff" | |- style = "background:#f8e8ff" | ||
|< | | <span style="color:#bb00bb;">α<sup>4</sup>-<span style="color:blue;">β<sup>7</sup></span> | ||
| <span style="color:green;">7.4</span> | |||
| <span style="color:#bb00bb;">04:01</span> || <span style="color:blue;">03:01</span> | |||
! style = "background:#fbfbfb" align = right | 0. | ! style = "background:#fbfbfb" align = right | 0. | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = left | 03 | ||
| style = "background:#fbfbfb" align = right | | | style = "background:#fbfbfb" align = right | | ||
|- style = "background:#f8e8ff" | |- style = "background:#f8e8ff" | ||
| | | <span style="color:#bb00bb;">α<sup>5</sup>-<span style="color:blue;">β<sup>7</sup></span> | ||
| < | | <span style="color:green;">7.5</span> | ||
! style = "background:#fbfbfb" align = right | | | <span style="color:#bb00bb;">05:05</span><sup>♣</sup> || <span style="color:blue;">03:01</span> | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = right | 11. | ||
| style = "background:#fbfbfb" align = right | <sup> | ! style = "background:#fbfbfb" align = left | 06 | ||
| style = "background:#fbfbfb" align = right | <sup>4th</sup> | |||
|- style = "background:#f8e8ff" | |- style = "background:#f8e8ff" | ||
| < | | <span style="color:#bb00bb;">α<sup>6</sup>-<span style="color:blue;">β<sup>7</sup></span> | ||
| <span style="color:green;">7.6</span> | |||
| <span style="color:#bb00bb;">06:01</span> || <span style="color:blue;">03:01</span> | |||
! style = "background:#fbfbfb" align = right | 0. | ! style = "background:#fbfbfb" align = right | 0. | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = left | 11 | ||
| style = "background:#fbfbfb" align = right | | | style = "background:#fbfbfb" align = right | | ||
|- style = "background:#ffe7e7" | |- style = "background:#ffe7e7" | ||
| style = "background:#f7dfdf" rowspan = " | | style = "background:#f7dfdf" rowspan = "2" | [[HLA-DQ8|DQ8]] | ||
| rowspan = " | | rowspan = "2" | <span style="color:#bb00bb;">α<sup>3</sup>-<span style="color:blue;">β<sup>8</sup></span> | ||
| < | | rowspan = 2 | <span style="color:green;">8.1</span> | ||
! style = "background:#fbfbfb" align = right | | | <span style="color:#bb00bb;">03:01</span> || <span style="color:blue;">03:02</span> | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = right | 9. | ||
| style = "background:#fbfbfb" align = right | <sup> | ! style = "background:#fbfbfb" align = left | 62 | ||
| style = "background:#fbfbfb" align = right |<sup>6th</sup> | |||
|- style = "background:#ffe8e8" | |- style = "background:#ffe8e8" | ||
| < | | <span style="color:#bb00bb;">03:02</span><sup>♠</sup> || <span style="color:blue;">03:02</span> | ||
| | |||
| < | |||
! style = "background:#fbfbfb" align = right | 0. | ! style = "background:#fbfbfb" align = right | 0. | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = left | 93 | ||
| style = "background:#fbfbfb" align = right | | | style = "background:#fbfbfb" align = right | | ||
|- style = "background:#fff7e7" | |- style = "background:#fff7e7" | ||
| style = "background:#f7efdf" rowspan = "2"| [[HLA-DQ9|DQ9]] | | style = "background:#f7efdf" rowspan = "2"| [[HLA-DQ9|DQ9]] | ||
|< | |<span style="color:#bb00bb;">α<sup>2</sup>-<span style="color:blue;">β<sup>9</sup></span> | ||
| <span style="color:green;">9.2</span> | |||
| <span style="color:#bb00bb;">02:01</span> || <span style="color:blue;">03:03</span> | |||
! style = "background:#fbfbfb" align = right | 3. | ! style = "background:#fbfbfb" align = right | 3. | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = left | 66 | ||
| style = "background:#fbfbfb" align = right | <sup> | | style = "background:#fbfbfb" align = right | <sup>9th</sup> | ||
|- style = "background:# | |- style = "background:#fff7e7" | ||
|< | |<span style="color:#bb00bb;">α<sup>3</sup>-<span style="color:blue;">β<sup>9</sup></span> | ||
| <span style="color:green;">9.3</span> | |||
| <span style="color:#bb00bb;">03:02</span> || <span style="color:blue;">03:03</span> | |||
! style = "background:#fbfbfb" align = right | 0. | ! style = "background:#fbfbfb" align = right | 0. | ||
! style = "background:#fbfbfb" align = left | | ! style = "background:#fbfbfb" align = left | 79 | ||
| style = "background:#fbfbfb" align = right | <sup> | | style = "background:#fbfbfb" align = right | | ||
|- | |||
| colspan = 8 | <small><sup>♠</sup>DQA1*03:02 & *03:03 not resolved; <sup>♣</sup>DQB1*05:01 & *05:05<br>, and some *03:03 are resolvable by haplotype</small> | |||
|} | |} | ||
'''Genetic Typing'''. With the exception of DQ2 (*02:01) which | |||
has a 98% detection capability, serotyping has drawbacks in relative accuracy. In addition, for many HLA studies genetic typing | |||
does not offer that much greater advantage over serotyping, but in the case of DQ there is a need for precise identification | |||
of HLA-DQB1 and HLA-DQA1 which cannot be provided by serotyping. | |||
Isoform functionality is dependent on αβ composition. Most studies indicate a chromosomal linkage between disease causing | |||
DQA1 and DQB1 genes. Therefore, the DQA1, α, component is as important as DQB1. An example of this is DQ2, DQ2 mediates [[Coeliac disease]] and [[Type 1 diabetes]] but only if the α<sup>5</sup> subunit is present. This subunit can be encoded by either DQA1*05:01 or DQA1*05:05. When the DQ2 encoding β-chain gene is on the same chromosome as the α<sup>5</sup> subunit isoform, then individuals | |||
who have this chromosome have a much higher risk of these two disease. When DQA1 and DQB1 alleles are linked in this way | |||
they form a haplotype. The DQA1*05:01-DQB1*02:01 haplotype is called the DQ2.5 haplotype, and the DQ that results α<sup>5</sup>β² is the "cis-haplotype" or "cis-chromosomal" isoform of DQ2.5 | |||
To detect these potential combinations one uses a technique called SSP-PCR (Sequence specific primer polymerase chain reaction). This techniques works because, outside of a few areas of Africa, we know the overwhelming majority of all DQ alleles in the world. The primers are specific for known DQ and thus, if a product is seen it means that gene motif is present. This results in nearly 100% accurate typing of DQA1 and DQB1 alleles. | |||
''' 'How does one know which isoforms are functionally unique and which isoforms are functionally synonymous with other isoforms'?'''. The IMGT/HLA database also provides alignments for various alleles, these alignments show the variable regions and conserved regions. By examining the structure of these variable regions with different ligands bound | |||
(such as the MMDB) one can see which residues come into close contact with peptides and those have side chains that are distal. Those changes more than 10 Angstoms away generally do not affect binding of peptides. The structure of [http://structure.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?Dopt=s&uid=17145 HLA-DQ8/insulin peptide at NCBI] can be view with [[Cn3D]] or [[Rasmol]]. In Cn3D one can highlight the peptide and then select for amino acids within 3 or more Angstroms of the peptide. Side chains that come close to the peptide can be identified and then examined on the sequence alignments at IMGT/HLA database. Anyone can download software and sequence. Have fun! | |||
====Effects of heterogeneity of isoform pairing==== | |||
As an MHC class II antigen-presenting receptor, DQ functions as a [[protein dimer|dimer]] containing two protein subunits, alpha (DQA1 gene product) and beta (DQB1 gene product), a DQ [[heterodimer]]. These receptors can be made from alpha+beta sets of two different DQ [[haplotype]]s, one set from the maternal and paternal [[chromosome]]. If one carries haplotype -A-B- from one parent and -a-b- from the other, that person makes 2 alpha [[isoform]]s (A and a) and 2 beta isoforms (B and b). This can produce 4 slightly different receptor heterodimers (or more simply, DQ isoforms). Two isoforms are in the [[wikt:cis-|cis]]-haplotype pairing (AB and ab) and 2 are in the [[:wikt:trans-|trans-]]haplotype pairing (Ab and aB). Such a person is a double [[heterozygote]] for these genes, for DQ the most popular situation. If a person carries haplotypes -A-'''B'''- and -A-'''b'''- then they can only make 2 DQ (AB and Ab), but if a person carries haplotypes -A-B- and -A-B- then they can only make DQ isoform AB, called a double [[homozygote]]. In coeliac disease, certain homozygotes and are at higher risk for disease and some specific complications of coeliac disease such as [[Gluten-sensitive enteropathy associated conditions#GSEA Lymphoma|Gluten-sensitive enteropathy associated T-cell lymphoma]] | |||
'''Homozygotes and double homozygotes'''<br> | |||
Homozygotes at DQ loci can change risk for disease. In mice for instance, mice with 2 copies of the DQ-like Ia<sup>b</sup> haplotype are more likely to progress toward fatal disease compared to mice that are heterozygotes only for the beta allele (MHC IA<sub>α</sub><sup>b</sup> / IA<sub>α</sub><sup>b</sup>, IA<sub>β</sub><sup>b</sup> / IA<sub>β</sub><sup>bm12</sup>). In humans, celiac disease DQ2.5/DQ2 homozygotes are several times more likely to have celiac disease versus DQ2.5/DQX individuals.<ref name="pmid17190762">{{cite journal |vauthors=Jores RD, Frau F, Cucca F, etal | title = HLA-DQB1*0201 homozygosis predisposes to severe intestinal damage in celiac disease | journal = Scand. J. Gastroenterol. | volume = 42 | issue = 1 | pages = 48–53 | year = 2007 | pmid = 17190762 | doi = 10.1080/00365520600789859}}</ref> DQ2/DQ2 homozygotes are at elevated risk for [[Gluten-sensitive enteropathy associated conditions#GSEA Cancers|severe complications]] of disease.<ref name="pmid17470479">{{cite journal |vauthors=Al-Toma A, Verbeek WH, Hadithi M, von Blomberg BM, Mulder CJ | title = Survival in refractory coeliac disease and enteropathy‐associated T‐cell lymphoma: retrospective evaluation of single‐centre experience | journal = Gut| volume = 56| issue = 10| pages = 1373–8| year = 2007 | pmid = 17470479 | doi = 10.1136/gut.2006.114512 | pmc=2000250}}</ref> For an explanation of the risk association see:[[Talk:HLA-DQ#Effects of heterogeneity of isoform pairing-Expanded]] | |||
'''Involvement of transhaplotypes in disease'''<br> | |||
There is some controversy in the literature whether trans-isoforms are relevant. Recent genetic studies into [[coeliac disease]] have revealed that the DQA1*05:05:X/Y:DQB1*02:02 gene products explain disease not linked to the haplotype that produces DQ8 and DQ2.5, strongly suggesting the trans-isoforms can be involved in disease. But, in this example, it is known that the transproduct is almost identical to a known cis-'isoform' produced by DQ2.5. There is other evidence that some haplotypes are linked to disease but show neutral linkage with other particular haplotypes are present. At present, the bias of relative isoform frequency toward cis pairing is unknown, it is known that some trans-isoforms occur. | |||
see:[[Talk:HLA-DQ#Effects of heterogeneity of isoform pairing-Expanded]] | |||
[[ | ===DQ Function in Autoimmunity === | ||
These | HLA D (-P,-Q,-R) genes are members of the [[Major histocompatibility complex]] (MHC) gene family and have analogs in other mammalian species. In mice the MHC locus known as IA is homologous to human HLA DQ. Several autoimmune diseases that occur in humans that are mediated by DQ also can be induced in mice and are mediated through IA. [[Myasthenia gravis]] is an example of one such disease.<ref name=Atassi>{{cite journal |vauthors=Atassi MZ, Oshima M, Deitiker P | title = n the initial trigger of myasthenia gravis and suppression of the disease by antibodies against the MHC peptide region involved in the presentation of a pathogenic T-cell epitope | journal = Crit Rev Immunol. | volume = 21 | issue = 1–3 | pages = 1–27 | year = 2001 | pmid = 11642597}}</ref> Linking specific sites on autoantigens is more difficult in humans due to the complex variation of heterologous humans, but subtle differences in T-cell stimulation associated with DQ-types has been observed.<ref name=Deitiker>{{cite journal |vauthors=Deitiker PR, Oshima M, Smith RG, Mosier DR, Atassi MZ | title = Subtle differences in HLA DQ haplotype-associated presentation of AChR alpha-chain peptides may suffice to mediate myasthenia gravis | journal = Autoimmunity | volume = 39 | issue = 4 | pages = 277–288 | year = 2006 | pmid = 16891216 | doi = 10.1080/08916930600738581}}</ref> These studies indicate that potentially a small change or increase in the presentation of a potential self-antigen can result in autoimmunity. This may explain | ||
why there is often linkage to DR or DQ, but the linkage is often weak. | |||
'''Regional Evolution''' | '''Regional Evolution''' | ||
{| border="0" cellpadding="2" cellspacing="0" align="left" style="text-align:center; margin-right: 1em; | {| border="0" cellpadding="2" cellspacing="0" align="left" style="text-align:center; margin-right: 1em; border:1px #b0ffff solid; background:#f7ffff;" | ||
|+ HLA DQ- subunit alleles, mature chains, contact variants | |+ HLA DQ- subunit alleles, mature chains, contact variants | ||
|- Style = "Background:#e7ffff" | |- Style = "Background:#e7ffff" | ||
| Line 439: | Line 360: | ||
|} | |} | ||
Many HLA DQ were under positive selection of 10,000s potentially 100,000s of years in some regions. As people moved they have | Many HLA DQ were under positive selection of 10,000s potentially 100,000s of years in some regions. As people moved they have tended to lose haplotypes and in the process lose | ||
allelic diversity. On the other hand, on arrival at new distal locations, selection | allelic diversity. On the other hand, on arrival at new distal locations, selection | ||
would offer unknown selective forces that would have initially favored diversity in arrivals. By an unknown process, rapid evolution occurs, as has been seen in South Americas | would offer unknown selective forces that would have initially favored diversity in arrivals. By an unknown process, rapid evolution occurs, as has been seen in South Americas | ||
| Line 445: | Line 366: | ||
appear. This process may be of immediate benefit of being positively selective in that new environment, but these new alleles might also be 'sloppy' in a selective perspective, having side effects if selection changed. The table to the left demonstrates how absolute diversity | appear. This process may be of immediate benefit of being positively selective in that new environment, but these new alleles might also be 'sloppy' in a selective perspective, having side effects if selection changed. The table to the left demonstrates how absolute diversity | ||
at the global level translates into relative diversity at the regional level. | at the global level translates into relative diversity at the regional level. | ||
- | - | ||
| Line 461: | Line 376: | ||
! width="60" | DQ8 | ! width="60" | DQ8 | ||
! width="60" | DQ2.5/8 | ! width="60" | DQ2.5/8 | ||
|- style = "background:# | |- style = "background:#fbf7ff" | | ||
! style = "background:#efdff8" | Sweden | ! style = "background:#efdff8" | Sweden | ||
|15.9 | |15.9 | ||
|18.7 | |18.7 | ||
!5.9 | !5.9 | ||
|- style = "background:# | |- style = "background:#fbf7ff" | | ||
! style = "background:#efdff8" | Jalisco | ! style = "background:#efdff8" | Jalisco | ||
|11.4 | |11.4 | ||
|22.8 | |22.8 | ||
!5.2 | !5.2 | ||
|- style = "background:# | |- style = "background:#fbf7ff" | | ||
! style = "background:#efdff8" | England | ! style = "background:#efdff8" | England | ||
|12.4 | |12.4 | ||
|16.8 | |16.8 | ||
!4.2 | !4.2 | ||
|- style = "background:# | |- style = "background:#fbf7ff" | | ||
! style = "background:#efdff8" | | ! style = "background:#efdff8" | Kazakh | ||
|13.1 | |13.1 | ||
|11 | |11 | ||
!2.9 | !2.9 | ||
|- style = "background:# | |- style = "background:#fbf7ff" | | ||
! style = "background:#efdff8" | Uygur | ! style = "background:#efdff8" | Uygur | ||
|12.6 | |12.6 | ||
|11.4 | |11.4 | ||
!2.9 | !2.9 | ||
|- style = "background:# | |- style = "background:#fbf7ff" | | ||
! style = "background:#efdff8" | Finland | ! style = "background:#efdff8" | Finland | ||
|9 | |9 | ||
|15.7 | |15.7 | ||
!2.8 | !2.8 | ||
|- style = "background:# | |- style = "background:#fbf7ff" | | ||
! style = "background:#efdff8" | Poland | ! style = "background:#efdff8" | Poland | ||
|10.7 | |10.7 | ||
| Line 498: | Line 413: | ||
|} | |} | ||
===DQ2.5/DQ8 Heterozygotes=== | |||
The distribution of this phenotype is largely the result of admixtures between peoples of eastern or central Asian origin and peoples of western or central Asian origin. The highest | The distribution of this phenotype is largely the result of admixtures between peoples of eastern or central Asian origin and peoples of western or central Asian origin. The highest | ||
frequencies, by random mating, are expected in Sweden, but pockets of high levels also occur in Mexico, and a larger range risk exists in Central Asia. | frequencies, by random mating, are expected in Sweden, but pockets of high levels also occur in Mexico, and a larger range risk exists in Central Asia. | ||
Diseases that appear to be increased in Heterozygotes are Type 1 Diabetes. New evidence is showing an increased risk for late onset Type 1 diabetes in Heterozygotes (which includes ambiguous Type I/Type II diabetes. Celiac Disease | Diseases that appear to be increased in Heterozygotes are Celiac Disease and Type 1 Diabetes. New evidence is showing an increased risk for late onset Type 1 diabetes in Heterozygotes (which includes ambiguous Type I/Type II diabetes). 95% of Celiac Disease patients are positive for DQ2 or DQ8.<ref name="Zonulin and Its Regulation of Intestinal Barrier Function: The Biological Door to Inflammation, Autoimmunity, and Cancer">{{cite journal|last=Fasano|first=Alessio|authorlink=Alessio Fasano|title=Dr|journal=Physiol Rev|year=2011|volume=91|url=http://physrev.physiology.org/content/91/1/151.full?sid=77927ed1-2ebe-4941-ad21-db9d0f6905fb}}</ref> | ||
==References== | ==References== | ||
{{reflist}} | |||
== External links == | == External links == | ||
* [http://www.allelefrequencies.net HLA Allele and Haplotype Frequency Database] | * [http://www.allelefrequencies.net HLA Allele and Haplotype Frequency Database] | ||
* [http://www.ebi.ac.uk/imgt/hla/ IMGT-HLA Database] | * [http://www.ebi.ac.uk/imgt/hla/ IMGT-HLA Database] | ||
{{Surface antigens}} | |||
{{DQ serotypes}} | |||
{{DEFAULTSORT:Hla-Dq}} | |||
[[Category:HLA-DQ haplotypes|0]] | [[Category:HLA-DQ haplotypes|0]] | ||
[[Category:Human MHC mediated diseases|Q]] | [[Category:Human MHC mediated diseases|Q]] | ||
[[Category:Genes on chromosome 6 | [[Category:Genes on human chromosome 6]] | ||
[[Category:MHC Class II]] | [[Category:MHC Class II]] | ||
Latest revision as of 13:29, 2 November 2017
MHC class II, DQ (heterodimer)
| ||
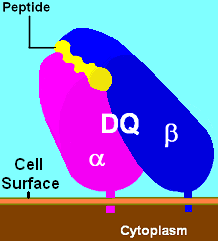
| ||
| DQ1 binding pocket with ligand | ||
| − | ||
| Protein type | cell surface receptor | |
| Function | Immune recognition and antigen presentation | |
| − | ||
| Subunit name |
Gene | Chromosomal locus |
| α | HLA-DQA1 | Chromosome 6p21.31 |
| β | HLA-DQB1 | Chromosome 6p21.31 |
HLA-DQ (DQ) is a cell surface receptor protein found on antigen presenting cells. It is an αβ heterodimer of type MHC class II. The α and β chains are encoded by two loci, HLA-DQA1 and HLA-DQB1, that are adjacent to each other on chromosome band 6p21.3. Both α-chain and β-chain vary greatly. A person often produces two α-chain and two β-chain variants and thus 4 isoforms of DQ. The DQ loci are in close genetic linkage to HLA-DR, and less closely linked to HLA-DP, HLA-A, HLA-B and HLA-C.
Different isoforms of DQ can bind to and present different antigens to T-cells. In this process T-cells are stimulated to grow and can signal B-cells to produce antibodies. DQ functions in recognizing and presenting foreign antigens (proteins derived from potential pathogens). But DQ is also involved in recognizing common self-antigens and presenting those antigens to the immune system in order to develop tolerance from a very young age.
When tolerance to self proteins is lost, DQ may become involved in autoimmune disease. Two autoimmune diseases in which HLA-DQ is involved are coeliac disease and diabetes mellitus type 1. DQ is one of several antigens involved in rejection of organ transplants. As a variable cell surface receptor on immune cells, these D antigens, originally HL-A4 antigens, are involved in graft versus host disease when lymphoid tissues are transplanted between people. Serological studies of DQ recognized that antibodies to DQ bind primarily to the β-chain. The currently used serotypes are HLA-DQ2, -DQ3, -DQ4, -DQ5, -DQ6, -DQ7, -DQ8, -DQ9. HLA-DQ1 is a weak reaction to the α-chain and was replaced by DQ5 and DQ6 serology. Serotyping is capable of identifying most aspects of DQ isoform structure and function, however sequence specific PCR is now the preferred method of determining HLA-DQA1 and HLA-DQB1 alleles, as serotyping cannot resolve, often, the critical contribution of the DQ α-chain. This can be compensated for by examining DR serotypes as well as DQ serotypes.
Structure, Functions, Genetics
Function
The name 'HLA DQ' originally describes a transplantation antigen of MHC class II category of the major histocompatibility complex of humans; however, this status is an artifact of the early era of organ transplantation.
HLA DQ functions as a cell surface receptor for foreign or self antigens. The immune system surveys antigens for foreign pathogens when presented by MHC receptors (like HLA DQ). The MHC Class II antigens are found on antigen presenting cells (APC) (macrophages, dendritic cells, and B-lymphocytes). Normally, these APC 'present' class II receptor/antigens to a great many T-cells, each with unique T-cell receptor (TCR) variants. A few TCR variants that recognize these DQ/antigen complexes are on CD4 positive (CD4+) T-cells. These T-cells, called T-helper cells, can promote the amplification of B-cells which, in turn recognize a different portion of the same antigen. Alternatively, macrophages and other megalocytes consume cells by apoptotic signaling and present self-antigens. Self antigens, in the right context, form a suppressor T-cell population that protects self tissues from immune attack or autoimmunity.
Genetics
HLA-DQ (DQ) is encoded on the HLA region of chromosome 6p21.3, in what was classically known as the "D" antigen region. This region encoded the subunits for DP,-Q and -R which are the major MHC class II antigens in humans. Each of these proteins have slightly different functions and are regulated in slightly different ways.
DQ is made up of two different subunits to form an αβ-heterodimer. Each subunit is encoded by its own "gene" (a coding locus). The DQ α subunit is encoded by the HLA-DQA1 gene and the DQ β subunit is encoded by the HLA-DQB1 gene. Both loci are variable in the human population (see regional evolution).
Detecting DQ isoforms
In the human population DQ is highly variable, the β subunit more so than the alpha chain. The variants are encoded by the HLA DQ genes and are the result of single nucleotide polymorphisms (SNP). Some SNP result in no change in amino-acid sequence. Others result in changes in regions that are removed when the proteins is processed to the cell surface, still others result in change in the non-functional regions of the protein, and some changes result in a change of function of the DQ isoform that is produced. The isoforms generally change in the peptides they bind and present to T-cells. Much of the isoform variation in DQ is within these 'functional' regions.
Serotyping. Antibodies raised against DQ tend to recognize these functional regions, in most cases the β-subunit. As a result, these antibodies can discriminate different classes of DQ based on the recognition similar DQβ proteins known as serotypes.
An example of a serotype is DQ2.
- Recognize HLA-DQB1*02 gene products which include gene products of the following alleles:
- HLA-DQB1*02:01
- HLA-DQB1*02:02
- HLA-DQB1*02:03
Sometimes DQ2 antibodies recognize other gene products, such as DQB1*03:03, resulting in serotyping errors. Because of this mistyping serotyping is not as reliable as gene sequencing or SSP-PCR.
While the DQ2 isoforms are recognized by the same antibodies, and all DQB1*02 are functionally similar, they can bind different α subunit and these αβ isoform variants can bind different sets of peptides. This difference in binding is an important feature that helps to understand autoimmune disease.
The first identified DQ were DQw1 to DQw3. DQw1 (DQ1) recognized the alpha chain of DQA1*01 alleles. This group was later split by beta chain recognition to DQ5 and DQ6. DQ3 is known as broad antigen serotypes, because they recognize a broad group of antigens. However, because of this broad antigen recognition their specificity and usefulness is somewhat less than desirable.
For most modern typing the DQ2, DQ4 - DQ9 set is used.
| DQ | DQ | DQ | Freq | |||||
|---|---|---|---|---|---|---|---|---|
| Serotype | cis-isoform | Subtype | A1 | B1 | %[1] | rank | ||
| DQ2 | α5-β2 | 2.5 | 05:01♣ | 02:01 | 13. | 16 | 2nd | |
| α2-β2 | 2.2 | 02:01 | 02:02 | 11. | 08 | 3rd | ||
| α3-β2 | 2.3 | 03:02♠ | 02:02 | 0. | 08 | |||
| DQ4 | α3-β4 | 4.3 | 03:01 | 04:02 | 0. | 03 | ||
| 03:02♠ | 04:02 | 0. | 11 | |||||
| α4-β4 | 4.4 | 04:01 | 04:02 | 2. | 26 | |||
| DQ5 | α1-β5.1 | 5.1 | 01:01 | 05:01 | 10. | 85 | 5th | |
| 01:02 | 05:01 | 0. | 03 | |||||
| 01:03 | 05:01 | 0. | 03 | |||||
| 01:04 | 05:01 | 0. | 71 | |||||
| α1-β5.2 | 5.2 | 01:02 | 05:02 | 1. | 20 | |||
| 01:03 | 05:02 | 0. | 05 | |||||
| α1-β5.3 | 5.3 | 01:04 | 05:03 | 2. | 03 | |||
| α1-β5.4 | 5.4 | 01:02 | 05:04 | 0. | 08 | |||
| DQ6 | α1-β6.1 | 6.1 | 01:03 | 06:01 | 0. | 66 | ||
| α1-β6.2 | 6.2 | 01:02 | 06:02 | 14. | 27 | 1st | ||
| 01:03 | 06:02 | 0. | 03 | |||||
| 01:04 | 06:02 | 0. | 03 | |||||
| α1-β6.3 | 6.3 | 01:02 | 06:03 | 0. | 27 | |||
| 01:03 | 06:03 | 5. | 66 | 8th | ||||
| α1-β6.4 | 6.4 | 01:02 | 06:04 | 3. | 40 | 10th | ||
| α1-β6.9 | 6.9 | 01:02 | 06:09 | 0. | 71 | |||
| DQ7 | α2-β7 | 7.2 | 02:01 | 03:01 | 0. | 05 | ||
| α3-β7 | 7.3 | 03:01 | 03:01 | 0. | 16 | |||
| 03:03♠ | 03:01 | 6. | 45 | 7th | ||||
| 03:01 | 03:04 | 0. | 09 | |||||
| 03:02♠ | 03:04 | 0. | 09 | |||||
| α4-β7 | 7.4 | 04:01 | 03:01 | 0. | 03 | |||
| α5-β7 | 7.5 | 05:05♣ | 03:01 | 11. | 06 | 4th | ||
| α6-β7 | 7.6 | 06:01 | 03:01 | 0. | 11 | |||
| DQ8 | α3-β8 | 8.1 | 03:01 | 03:02 | 9. | 62 | 6th | |
| 03:02♠ | 03:02 | 0. | 93 | |||||
| DQ9 | α2-β9 | 9.2 | 02:01 | 03:03 | 3. | 66 | 9th | |
| α3-β9 | 9.3 | 03:02 | 03:03 | 0. | 79 | |||
| ♠DQA1*03:02 & *03:03 not resolved; ♣DQB1*05:01 & *05:05 , and some *03:03 are resolvable by haplotype | ||||||||
Genetic Typing. With the exception of DQ2 (*02:01) which has a 98% detection capability, serotyping has drawbacks in relative accuracy. In addition, for many HLA studies genetic typing does not offer that much greater advantage over serotyping, but in the case of DQ there is a need for precise identification of HLA-DQB1 and HLA-DQA1 which cannot be provided by serotyping.
Isoform functionality is dependent on αβ composition. Most studies indicate a chromosomal linkage between disease causing DQA1 and DQB1 genes. Therefore, the DQA1, α, component is as important as DQB1. An example of this is DQ2, DQ2 mediates Coeliac disease and Type 1 diabetes but only if the α5 subunit is present. This subunit can be encoded by either DQA1*05:01 or DQA1*05:05. When the DQ2 encoding β-chain gene is on the same chromosome as the α5 subunit isoform, then individuals who have this chromosome have a much higher risk of these two disease. When DQA1 and DQB1 alleles are linked in this way they form a haplotype. The DQA1*05:01-DQB1*02:01 haplotype is called the DQ2.5 haplotype, and the DQ that results α5β² is the "cis-haplotype" or "cis-chromosomal" isoform of DQ2.5
To detect these potential combinations one uses a technique called SSP-PCR (Sequence specific primer polymerase chain reaction). This techniques works because, outside of a few areas of Africa, we know the overwhelming majority of all DQ alleles in the world. The primers are specific for known DQ and thus, if a product is seen it means that gene motif is present. This results in nearly 100% accurate typing of DQA1 and DQB1 alleles.
'How does one know which isoforms are functionally unique and which isoforms are functionally synonymous with other isoforms'?. The IMGT/HLA database also provides alignments for various alleles, these alignments show the variable regions and conserved regions. By examining the structure of these variable regions with different ligands bound (such as the MMDB) one can see which residues come into close contact with peptides and those have side chains that are distal. Those changes more than 10 Angstoms away generally do not affect binding of peptides. The structure of HLA-DQ8/insulin peptide at NCBI can be view with Cn3D or Rasmol. In Cn3D one can highlight the peptide and then select for amino acids within 3 or more Angstroms of the peptide. Side chains that come close to the peptide can be identified and then examined on the sequence alignments at IMGT/HLA database. Anyone can download software and sequence. Have fun!
Effects of heterogeneity of isoform pairing
As an MHC class II antigen-presenting receptor, DQ functions as a dimer containing two protein subunits, alpha (DQA1 gene product) and beta (DQB1 gene product), a DQ heterodimer. These receptors can be made from alpha+beta sets of two different DQ haplotypes, one set from the maternal and paternal chromosome. If one carries haplotype -A-B- from one parent and -a-b- from the other, that person makes 2 alpha isoforms (A and a) and 2 beta isoforms (B and b). This can produce 4 slightly different receptor heterodimers (or more simply, DQ isoforms). Two isoforms are in the cis-haplotype pairing (AB and ab) and 2 are in the trans-haplotype pairing (Ab and aB). Such a person is a double heterozygote for these genes, for DQ the most popular situation. If a person carries haplotypes -A-B- and -A-b- then they can only make 2 DQ (AB and Ab), but if a person carries haplotypes -A-B- and -A-B- then they can only make DQ isoform AB, called a double homozygote. In coeliac disease, certain homozygotes and are at higher risk for disease and some specific complications of coeliac disease such as Gluten-sensitive enteropathy associated T-cell lymphoma
Homozygotes and double homozygotes
Homozygotes at DQ loci can change risk for disease. In mice for instance, mice with 2 copies of the DQ-like Iab haplotype are more likely to progress toward fatal disease compared to mice that are heterozygotes only for the beta allele (MHC IAαb / IAαb, IAβb / IAβbm12). In humans, celiac disease DQ2.5/DQ2 homozygotes are several times more likely to have celiac disease versus DQ2.5/DQX individuals.[2] DQ2/DQ2 homozygotes are at elevated risk for severe complications of disease.[3] For an explanation of the risk association see:Talk:HLA-DQ#Effects of heterogeneity of isoform pairing-Expanded
Involvement of transhaplotypes in disease
There is some controversy in the literature whether trans-isoforms are relevant. Recent genetic studies into coeliac disease have revealed that the DQA1*05:05:X/Y:DQB1*02:02 gene products explain disease not linked to the haplotype that produces DQ8 and DQ2.5, strongly suggesting the trans-isoforms can be involved in disease. But, in this example, it is known that the transproduct is almost identical to a known cis-'isoform' produced by DQ2.5. There is other evidence that some haplotypes are linked to disease but show neutral linkage with other particular haplotypes are present. At present, the bias of relative isoform frequency toward cis pairing is unknown, it is known that some trans-isoforms occur.
see:Talk:HLA-DQ#Effects of heterogeneity of isoform pairing-Expanded
DQ Function in Autoimmunity
HLA D (-P,-Q,-R) genes are members of the Major histocompatibility complex (MHC) gene family and have analogs in other mammalian species. In mice the MHC locus known as IA is homologous to human HLA DQ. Several autoimmune diseases that occur in humans that are mediated by DQ also can be induced in mice and are mediated through IA. Myasthenia gravis is an example of one such disease.[4] Linking specific sites on autoantigens is more difficult in humans due to the complex variation of heterologous humans, but subtle differences in T-cell stimulation associated with DQ-types has been observed.[5] These studies indicate that potentially a small change or increase in the presentation of a potential self-antigen can result in autoimmunity. This may explain why there is often linkage to DR or DQ, but the linkage is often weak.
Regional Evolution
| Known | HLA-DQ | Potential | |
|---|---|---|---|
| Locus: | A1 | B1 | Combinations |
| Alleles | 33 | 78 | 2475 |
| Subunit: | α | β | isoforms |
| Mature Chains | 24 | 58 | 1392 |
| Contact Variants* | ~9 | 40 | 360 |
| Caucasian (USA) | |||
| Contact Variants (CV) | 7 | 12 | 84 |
| CV-haplotypes | 30 | ||
| *Subunits vary within 9Â of peptide in DQ2.5 or DQ8 | |||
Many HLA DQ were under positive selection of 10,000s potentially 100,000s of years in some regions. As people moved they have tended to lose haplotypes and in the process lose allelic diversity. On the other hand, on arrival at new distal locations, selection would offer unknown selective forces that would have initially favored diversity in arrivals. By an unknown process, rapid evolution occurs, as has been seen in South Americas indigeonous population (Parham and Ohta, 1996, Watkins 1995), and new alleles rapidly appear. This process may be of immediate benefit of being positively selective in that new environment, but these new alleles might also be 'sloppy' in a selective perspective, having side effects if selection changed. The table to the left demonstrates how absolute diversity at the global level translates into relative diversity at the regional level.
-
Heterozygous DQ Combinations and Disease
| DQ2.5 | DQ8 | DQ2.5/8 | |
|---|---|---|---|
| Sweden | 15.9 | 18.7 | 5.9 |
| Jalisco | 11.4 | 22.8 | 5.2 |
| England | 12.4 | 16.8 | 4.2 |
| Kazakh | 13.1 | 11 | 2.9 |
| Uygur | 12.6 | 11.4 | 2.9 |
| Finland | 9 | 15.7 | 2.8 |
| Poland | 10.7 | 9.9 | 2.1 |
DQ2.5/DQ8 Heterozygotes
The distribution of this phenotype is largely the result of admixtures between peoples of eastern or central Asian origin and peoples of western or central Asian origin. The highest frequencies, by random mating, are expected in Sweden, but pockets of high levels also occur in Mexico, and a larger range risk exists in Central Asia.
Diseases that appear to be increased in Heterozygotes are Celiac Disease and Type 1 Diabetes. New evidence is showing an increased risk for late onset Type 1 diabetes in Heterozygotes (which includes ambiguous Type I/Type II diabetes). 95% of Celiac Disease patients are positive for DQ2 or DQ8.[6]
References
- ↑ Klitz W, Maiers M, Spellman S, et al. (October 2003). "New HLA haplotype frequency reference standards: high-resolution and large sample typing of HLA DR-DQ haplotypes in a sample of European Americans". Tissue Antigens. 62 (4): 296–307. doi:10.1034/j.1399-0039.2003.00103.x. PMID 12974796.
- ↑ Jores RD, Frau F, Cucca F, et al. (2007). "HLA-DQB1*0201 homozygosis predisposes to severe intestinal damage in celiac disease". Scand. J. Gastroenterol. 42 (1): 48–53. doi:10.1080/00365520600789859. PMID 17190762.
- ↑ Al-Toma A, Verbeek WH, Hadithi M, von Blomberg BM, Mulder CJ (2007). "Survival in refractory coeliac disease and enteropathy‐associated T‐cell lymphoma: retrospective evaluation of single‐centre experience". Gut. 56 (10): 1373–8. doi:10.1136/gut.2006.114512. PMC 2000250. PMID 17470479.
- ↑ Atassi MZ, Oshima M, Deitiker P (2001). "n the initial trigger of myasthenia gravis and suppression of the disease by antibodies against the MHC peptide region involved in the presentation of a pathogenic T-cell epitope". Crit Rev Immunol. 21 (1–3): 1–27. PMID 11642597.
- ↑ Deitiker PR, Oshima M, Smith RG, Mosier DR, Atassi MZ (2006). "Subtle differences in HLA DQ haplotype-associated presentation of AChR alpha-chain peptides may suffice to mediate myasthenia gravis". Autoimmunity. 39 (4): 277–288. doi:10.1080/08916930600738581. PMID 16891216.
- ↑ Fasano, Alessio (2011). "Dr". Physiol Rev. 91.