Aconitase: Difference between revisions
(Rescuing 2 sources and tagging 0 as dead. #IABot (v2.0beta9)) |
Matt Pijoan (talk | contribs) m (1 revision imported) |
(No difference)
| |
Latest revision as of 12:19, 9 January 2019
| aconitate hydratase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| File:7ACN.jpg Illustration of pig aconitase in complex with the [Fe4S4] cluster. The protein is colored by secondary structure, and iron atoms are blue and the sulfur red.[1] | |||||||||
| Identifiers | |||||||||
| EC number | 4.2.1.3 | ||||||||
| CAS number | 9024-25-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Aconitase family (aconitate hydratase) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| File:PDB 1aco EBI.jpg Structure of aconitase.[2] | |||||||||
| Identifiers | |||||||||
| Symbol | Aconitase | ||||||||
| Pfam | PF00330 | ||||||||
| InterPro | IPR001030 | ||||||||
| PROSITE | PDOC00423 | ||||||||
| SCOP | 1aco | ||||||||
| SUPERFAMILY | 1aco | ||||||||
| |||||||||
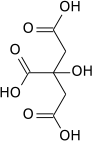
Aconitase (aconitate hydratase; EC 4.2.1.3) is an enzyme that catalyses the stereo-specific isomerization of citrate to isocitrate via cis-aconitate in the tricarboxylic acid cycle, a non-redox-active process.[3][4][5]
Structure
Aconitase, displayed in the structures in the right margin of this page, has two slightly different structures, depending on whether it is activated or inactivated.[6][7] In the inactive form, its structure is divided into four domains.[6] Counting from the N-terminus, only the first three of these domains are involved in close interactions with the [3Fe-4S] cluster, but the active site consists of residues from all four domains, including the larger C-terminal domain.[6] The Fe-S cluster and a SO42− anion also reside in the active site.[6] When the enzyme is activated, it gains an additional iron atom, creating a [4Fe-4S] cluster.[7][8] However, the structure of the rest of the enzyme is nearly unchanged; the conserved atoms between the two forms are in essentially the same positions, up to a difference of 0.1 angstroms.[7]
Function
In contrast with the majority of iron-sulfur proteins that function as electron carriers, the iron-sulfur cluster of aconitase reacts directly with an enzyme substrate. Aconitase has an active [Fe4S4]2+ cluster, which may convert to an inactive [Fe3S4]+ form. Three cysteine (Cys) residues have been shown to be ligands of the [Fe4S4] centre. In the active state, the labile iron ion of the [Fe4S4] cluster is not coordinated by Cys but by water molecules.
The iron-responsive element-binding protein (IRE-BP) and 3-isopropylmalate dehydratase (α-isopropylmalate isomerase; EC 4.2.1.33), an enzyme catalysing the second step in the biosynthesis of leucine, are known aconitase homologues. Iron regulatory elements (IREs) constitute a family of 28-nucleotide, non-coding, stem-loop structures that regulate iron storage, heme synthesis and iron uptake. They also participate in ribosome binding and control the mRNA turnover (degradation). The specific regulator protein, the IRE-BP, binds to IREs in both 5' and 3' regions, but only to RNA in the apo form, without the Fe-S cluster. Expression of IRE-BP in cultured cells has revealed that the protein functions either as an active aconitase, when cells are iron-replete, or as an active RNA-binding protein, when cells are iron-depleted. Mutant IRE-BPs, in which any or all of the three Cys residues involved in Fe-S formation are replaced by serine, have no aconitase activity, but retain RNA-binding properties.
Aconitase is inhibited by fluoroacetate, therefore fluoroacetate is poisonous. Fluoroacetate, in the citric acid cycle,can innocently enter as fluorocitrate. However, aconitase cannot bind this substrate and thus the citric acid cycle is halted. The iron sulfur cluster is highly sensitive to oxidation by superoxide.[9]
Mechanism
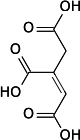
Aconitase employs a dehydration-hydration mechanism.[10] The catalytic residues involved are His-101 and Ser-642.[10] His-101 protonates the hydroxyl group on C3 of citrate, allowing it to leave as water, and Ser-642 concurrently abstracts the proton on C2, forming a double bond between C2 and C3, forming a cis-aconitate intermediate.[10][13] At this point, the intermediate is rotated 180°.[10] This rotation is referred to as a "flip."[11] Because of this flip, the intermediate is said to move from a "citrate mode" to a "isocitrate mode."[14]
How exactly this flip occurs is debatable. One theory is that, in the rate-limiting step of the mechanism, the cis-aconitate is released from the enzyme, then reattached in the isocitrate mode to complete the reaction.[14] This rate-liming step ensures that the right stereochemistry, specifically (2R,3S), is formed in the final product.[14][15] Another hypothesis is that cis-aconitate stays bound to the enzyme while it flips from the citrate to the isocitrate mode.[10]
In either case, flipping cis-aconitate allows the dehydration and hydration steps to occur on opposite faces of the intermediate.[10] Aconitase catalyzes trans elimination/addition of water, and the flip guarantees that the correct stereochemistry is formed in the product.[10][11] To complete the reaction, the serine and histidine residues reverse their original catalytic actions: the histidine, now basic, abstracts a proton from water, priming it as a nucleophile to attack at C2, and the protonated serine is deprotonated by the cis-aconitate double bond to complete the hydration, producing isocitrate.[10]
Family members
Aconitases are expressed in bacteria to humans. Humans express the following two aconitase isozymes:
|
| ||||||||||||||||||||||||||||||||||||||||||||||||
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
- ↑ The interactive pathway map can be edited at WikiPathways: "TCACycle_WP78".
References
- ↑ PDB: 7ACN; Lauble, H.; Kennedy, M. C.; Beinert, H.; Stout, C. D. (1992). "Crystal structures of aconitase with isocitrate and nitroisocitrate bound". Biochemistry. 31 (10): 2735–48. doi:10.1021/bi00125a014. PMID 1547214.
- ↑ PDB: 1ACO; Lauble, H; Kennedy, MC; Beinert, H; Stout, CD (1994). "Crystal Structures of Aconitase with Trans-aconitate and Nitrocitrate Bound". Journal of Molecular Biology. 237 (4): 437–51. doi:10.1006/jmbi.1994.1246. PMID 8151704.
- ↑ Beinert H, Kennedy MC (Dec 1993). "Aconitase, a two-faced protein: enzyme and iron regulatory factor". FASEB Journal. 7 (15): 1442–9. PMID 8262329.
- ↑ Flint, Dennis H.; Allen, Ronda M. (1996). "Iron−Sulfur Proteins with Nonredox Functions". Chemical Reviews. 96 (7): 2315–34. doi:10.1021/cr950041r.
- ↑ Beinert H, Kennedy MC, Stout CD (Nov 1996). "Aconitase as Ironminus signSulfur Protein, Enzyme, and Iron-Regulatory Protein". Chemical Reviews. 96 (7): 2335–2374. doi:10.1021/cr950040z. PMID 11848830.
- ↑ 6.0 6.1 6.2 6.3 Robbins AH, Stout CD (1989). "The structure of aconitase". Proteins. 5 (4): 289–312. doi:10.1002/prot.340050406. PMID 2798408.
- ↑ 7.0 7.1 7.2 Robbins AH, Stout CD (May 1989). "Structure of activated aconitase: formation of the [4Fe-4S] cluster in the crystal". Proceedings of the National Academy of Sciences of the United States of America. 86 (10): 3639–43. doi:10.1073/pnas.86.10.3639. PMC 287193. PMID 2726740.
- ↑ Lauble H, Kennedy MC, Beinert H, Stout CD (Mar 1992). "Crystal structures of aconitase with isocitrate and nitroisocitrate bound". Biochemistry. 31 (10): 2735–48. doi:10.1021/bi00125a014. PMID 1547214.
- ↑ Gardner, Paul R. (2002). "Aconitase: Sensitive target and measure of superoxide". Superoxide Dismutase. Methods in Enzymology. 349. pp. 9–23. doi:10.1016/S0076-6879(02)49317-2. ISBN 978-0-12-182252-1.
- ↑ 10.0 10.1 10.2 10.3 10.4 10.5 10.6 10.7 10.8 Takusagawa F. "Chapter 16: Citric Acid Cycle" (PDF). Takusagawa’s Note. The University of Kansas. Archived from the original (PDF) on 2012-03-24. Retrieved 2011-07-10.
- ↑ 11.0 11.1 11.2 Beinert H, Kennedy MC, Stout CD (Nov 1996). "Aconitase as Ironminus signSulfur Protein, Enzyme, and Iron-Regulatory Protein" (PDF). Chemical Reviews. 96 (7): 2335–2374. doi:10.1021/cr950040z. PMID 11848830. Archived from the original (PDF) on 2011-08-11. Retrieved 2011-05-16.
- ↑ 12.0 12.1 PDB: 1C96; Lloyd SJ, Lauble H, Prasad GS, Stout CD (December 1999). "The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex". Protein Sci. 8 (12): 2655–62. doi:10.1110/ps.8.12.2655. PMC 2144235. PMID 10631981.
- ↑ Han D, Canali R, Garcia J, Aguilera R, Gallaher TK, Cadenas E (Sep 2005). "Sites and mechanisms of aconitase inactivation by peroxynitrite: modulation by citrate and glutathione". Biochemistry. 44 (36): 11986–96. doi:10.1021/bi0509393. PMID 16142896.
- ↑ 14.0 14.1 14.2 Lauble H, Stout CD (May 1995). "Steric and conformational features of the aconitase mechanism". Proteins. 22 (1): 1–11. doi:10.1002/prot.340220102. PMID 7675781.
- ↑ "Aconitase family". The Prosthetic groups and Metal Ions in Protein Active Sites Database Version 2.0. The University of Leeds. 1999-02-02. Archived from the original on 2011-06-08. Retrieved 2011-07-10.
Further reading
- Frishman D, Hentze MW (Jul 1996). "Conservation of aconitase residues revealed by multiple sequence analysis. Implications for structure/function relationships". European Journal of Biochemistry / FEBS. 239 (1): 197–200. doi:10.1111/j.1432-1033.1996.0197u.x. PMID 8706708.
External links
- Aconitase at the US National Library of Medicine Medical Subject Headings (MeSH)
- Proteopedia Aconitase - the Aconitase structure in interactive 3D