C-Met: Difference between revisions
m (Robot: Automated text replacement (-{{WikiDoc Cardiology Network Infobox}} +, -<references /> +{{reflist|2}}, -{{reflist}} +{{reflist|2}})) |
imported>OAbot m (Open access bot: add pmc identifier to citation with #oabot.) |
||
| (One intermediate revision by one other user not shown) | |||
| Line 1: | Line 1: | ||
{{Use dmy dates|date=June 2013}} | |||
{{ | {{Infobox_gene}} | ||
'''c-Met''', also called '''tyrosine-protein kinase Met''' or '''hepatocyte growth factor receptor''' ('''HGFR'''),<ref name="pmid1846706">{{cite journal | vauthors = Bottaro DP, Rubin JS, Faletto DL, Chan AM, Kmiecik TE, Vande Woude GF, Aaronson SA | author-link7 = Stuart A. Aaronson | title = Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product | journal = Science | volume = 251 | issue = 4995 | pages = 802–4 | date = February 1991 | pmid = 1846706 | doi = 10.1126/science.1846706 }}</ref><ref name="pmid1611909">{{cite journal | vauthors = Galland F, Stefanova M, Lafage M, Birnbaum D | title = Localization of the 5' end of the MCF2 oncogene to human chromosome 15q15----q23 | journal = Cytogenet. Cell Genet. | volume = 60 | issue = 2 | pages = 114–6 | year = 1992 | pmid = 1611909 | doi = 10.1159/000133316 }}</ref> is a [[protein]] that in humans is encoded by the '''''MET''''' [[gene]]. The protein possesses [[tyrosine kinase]] activity.<ref name="pmid1531516">{{cite journal | vauthors = Cooper CS | title = The met oncogene: from detection by transfection to transmembrane receptor for hepatocyte growth factor | journal = Oncogene | volume = 7 | issue = 1 | pages = 3–7 | date = January 1992 | pmid = 1531516 | doi = }}</ref> The primary single chain precursor protein is post-translationally cleaved to produce the alpha and beta subunits, which are disulfide linked to form the mature receptor. | |||
| | |||
}} | |||
{{ | |||
| | |||
| | |||
}} | |||
MET is a single pass tyrosine kinase receptor essential for embryonic development, organogenesis and wound healing. [[Hepatocyte growth factor|Hepatocyte growth factor/Scatter Factor]] (HGF/SF) and its splicing isoform (NK1, NK2) are the only known ligands of the MET receptor. MET is normally expressed by cells of [[epithelium|epithelial]] origin, while expression of HGF/SF is restricted to cells of [[mesenchymal stem cell|mesenchymal]] origin. When HGF/SF binds its cognate receptor MET it induces its dimerization through a not yet completely understood mechanism leading to its activation. | |||
Abnormal MET activation in cancer correlates with poor prognosis, where aberrantly active MET triggers tumor growth, formation of new blood vessels ([[angiogenesis]]) that supply the tumor with nutrients, and cancer spread to other organs ([[metastasis]]). MET is deregulated in many types of human malignancies, including cancers of kidney, liver, stomach, breast, and brain. Normally, only [[stem cells]] and [[progenitor cells]] express MET, which allows these cells to grow invasively in order to generate new tissues in an embryo or regenerate damaged tissues in an adult. However, [[cancer stem cells]] are thought to hijack the ability of normal stem cells to express MET, and thus become the cause of cancer persistence and spread to other sites in the body. Both the overexpression of Met/HGFR, as well as its [[Autocrine signalling|autocrine]] activation by co-expression of its hepatocyte growth factor ligand, have been implicated in oncogenesis.<ref>{{cite journal | vauthors = Johnson M, Koukoulis G, Kochhar K, Kubo C, Nakamura T, Iyer A | title = Selective tumorigenesis in non-parenchymal liver epithelial cell lines by hepatocyte growth factor transfection | journal = Cancer Letters | volume = 96 | issue = 1 | pages = 37–48 | date = Sep 1995 | pmid = 7553606 | doi=10.1016/0304-3835(95)03915-j}}</ref><ref>{{cite journal | vauthors = Kochhar KS, Johnson ME, Volpert O, Iyer AP | title = Evidence for autocrine basis of transformation in NIH-3T3 cells transfected with met/HGF receptor gene | journal = Growth Factors | volume = 12 | issue = 4 | pages = 303–13 | date = 1995 | pmid = 8930021 | doi=10.3109/08977199509028968}}</ref> | |||
| | |||
== | |||
== | Various mutations in the MET gene are associated with [[Renal cell carcinoma#Classification|papillary renal carcinoma]].<ref>{{Cite web| title = Entrez Gene: MET met proto-oncogene (hepatocyte growth factor receptor)| url = https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=4233| accessdate = }}</ref> | ||
== Gene == | |||
MET proto-oncogene ([https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=4233&ordinalpos=1&itool=EntrezSystem2.PEntrez.Gene.Gene_ResultsPanel.Gene_RVDocSum GeneID: 4233]) has a total length of 125,982 bp, and it is located in the 7q31 locus of chromosome 7.<ref name="pmid4069211">{{cite journal | vauthors = Dean M, Park M, Le Beau MM, Robins TS, Diaz MO, Rowley JD, Blair DG, Vande Woude GF | title = The human met oncogene is related to the tyrosine kinase oncogenes | journal = Nature | volume = 318 | issue = 6044 | pages = 385–8 | year = 1985 | pmid = 4069211 | doi = 10.1038/318385a0 }}</ref> MET is transcribed into a 6,641 bp mature mRNA, which is then translated into a 1,390 amino-acid MET protein. | |||
== | <!-- Deleted image removed: [[File:Met Gene.jpg|thumb|780px|left|Structure of human ''MET'' proto-oncogene ([https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=4233&ordinalpos=1&itool=EntrezSystem2.PEntrez.Gene.Gene_ResultsPanel.Gene_RVDocSum NCBI])|{{Deletable image-caption|1=Wednesday, 20 August 2008|date=May 2012}}]] --> | ||
== Protein == | |||
[[File:Met Structure.jpg|thumb|400px|Schematic structure of MET protein <ref name="Gentile_2008"/>]] | |||
- | MET is a [[receptor tyrosine kinase]] (RTK) that is produced as a single-chain precursor. The precursor is proteolytically cleaved at a [[furin]] site to yield a highly glycosylated extracellular α-subunit and a transmembrane β-subunit, which are linked together by a [[disulfide bridge]].<ref name="Birchmeier_2003">{{cite journal | vauthors = Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF | title = Met, metastasis, motility and more | journal = Nat. Rev. Mol. Cell Biol. | volume = 4 | issue = 12 | pages = 915–25 | date = December 2003 | pmid = 14685170 | doi = 10.1038/nrm1261 }}</ref> | ||
=== Extracellular === | |||
==See also== | * Region of homology to [[semaphorin]]s (Sema domain), which includes the full α-chain and the N-terminal part of the β-chain | ||
* Cysteine-rich MET-related sequence (MRS domain) | |||
* Glycine-proline-rich repeats (G-P repeats) | |||
* Four immunoglobulin-like structures (Ig domains), a typical protein-protein interaction region.<ref name="Birchmeier_2003"/> | |||
=== Intracellular === | |||
A Juxtamembrane segment that contains: | |||
* a serine residue (Ser 985), which inhibits the receptor kinase activity upon phosphorylation<ref name="pmid8294430">{{cite journal | vauthors = Gandino L, Longati P, Medico E, Prat M, Comoglio PM | title = Phosphorylation of serine 985 negatively regulates the hepatocyte growth factor receptor kinase | journal = J. Biol. Chem. | volume = 269 | issue = 3 | pages = 1815–20 | date = January 1994 | pmid = 8294430 | doi = }}</ref> | |||
* a tyrosine (Tyr 1003), which is responsible for MET [[polyubiquitination]], [[endocytosis]], and degradation upon interaction with the [[ubiquitin ligase]] CBL<ref name="pmid11741535">{{cite journal | vauthors = Peschard P, Fournier TM, Lamorte L, Naujokas MA, Band H, Langdon WY, Park M | title = Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein | journal = Mol. Cell | volume = 8 | issue = 5 | pages = 995–1004 | date = November 2001 | pmid = 11741535 | doi = 10.1016/S1097-2765(01)00378-1 }}</ref> | |||
* Tyrosine kinase domain, which mediates MET biological activity. Following MET activation, transphosphorylation occurs on Tyr 1234 and Tyr 1235 | |||
* C-terminal region contains two crucial tyrosines (Tyr 1349 and Tyr 1356), which are inserted into the multisubstrate docking site, capable of recruiting downstream adapter proteins with [[SH2 domain|Src homology-2]] (SH2) domains.<ref name="pmid7513258">{{cite journal | vauthors = Ponzetto C, Bardelli A, Zhen Z, Maina F, dalla Zonca P, Giordano S, Graziani A, Panayotou G, Comoglio PM | title = A multifunctional docking site mediates signaling and transformation by the hepatocyte growth factor/scatter factor receptor family | journal = Cell | volume = 77 | issue = 2 | pages = 261–71 | date = April 1994 | pmid = 7513258 | doi = 10.1016/0092-8674(94)90318-2 }}</ref> The two tyrosines of the docking site have been reported to be necessary and sufficient for the signal transduction both ''in vitro''.<ref name="pmid7513258"/><ref name="b">{{cite journal | vauthors = Maina F, Casagranda F, Audero E, Simeone A, Comoglio PM, Klein R, Ponzetto C | title = Uncoupling of Grb2 from the Met receptor in vivo reveals complex roles in muscle development | journal = Cell | volume = 87 | issue = 3 | pages = 531–42 | date = November 1996 | pmid = 8898205 | doi = 10.1016/S0092-8674(00)81372-0 }}</ref> | |||
== MET signaling pathway == | |||
[[File:MET Signalosome.jpg|thumb|400px|MET signaling complex<ref name="Abounader_2004"/>]] | |||
<!-- Deleted image removed: [[File:Met Pathway.gif|thumb|520px|Human MET signaling pathway, according to the Cancer Genome Anatomy Project [http://cgap.nci.nih.gov/Pathways/BioCarta/h_metPathway (CGAP)]]] --> | |||
MET activation by its ligand [[Hepatocyte growth factor|HGF]] induces MET kinase catalytic activity, which triggers transphosphorylation of the tyrosines Tyr 1234 and Tyr 1235. These two tyrosines engage various signal transducers,<ref>{{cite journal | vauthors = Johnson M, Kochhar K, Nakamura T, Iyer A | title = Hepatocyte growth factor-induced signal transduction in two normal mouse epithelial cell lines | journal = Biochemistry and Molecular Biology International | volume = 36 | issue = 3 | pages = 465–74 | date = Jul 1995 | pmid = 7549943 | url = https://www.ncbi.nlm.nih.gov/pubmed/?term=7549943 }}</ref> thus initiating a whole spectrum of biological activities driven by MET, collectively known as the invasive growth program. The transducers interact with the intracellular multisubstrate docking site of MET either directly, such as [[GRB2]], [[Src homology 2 domain-containing|SHC]],<ref name="B">{{cite journal | vauthors = Pelicci G, Giordano S, Zhen Z, Salcini AE, Lanfrancone L, Bardelli A, Panayotou G, Waterfield MD, Ponzetto C, Pelicci PG | title = The motogenic and mitogenic responses to HGF are amplified by the Shc adaptor protein | journal = Oncogene | volume = 10 | issue = 8 | pages = 1631–8 | date = April 1995 | pmid = 7731718 | doi = }}</ref> [[Src (gene)|SRC]], and the p85 regulatory subunit of phosphatidylinositol-3 kinase ([[PI3K]]),<ref name=B/> or indirectly through the scaffolding protein Gab1<ref name="pmid8906793">{{cite journal | vauthors = Weidner KM, Di Cesare S, Sachs M, Brinkmann V, Behrens J, Birchmeier W | title = Interaction between Gab1 and the c-Met receptor tyrosine kinase is responsible for epithelial morphogenesis | journal = Nature | volume = 384 | issue = 6605 | pages = 173–6 | date = November 1996 | pmid = 8906793 | doi = 10.1038/384173a0 }}</ref> | |||
Tyr 1349 and Tyr 1356 of the multisubstrate docking site are both involved in the interaction with GAB1, SRC, and SHC, while only Tyr 1356 is involved in the recruitment of GRB2, phospholipase C γ (PLC-γ), p85, and SHP2.<ref name="pmid11114738">{{cite journal | vauthors = Furge KA, Zhang YW, Vande Woude GF | title = Met receptor tyrosine kinase: enhanced signaling through adapter proteins | journal = Oncogene | volume = 19 | issue = 49 | pages = 5582–9 | date = November 2000 | pmid = 11114738 | doi = 10.1038/sj.onc.1203859 }}</ref> | |||
GAB1 is a key coordinator of the cellular responses to MET and binds the MET intracellular region with high [[avidity]], but low [[Receptor affinity|affinity]].<ref name="pmid11313945">{{cite journal | vauthors = Gual P, Giordano S, Anguissola S, Parker PJ, Comoglio PM | title = Gab1 phosphorylation: a novel mechanism for negative regulation of HGF receptor signaling | journal = Oncogene | volume = 20 | issue = 2 | pages = 156–66 | date = January 2001 | pmid = 11313945 | doi = 10.1038/sj.onc.1204047 }}</ref> Upon interaction with MET, GAB1 becomes phosphorylated on several tyrosine residues which, in turn, recruit a number of signalling effectors, including [[PI3K]], SHP2, and PLC-γ. GAB1 phosphorylation by MET results in a sustained signal that mediates most of the downstream signaling pathways.<ref name="pmid10734310">{{cite journal | vauthors = Gual P, Giordano S, Williams TA, Rocchi S, Van Obberghen E, Comoglio PM | title = Sustained recruitment of phospholipase C-gamma to Gab1 is required for HGF-induced branching tubulogenesis | journal = Oncogene | volume = 19 | issue = 12 | pages = 1509–18 | date = March 2000 | pmid = 10734310 | doi = 10.1038/sj.onc.1203514 }}</ref> | |||
===Activation of signal transduction=== | |||
MET engagement activates multiple [[signal transduction]] pathways: | |||
* The [[Ras (protein)|RAS]] pathway mediates HGF-induced [[scattering]] and [[cell growth|proliferation]] signals, which lead to [[branching morphogenesis]].<ref name="pmid15239951">{{cite journal | vauthors = O'Brien LE, Tang K, Kats ES, Schutz-Geschwender A, Lipschutz JH, Mostov KE | title = ERK and MMPs sequentially regulate distinct stages of epithelial tubule development | journal = Dev. Cell | volume = 7 | issue = 1 | pages = 21–32 | date = July 2004 | pmid = 15239951 | doi = 10.1016/j.devcel.2004.06.001 }}</ref> Of note, HGF, differently from most [[mitogens]], induces sustained RAS activation, and thus prolonged [[mitogen-activated protein kinase|MAPK]] activity.<ref name="pmid7834738">{{cite journal | vauthors = Marshall CJ | title = Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation | journal = Cell | volume = 80 | issue = 2 | pages = 179–85 | date = January 1995 | pmid = 7834738 | doi = 10.1016/0092-8674(95)90401-8 }}</ref> | |||
* The [[phosphoinositide 3-kinase|PI3K]] pathway is activated in two ways: PI3K can be either downstream of RAS, or it can be recruited directly through the multifunctional docking site.<ref name="pmid1718989">{{cite journal | vauthors = Graziani A, Gramaglia D, Cantley LC, Comoglio PM | title = The tyrosine-phosphorylated hepatocyte growth factor/scatter factor receptor associates with phosphatidylinositol 3-kinase | journal = J. Biol. Chem. | volume = 266 | issue = 33 | pages = 22087–90 | date = November 1991 | pmid = 1718989 | doi = }}</ref> Activation of the PI3K pathway is currently associated with [[cell motility]] through remodeling of adhesion to the extracellular matrix as well as localized recruitment of transducers involved in cytoskeletal reorganization, such as [[RAC1]] and [[PAK1|PAK]]. PI3K activation also triggers a [[wiktionary:survival|survival]] signal due to activation of the [[AKT]] pathway.<ref name="Gentile_2008">{{cite journal | vauthors = Gentile A, Trusolino L, Comoglio PM | title = The Met tyrosine kinase receptor in development and cancer | journal = Cancer Metastasis Rev. | volume = 27 | issue = 1 | pages = 85–94 | date = March 2008 | pmid = 18175071 | doi = 10.1007/s10555-007-9107-6 }}</ref> | |||
* The [[STAT protein|STAT]] pathway, together with the sustained MAPK activation, is necessary for the HGF-induced [[branching morphogenesis]]. MET activates the [[STAT3]] [[transcription factor]] directly, through an [[SH2 domain]].<ref name="pmid9440692">{{cite journal | vauthors = Boccaccio C, Andò M, Tamagnone L, Bardelli A, Michieli P, Battistini C, Comoglio PM | title = Induction of epithelial tubules by growth factor HGF depends on the STAT pathway | journal = Nature | volume = 391 | issue = 6664 | pages = 285–8 | date = January 1998 | pmid = 9440692 | doi = 10.1038/34657 }}</ref> | |||
* The [[beta-catenin]] pathway, a key component of the [[Wnt signaling pathway]], translocates into the nucleus following MET activation and participates in transcriptional regulation of numerous genes.<ref name="pmid11929826">{{cite journal | vauthors = Monga SP, Mars WM, Pediaditakis P, Bell A, Mulé K, Bowen WC, Wang X, Zarnegar R, Michalopoulos GK | title = Hepatocyte growth factor induces Wnt-independent nuclear translocation of beta-catenin after Met-beta-catenin dissociation in hepatocytes | journal = Cancer Res. | volume = 62 | issue = 7 | pages = 2064–71 | date = April 2002 | pmid = 11929826 | doi = }}</ref> | |||
* The [[Notch signaling pathway|Notch]] pathway, through transcriptional activation of [[Delta (ligand)|Delta ligand]] (see [[DLL3]]).<ref name="Abounader_2004"/><ref name="pmid18369158">{{cite journal | vauthors = Gude NA, Emmanuel G, Wu W, Cottage CT, Fischer K, Quijada P, Muraski JA, Alvarez R, Rubio M, Schaefer E, Sussman MA | title = Activation of Notch-mediated protective signaling in the myocardium | journal = Circ. Res. | volume = 102 | issue = 9 | pages = 1025–35 | date = May 2008 | pmid = 18369158 | doi = 10.1161/CIRCRESAHA.107.164749 | pmc = 3760732 }}</ref> | |||
[[File:MET Interplay.jpg|thumb|400px|Interplay between MET, beta catenin, Wnt, and Notch signaling pathways<ref name="Abounader_2004"/>]] | |||
== Role in development == | |||
MET mediates a complex program known as invasive growth.<ref name="Gentile_2008"/> Activation of MET triggers [[mitogenesis]], and [[morphogenesis]].<ref>{{cite journal | vauthors = Johnson M, Koukoulis G, Matsumoto K, Nakamura T, Iyer A | title = Hepatocyte growth factor induces proliferation and morphogenesis in nonparenchymal epithelial liver cells | journal = Hepatology | volume = 17 | issue = 6 | pages = 1052–61 | date = Jun 1993 | pmid = 8514254 | doi=10.1016/0270-9139(93)90122-4}}</ref><ref name="urlcmetfields">{{Cite web| url = http://www.healthvalue.net/cmetfields.html | title = he fields of HGF/c-Met involvement | authorlink = | coauthors = | date = | work = | publisher = HealthValue | pages = | archiveurl = | archivedate = | quote = | accessdate = 2009-06-13}}</ref> | |||
During [[embryonic development]], transformation of the flat, two-layer [[germinal disc]] into a three-dimensional body depends on transition of some cells from an [[epithelium|epithelial]] [[phenotype]] to spindle-shaped cells with motile behaviour, a [[Mesenchymal stem cell|mesenchymal]] [[phenotype]]. This process is referred to as [[epithelial-mesenchymal transition]] (EMT).<ref name="Boccaccio_2006">{{cite journal | vauthors = Boccaccio C, Comoglio PM | title = Invasive growth: a MET-driven genetic programme for cancer and stem cells | journal = Nat. Rev. Cancer | volume = 6 | issue = 8 | pages = 637–45 | date = August 2006 | pmid = 16862193 | doi = 10.1038/nrc1912 }}</ref> Later in embryonic development, MET is crucial for [[gastrulation]], [[angiogenesis]], [[myoblast]] migration, [[bone remodeling]], and [[nerve]] sprouting among others.<ref name="pmid9789329">{{cite journal | vauthors = Birchmeier C, Gherardi E | title = Developmental roles of HGF/SF and its receptor, the c-Met tyrosine kinase | journal = Trends Cell Biol. | volume = 8 | issue = 10 | pages = 404–10 | date = October 1998 | pmid = 9789329 | doi = 10.1016/S0962-8924(98)01359-2 }}</ref> MET is essential for [[embryogenesis]], because ''MET'' <sup>−/−</sup> mice die ''in utero'' due to severe defects in placental development.<ref name="pmid7854453">{{cite journal | vauthors = Uehara Y, Minowa O, Mori C, Shiota K, Kuno J, Noda T, Kitamura N | title = Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor | journal = Nature | volume = 373 | issue = 6516 | pages = 702–5 | date = February 1995 | pmid = 7854453 | doi = 10.1038/373702a0 }}</ref> Along with [[Ectodysplasin A]], it has been shown to be involved in the differentiation of anatomical placodes, precursors of scales, feathers and hair follicles in vertebrates.<ref name="pmid27336951">{{cite journal | vauthors = Barrow-McGee R, Kishi N, Joffre C, Ménard L, Hervieu A, Bakhouche BA, Noval AJ, Mai A, Guzmán C, Robert-Masson L, Iturrioz X, Hulit J, Brennan CH, Hart IR, Parker PJ, Ivaska J, Kermorgant S | display-authors = 6 | title = Beta 1-integrin-c-Met cooperation reveals an inside-in survival signalling on autophagy-related endomembranes | journal = Nature Communications | volume = 7 | issue = | pages = 11942 | year = 2016 | pmid = 27336951 | doi = 10.1038/ncomms11942 | pmc=4931016}}</ref> Furthermore, MET is required for such critical processes as [[liver]] regeneration and [[wound healing]] during adulthood.<ref name="Gentile_2008"/> | |||
HGF/MET axis is also involved in myocardial development. Both HGF and MET receptor mRNAs are co-expressed in cardiomyocytes from E7.5, soon after the heart has been determined, to E9.5. Transcripts for HGF ligand and receptor are first detected before the occurrence of cardiac beating and looping, and persist throughout the looping stage, when heart morphology begins to elaborate.<ref>{{cite journal | vauthors = Rappolee DA, Iyer A, Patel Y | title = Hepatocyte growth factor and its receptor are expressed in cardiac myocytes during early cardiogenesis | journal = Circulation Research | volume = 78 | issue = 6 | pages = 1028–36 | date = Jun 1996 | pmid = 8635233 | doi = 10.1161/01.RES.78.6.1028 }}</ref> In avian studies, HGF was found in the myocardial layer of the atrioventricular canal, in a developmental stage in which the epithelial to mesenchymal transformation (EMT) of the endocardial cushion occurs.<ref name="pmid9915579">{{cite journal | vauthors = Song W, Majka SM, McGuire PG | title = Hepatocyte growth factor expression in the developing myocardium: evidence for a role in the regulation of the mesenchymal cell phenotype and urokinase expression | journal = Developmental Dynamics | volume = 214 | issue = 1 | pages = 92–100 | year = 1999 | pmid = 9915579 | doi = 10.1002/(SICI)1097-0177(199901)214:1<92::AID-DVDY9>3.0.CO;2-X }}</ref> However, MET is not essential for heart development, since α-MHCMet-KO mice show normal heart development.<ref name=":0">{{cite journal | vauthors = Arechederra M, Carmona R, González-Nuñez M, Gutiérrez-Uzquiza A, Bragado P, Cruz-González I, Cano E, Guerrero C, Sánchez A, López-Novoa JM, Schneider MD, Maina F, Muñoz-Chápuli R, Porras A | title = Met signaling in cardiomyocytes is required for normal cardiac function in adult mice | journal = Biochimica et Biophysica Acta | volume = 1832 | issue = 12 | pages = 2204–15 | date = Dec 2013 | pmid = 23994610 | doi = 10.1016/j.bbadis.2013.08.008 }}</ref> | |||
==Expression== | |||
=== Tissue distribution === | |||
MET is normally expressed by [[epithelial cells]].<ref name="Gentile_2008"/> However, MET is also found on [[endothelial cells]], [[neurons]], [[hepatocytes]], [[hematopoietic]] cells, [[melanocytes]] and neonatal cardiomyocytes.<ref name="urlcmetfields"/><ref>{{cite journal | vauthors = Leo C, Sala V, Morello M, Chiribiri A, Riess I, Mancardi D, Schiaffino S, Ponzetto C, Crepaldi T | title = Activated Met signalling in the developing mouse heart leads to cardiac disease | journal = PLOS ONE | volume = 6 | issue = 2 | pages = e14675 | date = 2011-02-09 | pmid = 21347410 | pmc = 3036588 | doi = 10.1371/journal.pone.0014675 }}</ref> HGF expression is restricted to cells of [[mesenchymal]] origin.<ref name="Boccaccio_2006"/> | |||
=== Transcriptional control === | |||
MET transcription is activated by HGF and several [[growth factors]].<ref name="Shirasaki_1999">{{cite journal | vauthors = Shirasaki F, Makhluf HA, LeRoy C, Watson DK, Trojanowska M | title = Ets transcription factors cooperate with Sp1 to activate the human tenascin-C promoter | journal = Oncogene | volume = 18 | issue = 54 | pages = 7755–64 | date = December 1999 | pmid = 10618716 | doi = 10.1038/sj.onc.1203360 }}</ref> ''MET'' promoter has four putative binding sites for [[ETS transcription factor family|Ets]], a family of [[transcription factors]] that control several invasive growth genes.<ref name="Shirasaki_1999"/> [[ETS1]] activates MET transcription ''in vitro''.<ref name="pmid8934537">{{cite journal | vauthors = Gambarotta G, Boccaccio C, Giordano S, Andŏ M, Stella MC, Comoglio PM | title = Ets up-regulates MET transcription | journal = Oncogene | volume = 13 | issue = 9 | pages = 1911–7 | date = November 1996 | pmid = 8934537 | doi = }}</ref> MET transcription is activated by [[hypoxia-inducible factor 1]] (HIF1), which is activated by low concentration of intracellular oxygen.<ref name="pmid12726861">{{cite journal | vauthors = Pennacchietti S, Michieli P, Galluzzo M, Mazzone M, Giordano S, Comoglio PM | title = Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene | journal = Cancer Cell | volume = 3 | issue = 4 | pages = 347–61 | date = April 2003 | pmid = 12726861 | doi = 10.1016/S1535-6108(03)00085-0 }}</ref> HIF1 can bind to one of the several [[hypoxia (medical)|hypoxia]] response elements (HREs) in the MET promoter.<ref name="Boccaccio_2006"/> Hypoxia also activates transcription factor [[AP-1 transcription factor|AP-1]], which is involved in MET transcription.<ref name="Boccaccio_2006"/> | |||
== Clinical significance == | |||
=== Role in cancer === | |||
MET pathway plays an important role in the development of cancer through: | |||
* activation of key oncogenic pathways ([[Ras (protein)|RAS]], [[Phosphoinositide 3-kinase|PI3K]], [[STAT3]], [[beta-catenin]]); | |||
* [[angiogenesis]] (sprouting of new blood vessels from pre-existing ones to supply a tumor with nutrients); | |||
* scatter (cells dissociation due to [[metalloprotease]] production), which often leads to [[metastasis]].<ref name="urlc-metandcancer">{{Cite web| url = http://www.healthvalue.net/c-metandcancer.html | title = HGF/c-Met and cancer | authorlink = | coauthors = | date = | work = | publisher = HealthValue | pages = | archiveurl = | archivedate = | quote = | accessdate = 2009-06-13}}</ref> | |||
Coordinated down-regulation of both MET and its downstream effector extracellular signal-regulated kinase 2 (ERK2) by [[Mir-199 microRNA precursor|miR-199a*]] may be effective in inhibiting not only cell proliferation but also motility and invasive capabilities of tumor cells.<ref name="pmid18456660">{{cite journal | vauthors = Kim S, Lee UJ, Kim MN, Lee EJ, Kim JY, Lee MY, Choung S, Kim YJ, Choi YC | title = MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2) | journal = J. Biol. Chem. | volume = 283 | issue = 26 | pages = 18158–66 | date = June 2008 | pmid = 18456660 | doi = 10.1074/jbc.M800186200 }}</ref> | |||
MET amplification has emerged as a potential biomarker of the [[clear cell adenocarcinoma|clear cell tumor]] subtype.<ref name="pmid22525820">{{cite journal | vauthors = del Carmen MG, Birrer M, Schorge JO | title = Clear cell carcinoma of the ovary: a review of the literature | journal = Gynecol. Oncol. | volume = 126 | issue = 3 | pages = 481–90 | date = September 2012 | pmid = 22525820 | doi = 10.1016/j.ygyno.2012.04.021 }}</ref> | |||
The amplification of the [[cell surface receptor]] MET often drives resistance to [[EGFR inhibitors|anti-EGFR therapies]] in [[colorectal cancer]].<ref name="pmid23729478">{{cite journal | vauthors = Bardelli A, Corso S, Bertotti A, Hobor S, Valtorta E, Siravegna G, Sartore-Bianchi A, Scala E, Cassingena A, Zecchin D, Apicella M, Migliardi G, Galimi F, Lauricella C, Zanon C, Perera T, Veronese S, Corti G, Amatu A, Gambacorta M, Diaz LA, Sausen M, Velculescu VE, Comoglio P, Trusolino L, Di Nicolantonio F, Giordano S, Siena S | title = Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer | journal = Cancer Discov | volume = 3 | issue = 6 | pages = 658–73 | date = June 2013 | pmid = 23729478 | pmc = 4078408 | doi = 10.1158/2159-8290.CD-12-0558 }}</ref> | |||
=== Role in autism === | |||
The SFARIgene database lists MET with an [[autism]] score of 2.0, which indicates that it is a strong candidate for playing a role in cases of autism. The database also identifies at least one study that found a role for MET in cases of [[schizophrenia]]. The gene was first implicated in autism in a study that identified a polymorphism in the promoter of the MET gene.<ref name="pmid17053076">{{cite journal | vauthors = Campbell DB, Sutcliffe JS, Ebert PJ, Militerni R, Bravaccio C, Trillo S, Elia M, Schneider C, Melmed R, Sacco R, Persico AM, Levitt P | title = A genetic variant that disrupts MET transcription is associated with autism | journal = Proc. Natl. Acad. Sci. U.S.A. | volume = 103 | issue = 45 | pages = 16834–9 | year = 2006 | pmid = 17053076 | doi = 10.1073/pnas.0605296103 | pmc=1838551}}</ref> The polymorphism reduces transcription by 50%. Further, the variant as an autism risk polymorphism has been replicated, and shown to be enriched in children with autism and gastrointestinal disturbances.<ref name="pmid19255034">{{cite journal | vauthors = Campbell DB, Buie TM, Winter H, Bauman M, Sutcliffe JS, Perrin JM, Levitt P | title = Distinct genetic risk based on association of MET in families with co-occurring autism and gastrointestinal conditions | journal = Pediatrics | volume = 123 | issue = 3 | pages = 1018–24 | year = 2009 | pmid = 19255034 | doi = 10.1542/peds.2008-0819 }}</ref> A rare mutation that appears in two family members, one with autism and the other with a social and communication disorder.<ref name="pmid24909855">{{cite journal | vauthors = Lambert N, Wermenbol V, Pichon B, Acosta S, van den Ameele J, Perazzolo C, Messina D, Musumeci MF, Dessars B, De Leener A, Abramowicz M, Vilain C | title = A familial heterozygous null mutation of MET in autism spectrum disorder | journal = Autism Res | volume = 7 | issue = 5 | pages = 617–22 | year = 2014 | pmid = 24909855 | doi = 10.1002/aur.1396 }}</ref> The role of the receptor in brain development is distinct from its role in other developmental processes. Activation of the MET receptor regulates synapse formation<ref name="pmid25471559">{{cite journal | vauthors = Qiu S, Lu Z, Levitt P | title = MET receptor tyrosine kinase controls dendritic complexity, spine morphogenesis, and glutamatergic synapse maturation in the hippocampus | journal = J. Neurosci. | volume = 34 | issue = 49 | pages = 16166–79 | year = 2014 | pmid = 25471559 | doi = 10.1523/JNEUROSCI.2580-14.2014 | pmc=4252539}}</ref><ref name="pmid23787772">{{cite journal | vauthors = Eagleson KL, Milner TA, Xie Z, Levitt P | title = Synaptic and extrasynaptic location of the receptor tyrosine kinase met during postnatal development in the mouse neocortex and hippocampus | journal = J. Comp. Neurol. | volume = 521 | issue = 14 | pages = 3241–59 | year = 2013 | pmid = 23787772 | pmc = 3942873 | doi = 10.1002/cne.23343 }}</ref><ref name="pmid21509596">{{cite journal | vauthors = Judson MC, Eagleson KL, Levitt P | title = A new synaptic player leading to autism risk: Met receptor tyrosine kinase | journal = J Neurodev Disord | volume = 3 | issue = 3 | pages = 282–92 | year = 2011 | pmid = 21509596 | pmc = 3261279 | doi = 10.1007/s11689-011-9081-8 }}</ref><ref name="pmid21490227">{{cite journal | vauthors = Qiu S, Anderson CT, Levitt P, Shepherd GM | title = Circuit-specific intracortical hyperconnectivity in mice with deletion of the autism-associated Met receptor tyrosine kinase | journal = J. Neurosci. | volume = 31 | issue = 15 | pages = 5855–64 | year = 2011 | pmid = 21490227 | pmc = 3086026 | doi = 10.1523/JNEUROSCI.6569-10.2011 }}</ref><ref name="pmid20853516">{{cite journal | vauthors = Judson MC, Eagleson KL, Wang L, Levitt P | title = Evidence of cell-nonautonomous changes in dendrite and dendritic spine morphology in the met-signaling-deficient mouse forebrain | journal = J. Comp. Neurol. | volume = 518 | issue = 21 | pages = 4463–78 | year = 2010 | pmid = 20853516 | pmc = 2952412 | doi = 10.1002/cne.22467 }}</ref> and can impact the development and function of circuits involved in social and emotional behavior.<ref>{{cite journal | pmid = 22958829 | doi=10.1016/j.neuron.2012.07.010 | volume=75 | title=Autism-associated promoter variant in MET impacts functional and structural brain networks | pmc=3454529 | journal=Neuron | pages=904–15 | vauthors=Rudie JD, Hernandez LM, Brown JA, Beck-Pancer D, Colich NL, Gorrindo P, Thompson PM, Geschwind DH, Bookheimer SY, Levitt P, Dapretto M}}</ref> | |||
=== Role in heart function === | |||
In adult mice, MET is required to protect cardiomyocytes by preventing age-related oxidative stress, apoptosis, fibrosis and cardiac dysfunction.<ref name=":0" /> Moreover, MET inhibitors, such as Crizotinib or PF-04254644, have been tested by short-term treatments in cellular and preclinical models, and have been shown to induce cardiomyocytes death through ROS production, activation of caspases, metabolism alteration and blockage of ion channels.<ref>{{cite journal | vauthors = Doherty KR, Wappel RL, Talbert DR, Trusk PB, Moran DM, Kramer JW, Brown AM, Shell SA, Bacus S | title = Multi-parameter in vitro toxicity testing of crizotinib, sunitinib, erlotinib, and nilotinib in human cardiomyocytes | journal = Toxicology and Applied Pharmacology | volume = 272 | issue = 1 | pages = 245–55 | date = October 2013 | pmid = 23707608 | doi = 10.1016/j.taap.2013.04.027 }}</ref><ref>{{cite journal | vauthors = Aguirre SA, Heyen JR, Collette W, Bobrowski W, Blasi ER | title = Cardiovascular effects in rats following exposure to a receptor tyrosine kinase inhibitor | journal = Toxicologic Pathology | volume = 38 | issue = 3 | pages = 416–28 | date = April 2010 | pmid = 20231546 | doi = 10.1177/0192623310364027 }}</ref> | |||
In the injured heart, HGF/MET axis plays important roles in cardioprotection by promoting pro-survival (anti-apoptotic and anti-autophagic) effects in cardiomyocytes, angiogenesis, inhibition of fibrosis, anti-inflammatory and immunomodulatory signals, and regeneration through activation of cardiac stem cells.<ref>{{cite journal | vauthors = Schmoldt A, Benthe HF, Haberland G, Scott WA, Mahoney E, Pounds JG, Long GJ, Rosen JF | title = Cellular and molecular toxicity of lead in bone | journal = Environmental Health Perspectives | volume = 91 | issue = 17 | pages = 17–32 | date = February 1991 | pmid = 2040247 | doi = 10.3390/biomedicines2040247 | pmc=1519349}}</ref><ref>{{cite journal | vauthors = Sala V, Crepaldi T | title = Novel therapy for myocardial infarction: can HGF/Met be beneficial? | journal = Cellular and Molecular Life Sciences | volume = 68 | issue = 10 | pages = 1703–17 | date = May 2011 | pmid = 21327916 | doi = 10.1007/s00018-011-0633-6 }}</ref> | |||
== Interaction with tumour suppressor genes == | |||
=== PTEN === | |||
[[PTEN (gene)|PTEN]] (phosphatase and tensin homolog) is a [[tumor suppressor gene]] encoding a protein PTEN, which possesses lipid and protein phosphatase-dependent as well as phosphatase-independent activities.<ref name="pmid9593664">{{cite journal | vauthors = Maehama T, Dixon JE | title = The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate | journal = J. Biol. Chem. | volume = 273 | issue = 22 | pages = 13375–8 | date = May 1998 | pmid = 9593664 | doi = 10.1074/jbc.273.22.13375 }}</ref> PTEN protein [[phosphatase]] is able to interfere with MET signaling by dephosphorylating either PIP<sub>3</sub> generated by [[PI3K]], or the p52 isoform of [[SHC1|SHC]]. SHC dephosphorylation inhibits recruitment of the [[GRB2]] adapter to activated MET.<ref name="Abounader_2004">{{cite journal | vauthors = Abounader R, Reznik T, Colantuoni C, Martinez-Murillo F, Rosen EM, Laterra J | title = Regulation of c-Met-dependent gene expression by PTEN | journal = Oncogene | volume = 23 | issue = 57 | pages = 9173–82 | date = December 2004 | pmid = 15516982 | doi = 10.1038/sj.onc.1208146 }}</ref> | |||
=== VHL === | |||
There is evidence of correlation between inactivation of [[VHL]] [[tumor suppressor gene]] and increased MET signaling in [[renal cell carcinoma]] (RCC) and also in malignant transformations of the heart.<ref name="pmid15930277">{{cite journal | vauthors = Morris MR, Gentle D, Abdulrahman M, Maina EN, Gupta K, Banks RE, Wiesener MS, Kishida T, Yao M, Teh B, Latif F, Maher ER | title = Tumor suppressor activity and epigenetic inactivation of hepatocyte growth factor activator inhibitor type 2/SPINT2 in papillary and clear cell renal cell carcinoma | journal = Cancer Res. | volume = 65 | issue = 11 | pages = 4598–606 | date = June 2005 | pmid = 15930277 | doi = 10.1158/0008-5472.CAN-04-3371 }}</ref><ref>{{cite journal | vauthors = Lei L, Mason S, Liu D, Huang Y, Marks C, Hickey R, Jovin IS, Pypaert M, Johnson RS, Giordano FJ | title = Hypoxia-inducible factor-dependent degeneration, failure, and malignant transformation of the heart in the absence of the von Hippel-Lindau protein | journal = Molecular and Cellular Biology | volume = 28 | issue = 11 | pages = 3790–803 | date = June 2008 | pmid = 18285456 | pmc = 2423296 | doi = 10.1128/MCB.01580-07 }}</ref> | |||
== Cancer therapies targeting HGF/MET == | |||
[[File:Targeting Met.gif|thumb|400px|Strategies to inhibit biological activity of MET <ref name="Gentile_2008"/>]] | |||
Since tumor invasion and metastasis are the main cause of death in cancer patients, interfering with MET signaling appears to be a promising therapeutic approach. A comprehensive list of HGF and MET targeted experimental therapeutics for oncology now in human clinical trials can be found [https://ccrod.cancer.gov/confluence/display/CCRHGF/Home here]. | |||
=== MET kinase inhibitors === | |||
{{details|c-Met inhibitor}} | |||
Kinase inhibitors are low molecular weight molecules that prevent [[Adenosine triphosphate|ATP]] binding to MET, thus inhibiting receptor transphosphorylation and recruitment of the downstream effectors. The limitations of kinase inhibitors include the facts that they only inhibit kinase-dependent MET activation, and that none of them is fully specific for MET. | |||
* [[K252a]] ([[Fermentek]] Biotechnology) is a [[staurosporine]] analogue isolated from ''Nocardiopsis'' sp. soil [[fungi]], and it is a potent inhibitor of all [[receptor tyrosine kinases]] (RTKs). At nanomolar concentrations, K252a inhibits both the [[wild type]] and the mutant (M1268T) MET function.<ref name="pmid12118367">{{cite journal | vauthors = Morotti A, Mila S, Accornero P, Tagliabue E, Ponzetto C | title = K252a inhibits the oncogenic properties of Met, the HGF receptor | journal = Oncogene | volume = 21 | issue = 32 | pages = 4885–93 | date = July 2002 | pmid = 12118367 | doi = 10.1038/sj.onc.1205622 }}</ref> | |||
* SU11274 ([[SUGEN]]) specifically inhibits MET kinase activity and its subsequent signaling. SU11274 is also an effective inhibitor of the M1268T and H1112Y MET mutants, but not the L1213V and Y1248H mutants.<ref name="pmid15064724">{{cite journal | vauthors = Berthou S, Aebersold DM, Schmidt LS, Stroka D, Heigl C, Streit B, Stalder D, Gruber G, Liang C, Howlett AR, Candinas D, Greiner RH, Lipson KE, Zimmer Y | title = The Met kinase inhibitor SU11274 exhibits a selective inhibition pattern toward different receptor mutated variants | journal = Oncogene | volume = 23 | issue = 31 | pages = 5387–93 | date = July 2004 | pmid = 15064724 | doi = 10.1038/sj.onc.1207691 }}</ref> SU11274 has been demonstrated to inhibit HGF-induced motility and invasion of epithelial and carcinoma cells.<ref name="pmid14617781">{{cite journal | vauthors = Wang X, Le P, Liang C, Chan J, Kiewlich D, Miller T, Harris D, Sun L, Rice A, Vasile S, Blake RA, Howlett AR, Patel N, McMahon G, Lipson KE | title = Potent and selective inhibitors of the Met [hepatocyte growth factor/scatter factor (HGF/SF) receptor] tyrosine kinase block HGF/SF-induced tumor cell growth and invasion | journal = Mol. Cancer Ther. | volume = 2 | issue = 11 | pages = 1085–92 | date = November 2003 | pmid = 14617781 | doi = }}</ref> | |||
* PHA-665752 ([[Pfizer]]) specifically inhibits MET kinase activity, and it has been demonstrated to represses both HGF-dependent and constitutive MET phosphorylation.<ref name="pmid14612533">{{cite journal | vauthors = Christensen JG, Schreck R, Burrows J, Kuruganti P, Chan E, Le P, Chen J, Wang X, Ruslim L, Blake R, Lipson KE, Ramphal J, Do S, Cui JJ, Cherrington JM, Mendel DB | title = A selective small molecule inhibitor of c-Met kinase inhibits c-Met-dependent phenotypes in vitro and exhibits cytoreductive antitumor activity in vivo | journal = Cancer Res. | volume = 63 | issue = 21 | pages = 7345–55 | date = November 2003 | pmid = 14612533 | doi = }}</ref> Furthermore, some tumors harboring ''MET'' amplifications are highly sensitive to treatment with PHA-665752.<ref name="pmid16461907">{{cite journal | vauthors = Smolen GA, Sordella R, Muir B, Mohapatra G, Barmettler A, Archibald H, Kim WJ, Okimoto RA, Bell DW, Sgroi DC, Christensen JG, Settleman J, Haber DA | title = Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752 | journal = Proc. Natl. Acad. Sci. U.S.A. | volume = 103 | issue = 7 | pages = 2316–21 | date = February 2006 | pmid = 16461907 | pmc = 1413705 | doi = 10.1073/pnas.0508776103 }}</ref> | |||
* [[ARQ197]] (ArQule) is a promising selective inhibitor of MET, which entered a phase 2 clinical trial in 2008. (Failed a phase 3 in 2017) | |||
* [[Foretinib]] (XL880, Exelixis) targets multiple [[receptor tyrosine kinases]] (RTKs) with growth-promoting and angiogenic properties. The primary targets of foretinib are MET, [[VEGFR2]], and [[Kinase insert domain receptor|KDR]]. Foretinib has completed a phase 2 clinical trials with indications for papillary [[renal cell carcinoma]], [[gastric cancer]], and [[head and neck cancer]].<ref>[http://www.exelixis.com/eortc/posters/AACR_NCI_EORTC2007_XL880_phase2.pdf Poster]</ref> | |||
* SGX523 (SGX Pharmaceuticals) specifically inhibits MET at low nanomolar concentrations. | |||
* MP470 (SuperGen) is a novel inhibitor of [[c-KIT]], MET, [[PDGFR]], Flt3, and [[AXL receptor tyrosine kinase|AXL]]. Phase I clinical trial of MP470 had been announced in 2007. | |||
=== HGF inhibitors === | |||
Since HGF is the only known ligand of MET, formation of a HGF:MET complex blocks MET [[biological activity]]. For this purpose, truncated HGF, anti-HGF neutralizing antibodies, and an uncleavable form of HGF have been utilized so far. The major limitation of HGF inhibitors is that they block only HGF-dependent MET activation. | |||
* NK4 competes with HGF as it binds MET without inducing receptor activation, thus behaving as a full [[antagonist]]. NK4 is a molecule bearing the N-terminal hairpin and the four kringle domains of HGF. Moreover, NK4 is structurally similar to angiostatins, which is why it possesses anti-angiogenic activity.<ref name="pmid12824898">{{cite journal | vauthors = Matsumoto K, Nakamura T | title = NK4 (HGF-antagonist/angiogenesis inhibitor) in cancer biology and therapeutics | journal = Cancer Sci. | volume = 94 | issue = 4 | pages = 321–7 | date = April 2003 | pmid = 12824898 | doi = 10.1111/j.1349-7006.2003.tb01440.x }}</ref> | |||
* Neutralizing anti-HGF antibodies were initially tested in combination, and it was shown that at least three [[antibodies]], acting on different HGF [[epitopes]], are necessary to prevent MET tyrosine kinase activation.<ref name="pmid11416216">{{cite journal | vauthors = Cao B, Su Y, Oskarsson M, Zhao P, Kort EJ, Fisher RJ, Wang LM, Vande Woude GF | title = Neutralizing monoclonal antibodies to hepatocyte growth factor/scatter factor (HGF/SF) display antitumor activity in animal models | journal = Proc. Natl. Acad. Sci. U.S.A. | volume = 98 | issue = 13 | pages = 7443–8 | date = June 2001 | pmid = 11416216 | pmc = 34688 | doi = 10.1073/pnas.131200498 }}</ref> More recently, it has been demonstrated that fully human [[monoclonal antibodies]] can individually bind and neutralize human HGF, leading to regression of tumors in mouse models.<ref name="pmid16452232">{{cite journal | vauthors = Burgess T, Coxon A, Meyer S, Sun J, Rex K, Tsuruda T, Chen Q, Ho SY, Li L, Kaufman S, McDorman K, Cattley RC, Sun J, Elliott G, Zhang K, Feng X, Jia XC, Green L, Radinsky R, Kendall R | title = Fully human monoclonal antibodies to hepatocyte growth factor with therapeutic potential against hepatocyte growth factor/c-Met-dependent human tumors | journal = Cancer Res. | volume = 66 | issue = 3 | pages = 1721–9 | date = February 2006 | pmid = 16452232 | doi = 10.1158/0008-5472.CAN-05-3329 }}</ref> Two anti-HGF antibodies are currently available: the humanized AV299 (AVEO), and the fully human AMG102 ([[Amgen]]). | |||
* Uncleavable HGF is an engineered form of pro-HGF carrying a single amino-acid substitution, which prevents the maturation of the molecule. Uncleavable HGF is capable of blocking MET-induced biological responses by binding MET with high affinity and displacing mature HGF. Moreover, uncleavable HGF competes with the wild-type endogenous pro-HGF for the catalytic domain of [[proteases]] that cleave HGF precursors. Local and systemic expression of uncleavable HGF inhibits tumor growth and, more importantly, prevents [[metastasis]].<ref name="pmid15545993">{{cite journal | vauthors = Mazzone M, Basilico C, Cavassa S, Pennacchietti S, Risio M, Naldini L, Comoglio PM, Michieli P | title = An uncleavable form of pro-scatter factor suppresses tumor growth and dissemination in mice | journal = J. Clin. Invest. | volume = 114 | issue = 10 | pages = 1418–32 | date = November 2004 | pmid = 15545993 | pmc = 525743 | doi = 10.1172/JCI22235 }}</ref> | |||
=== Decoy MET === | |||
Decoy MET refers to a soluble truncated MET receptor. Decoys are able to inhibit MET activation mediated by both HGF-dependent and independent mechanisms, as decoys prevent both the ligand binding and the MET receptor homodimerization. CGEN241 ([[Compugen (Israeli company)|Compugen]]) is a decoy MET that is highly efficient in inhibiting tumor growth and preventing [[metastasis]] in animal models.<ref name="pmid15261142">{{cite journal | vauthors = Michieli P, Mazzone M, Basilico C, Cavassa S, Sottile A, Naldini L, Comoglio PM | title = Targeting the tumor and its microenvironment by a dual-function decoy Met receptor | journal = Cancer Cell | volume = 6 | issue = 1 | pages = 61–73 | date = July 2004 | pmid = 15261142 | doi = 10.1016/j.ccr.2004.05.032 }}</ref> | |||
=== Immunotherapy targeting MET === | |||
Drugs used for [[immunotherapy]] can act either passively by enhancing the immunologic response to MET-expressing tumor cells, or actively by stimulating [[immune cells]] and altering differentiation/growth of tumor cells.<ref name="Reang_2006">{{cite journal | vauthors = Reang P, Gupta M, Kohli K | title = Biological response modifiers in cancer | journal = MedGenMed | volume = 8 | issue = 4 | pages = 33 | year = 2006 | pmid = 17415315 | pmc = 1868326 | doi = }}</ref> | |||
==== Passive immunotherapy ==== | |||
Administering [[monoclonal antibodies]] (mAbs) is a form of passive immunotherapy. MAbs facilitate destruction of tumor cells by [[complement-dependent cytotoxicity]] (CDC) and cell-mediated cytotoxicity ([[Antibody-dependent cell-mediated cytotoxicity|ADCC]]). In CDC, mAbs bind to specific [[antigen]], leading to activation of the [[complement cascade]], which in turn leads to formation of pores in tumor cells. In ADCC, the Fab domain of a mAb binds to a [[tumor antigen]], and Fc domain binds to Fc receptors present on effector cells ([[phagocytes]] and [[NK cells]]), thus forming a bridge between an effector and a target cells. This induces the effector cell activation, leading to [[phagocytosis]] of the tumor cell by [[neutrophils]] and [[macrophages]]. Furthermore, [[NK cells]] release [[cytotoxic]] molecules, which lyse tumor cells.<ref name="Reang_2006"/> | |||
* DN30 is monoclonal anti-MET antibody that recognizes the extracellular portion of MET. DN30 induces both [[Proteases in angiogenesis#Ectodomain shedding|shedding]] of the MET [[ectodomain]] as well as cleavage of the intracellular domain, which is successively degraded by [[proteasome]] machinery. As a consequence, on one side MET is inactivated, and on the other side the shed portion of extracellular MET hampers activation of other MET receptors, acting as a decoy. DN30 inhibits tumour growth and prevents [[metastasis]] in animal models.<ref name="pmid16547140">{{cite journal | vauthors = Petrelli A, Circosta P, Granziero L, Mazzone M, Pisacane A, Fenoglio S, Comoglio PM, Giordano S | title = Ab-induced ectodomain shedding mediates hepatocyte growth factor receptor down-regulation and hampers biological activity | journal = Proc. Natl. Acad. Sci. U.S.A. | volume = 103 | issue = 13 | pages = 5090–5 | date = March 2006 | pmid = 16547140 | pmc = 1458799 | doi = 10.1073/pnas.0508156103 }}</ref> | |||
* OA-5D5 is one-armed monoclonal anti-MET antibody that was demonstrated to inhibit orthotopic pancreatic<ref name="pmid18519697">{{cite journal | vauthors = Jin H, Yang R, Zheng Z, Romero M, Ross J, Bou-Reslan H, Carano RA, Kasman I, Mai E, Young J, Zha J, Zhang Z, Ross S, Schwall R, Colbern G, Merchant M | title = MetMAb, the one-armed 5D5 anti-c-Met antibody, inhibits orthotopic pancreatic tumor growth and improves survival | journal = Cancer Res. | volume = 68 | issue = 11 | pages = 4360–8 | date = June 2008 | pmid = 18519697 | doi = 10.1158/0008-5472.CAN-07-5960 }}</ref> and glioblastoma<ref name="pmid17062691">{{cite journal | vauthors = Martens T, Schmidt NO, Eckerich C, Fillbrandt R, Merchant M, Schwall R, Westphal M, Lamszus K | title = A novel one-armed anti-c-Met antibody inhibits glioblastoma growth in vivo | journal = Clin. Cancer Res. | volume = 12 | issue = 20 Pt 1 | pages = 6144–52 | date = October 2006 | pmid = 17062691 | doi = 10.1158/1078-0432.CCR-05-1418 }}</ref> tumor growth and to improve survival in tumor xenograft models. OA-5D5 is produced as a recombinant protein in ''Escherichia coli''. It is composed of murine variable domains for the heavy and light chains with human IgG1 constant domains. The antibody blocks HGF binding to MET in a competitive fashion. | |||
==== Active immunotherapy ==== | |||
[[Active immunotherapy]] to MET-expressing tumors can be achieved by administering [[cytokines]], such as [[interferons]] (IFNs) and [[interleukins]] ([[Interleukin 2|IL-2]]), which triggers non-specific stimulation of numerous immune cells. IFNs have been tested as therapies for many types of cancers and have demonstrated therapeutic benefits. IL-2 has been approved by the [[Food and Drug Administration (United States)|U.S. Food and Drug Administration]] (FDA) for the treatment of [[renal cell carcinoma]] and metastatic melanoma, which often have deregulated MET activity.<ref name="Reang_2006"/> | |||
== Interactions == | |||
Met has been shown to [[Protein-protein interaction|interact]] with: | |||
{{div col|colwidth=20em}} | |||
* [[CDH1 (gene)|CDH1]],<ref name = pmid11254878>{{cite journal | vauthors = Davies G, Jiang WG, Mason MD | title = HGF/SF modifies the interaction between its receptor c-Met, and the E-cadherin/catenin complex in prostate cancer cells | journal = Int. J. Mol. Med. | volume = 7 | issue = 4 | pages = 385–8 | year = 2001 | pmid = 11254878 | doi = 10.3892/ijmm.7.4.385 }}</ref> | |||
* [[Cbl gene]],<ref name = pmid11894096>{{cite journal | vauthors = Petrelli A, Gilestro GF, Lanzardo S, Comoglio PM, Migone N, Giordano S | title = The endophilin-CIN85-Cbl complex mediates ligand-dependent downregulation of c-Met | journal = Nature | volume = 416 | issue = 6877 | pages = 187–90 | year = 2002 | pmid = 11894096 | doi = 10.1038/416187a }}</ref><ref name = pmid18273061>{{cite journal | vauthors = Ng C, Jackson RA, Buschdorf JP, Sun Q, Guy GR, Sivaraman J | title = Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates | journal = EMBO J. | volume = 27 | issue = 5 | pages = 804–16 | year = 2008 | pmid = 18273061 | pmc = 2265755 | doi = 10.1038/emboj.2008.18 }}</ref> | |||
* [[GLMN]],<ref name = pmid11571281>{{cite journal | vauthors = Grisendi S, Chambraud B, Gout I, Comoglio PM, Crepaldi T | title = Ligand-regulated binding of FAP68 to the hepatocyte growth factor receptor | journal = J. Biol. Chem. | volume = 276 | issue = 49 | pages = 46632–8 | year = 2001 | pmid = 11571281 | doi = 10.1074/jbc.M104323200 }}</ref> | |||
* [[Grb2]],<ref name = pmid8662889>{{cite journal | vauthors = Ponzetto C, Zhen Z, Audero E, Maina F, Bardelli A, Basile ML, Giordano S, Narsimhan R, Comoglio P | title = Specific uncoupling of GRB2 from the Met receptor. Differential effects on transformation and motility | journal = J. Biol. Chem. | volume = 271 | issue = 24 | pages = 14119–23 | year = 1996 | pmid = 8662889 | doi = 10.1074/jbc.271.24.14119 }}</ref><ref name = pmid9660480>{{cite journal | vauthors = Liang Q, Mohan RR, Chen L, Wilson SE | title = Signaling by HGF and KGF in corneal epithelial cells: Ras/MAP kinase and Jak-STAT pathways | journal = Invest. Ophthalmol. Vis. Sci. | volume = 39 | issue = 8 | pages = 1329–38 | year = 1998 | pmid = 9660480 | doi = }}</ref> | |||
* [[Hepatocyte growth factor]],<ref name = pmid8380735>{{cite journal | vauthors = Comoglio PM | title = Structure, biosynthesis and biochemical properties of the HGF receptor in normal and malignant cells | journal = EXS | volume = 65 | issue = | pages = 131–65 | year = 1993 | pmid = 8380735 | doi = }}</ref><ref name = pmid1655405>{{cite journal | vauthors = Naldini L, Weidner KM, Vigna E, Gaudino G, Bardelli A, Ponzetto C, Narsimhan RP, Hartmann G, Zarnegar R, Michalopoulos GK | title = Scatter factor and hepatocyte growth factor are indistinguishable ligands for the MET receptor | journal = EMBO J. | volume = 10 | issue = 10 | pages = 2867–78 | year = 1991 | pmid = 1655405 | pmc = 452997 | doi = }}</ref> | |||
* [[PTPRM|PTPmu]],<ref name = "pmid10425198">{{cite journal | vauthors = Hiscox S, Jiang WG | title = Association of the HGF/SF receptor, c-met, with the cell-surface adhesion molecule, E-cadherin, and catenins in human tumor cells | journal = Biochem. Biophys. Res. Commun. | volume = 261 | issue = 2 | pages = 406–11 | year = 1999 | pmid = 10425198 | doi = 10.1006/bbrc.1999.1002 }}</ref> and | |||
* [[RANBP9]]<ref name = pmid12147692>{{cite journal | vauthors = Wang D, Li Z, Messing EM, Wu G | title = Activation of Ras/Erk pathway by a novel MET-interacting protein RanBPM | journal = J. Biol. Chem. | volume = 277 | issue = 39 | pages = 36216–22 | year = 2002 | pmid = 12147692 | doi = 10.1074/jbc.M205111200 }}</ref> | |||
{{Div col end}} | |||
== See also == | |||
* [[c-Met inhibitors]] | |||
* [[Tpr-met fusion protein]] | * [[Tpr-met fusion protein]] | ||
==References== | == References == | ||
{{ | {{Reflist|2}} | ||
==Further reading== | == Further reading == | ||
{{ | {{Refbegin| 2}} | ||
* {{cite journal | | * {{cite journal | vauthors = Peruzzi B, Bottaro DP | title = Targeting the c-Met signaling pathway in cancer | journal = Clin. Cancer Res. | volume = 12 | issue = 12 | pages = 3657–60 | year = 2006 | pmid = 16778093 | doi = 10.1158/1078-0432.CCR-06-0818 }} | ||
* Birchmeier | * {{cite journal | vauthors = Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF | title = Met, metastasis, motility and more | journal = Nat. Rev. Mol. Cell Biol. | volume = 4 | issue = 12 | pages = 915–25 | date = December 2003 | pmid = 14685170 | doi = 10.1038/nrm1261 }} | ||
* Zhang, | * {{cite journal | vauthors = Zhang YW, Vande Woude GF | title = HGF/SF-met signaling in the control of branching morphogenesis and invasion | journal = J. Cell. Biochem. | volume = 88 | issue = 2 | pages = 408–17 | date = February 2003 | pmid = 12520544 | doi = 10.1002/jcb.10358 }} | ||
* Paumelle | * {{cite journal | vauthors = Paumelle R, Tulasne D, Kherrouche Z, Plaza S, Leroy C, Reveneau S, Vandenbunder B, Fafeur V, Tulashe D, Reveneau S | title = Hepatocyte growth factor/scatter factor activates the ETS1 transcription factor by a RAS-RAF-MEK-ERK signaling pathway | journal = Oncogene | volume = 21 | issue = 15 | pages = 2309–19 | date = April 2002 | pmid = 11948414 | doi = 10.1038/sj.onc.1205297 }} | ||
* {{cite journal | vauthors = Comoglio PM | title = Structure, biosynthesis and biochemical properties of the HGF receptor in normal and malignant cells | journal = EXS | volume = 65 | issue = | pages = 131–65 | year = 1993 | pmid = 8380735 | doi = }} | |||
* {{cite journal | vauthors = Maulik G, Shrikhande A, Kijima T, Ma PC, Morrison PT, Salgia R | title = Role of the hepatocyte growth factor receptor, c-Met, in oncogenesis and potential for therapeutic inhibition | journal = Cytokine Growth Factor Rev. | volume = 13 | issue = 1 | pages = 41–59 | year = 2002 | pmid = 11750879 | doi = 10.1016/S1359-6101(01)00029-6 }} | |||
*{{cite journal | * {{cite journal | vauthors = Ma PC, Maulik G, Christensen J, Salgia R | title = c-Met: structure, functions and potential for therapeutic inhibition | journal = Cancer Metastasis Rev. | volume = 22 | issue = 4 | pages = 309–25 | year = 2003 | pmid = 12884908 | doi = 10.1023/A:1023768811842 }} | ||
*{{cite journal | * {{cite journal | vauthors = Knudsen BS, Edlund M | title = Prostate cancer and the met hepatocyte growth factor receptor | journal = Adv. Cancer Res. | volume = 91 | issue = | pages = 31–67 | year = 2004 | pmid = 15327888 | doi = 10.1016/S0065-230X(04)91002-0 | isbn = 978-0-12-006691-9 | series = Advances in Cancer Research }} | ||
*{{cite journal | * {{cite journal | vauthors = Dharmawardana PG, Giubellino A, Bottaro DP | title = Hereditary papillary renal carcinoma type I | journal = Curr. Mol. Med. | volume = 4 | issue = 8 | pages = 855–68 | year = 2004 | pmid = 15579033 | doi = 10.2174/1566524043359674 }} | ||
*{{cite journal | * {{cite journal | vauthors = Kemp LE, Mulloy B, Gherardi E | title = Signalling by HGF/SF and Met: the role of heparan sulphate co-receptors | journal = Biochem. Soc. Trans. | volume = 34 | issue = Pt 3 | pages = 414–7 | year = 2006 | pmid = 16709175 | doi = 10.1042/BST0340414 }} | ||
*{{cite journal | {{Refend}} | ||
*{{cite journal | |||
}} | |||
{{ | |||
==External links== | == External links == | ||
* {{MeshName|Proto-Oncogene+Proteins+c-met}} | * {{MeshName|Proto-Oncogene+Proteins+c-met}} | ||
* [http://www.expasy.org/uniprot/P08581 UniProtKB/Swiss-Prot entry P08581: MET_HUMAN], ExPASy (Expert Protein Analysis System) proteomics server of the Swiss Institute of Bioinformatics (SIB). | * [http://www.expasy.org/uniprot/P08581 UniProtKB/Swiss-Prot entry P08581: MET_HUMAN], ExPASy (Expert Protein Analysis System) proteomics server of the [[Swiss Institute of Bioinformatics]] (SIB) | ||
* A [http://www.vai.org/met/ table] with references to significant roles of MET in cancer | |||
* {{UCSC gene info|MET}} | |||
{{PDB Gallery|geneid=4233}} | |||
{{Growth factor receptors}} | |||
{{Oncogenes}} | {{Oncogenes}} | ||
{{Tyrosine kinases}} | {{Tyrosine kinases}} | ||
{{Growth factor | {{Enzymes}} | ||
{{Growth factor receptor modulators}} | |||
{{Portal bar|Molecular and Cellular Biology|border=no}} | |||
[[Category:Tyrosine kinase receptors]] | |||
[[Category:EC 2.7.10]] | |||
Latest revision as of 18:30, 15 May 2018
| VALUE_ERROR (nil) | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Aliases | |||||||
| External IDs | GeneCards: [1] | ||||||
| Orthologs | |||||||
| Species | Human | Mouse | |||||
| Entrez |
|
| |||||
| Ensembl |
|
| |||||
| UniProt |
|
| |||||
| RefSeq (mRNA) |
|
| |||||
| RefSeq (protein) |
|
| |||||
| Location (UCSC) | n/a | n/a | |||||
| PubMed search | n/a | n/a | |||||
| Wikidata | |||||||
| |||||||
c-Met, also called tyrosine-protein kinase Met or hepatocyte growth factor receptor (HGFR),[1][2] is a protein that in humans is encoded by the MET gene. The protein possesses tyrosine kinase activity.[3] The primary single chain precursor protein is post-translationally cleaved to produce the alpha and beta subunits, which are disulfide linked to form the mature receptor.
MET is a single pass tyrosine kinase receptor essential for embryonic development, organogenesis and wound healing. Hepatocyte growth factor/Scatter Factor (HGF/SF) and its splicing isoform (NK1, NK2) are the only known ligands of the MET receptor. MET is normally expressed by cells of epithelial origin, while expression of HGF/SF is restricted to cells of mesenchymal origin. When HGF/SF binds its cognate receptor MET it induces its dimerization through a not yet completely understood mechanism leading to its activation.
Abnormal MET activation in cancer correlates with poor prognosis, where aberrantly active MET triggers tumor growth, formation of new blood vessels (angiogenesis) that supply the tumor with nutrients, and cancer spread to other organs (metastasis). MET is deregulated in many types of human malignancies, including cancers of kidney, liver, stomach, breast, and brain. Normally, only stem cells and progenitor cells express MET, which allows these cells to grow invasively in order to generate new tissues in an embryo or regenerate damaged tissues in an adult. However, cancer stem cells are thought to hijack the ability of normal stem cells to express MET, and thus become the cause of cancer persistence and spread to other sites in the body. Both the overexpression of Met/HGFR, as well as its autocrine activation by co-expression of its hepatocyte growth factor ligand, have been implicated in oncogenesis.[4][5]
Various mutations in the MET gene are associated with papillary renal carcinoma.[6]
Gene
MET proto-oncogene (GeneID: 4233) has a total length of 125,982 bp, and it is located in the 7q31 locus of chromosome 7.[7] MET is transcribed into a 6,641 bp mature mRNA, which is then translated into a 1,390 amino-acid MET protein.
Protein
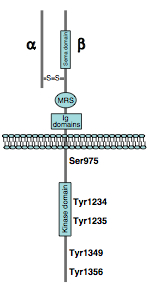
MET is a receptor tyrosine kinase (RTK) that is produced as a single-chain precursor. The precursor is proteolytically cleaved at a furin site to yield a highly glycosylated extracellular α-subunit and a transmembrane β-subunit, which are linked together by a disulfide bridge.[9]
Extracellular
- Region of homology to semaphorins (Sema domain), which includes the full α-chain and the N-terminal part of the β-chain
- Cysteine-rich MET-related sequence (MRS domain)
- Glycine-proline-rich repeats (G-P repeats)
- Four immunoglobulin-like structures (Ig domains), a typical protein-protein interaction region.[9]
Intracellular
A Juxtamembrane segment that contains:
- a serine residue (Ser 985), which inhibits the receptor kinase activity upon phosphorylation[10]
- a tyrosine (Tyr 1003), which is responsible for MET polyubiquitination, endocytosis, and degradation upon interaction with the ubiquitin ligase CBL[11]
- Tyrosine kinase domain, which mediates MET biological activity. Following MET activation, transphosphorylation occurs on Tyr 1234 and Tyr 1235
- C-terminal region contains two crucial tyrosines (Tyr 1349 and Tyr 1356), which are inserted into the multisubstrate docking site, capable of recruiting downstream adapter proteins with Src homology-2 (SH2) domains.[12] The two tyrosines of the docking site have been reported to be necessary and sufficient for the signal transduction both in vitro.[12][13]
MET signaling pathway
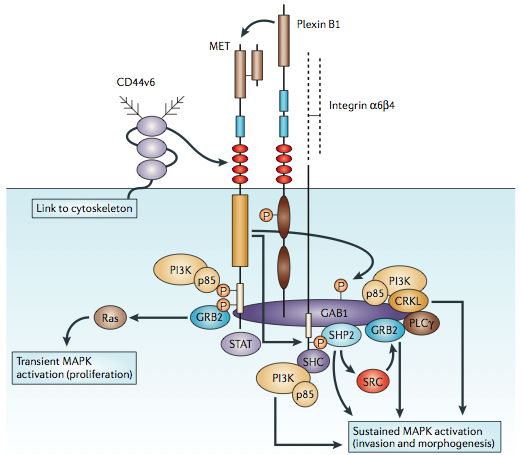
MET activation by its ligand HGF induces MET kinase catalytic activity, which triggers transphosphorylation of the tyrosines Tyr 1234 and Tyr 1235. These two tyrosines engage various signal transducers,[15] thus initiating a whole spectrum of biological activities driven by MET, collectively known as the invasive growth program. The transducers interact with the intracellular multisubstrate docking site of MET either directly, such as GRB2, SHC,[16] SRC, and the p85 regulatory subunit of phosphatidylinositol-3 kinase (PI3K),[16] or indirectly through the scaffolding protein Gab1[17]
Tyr 1349 and Tyr 1356 of the multisubstrate docking site are both involved in the interaction with GAB1, SRC, and SHC, while only Tyr 1356 is involved in the recruitment of GRB2, phospholipase C γ (PLC-γ), p85, and SHP2.[18]
GAB1 is a key coordinator of the cellular responses to MET and binds the MET intracellular region with high avidity, but low affinity.[19] Upon interaction with MET, GAB1 becomes phosphorylated on several tyrosine residues which, in turn, recruit a number of signalling effectors, including PI3K, SHP2, and PLC-γ. GAB1 phosphorylation by MET results in a sustained signal that mediates most of the downstream signaling pathways.[20]
Activation of signal transduction
MET engagement activates multiple signal transduction pathways:
- The RAS pathway mediates HGF-induced scattering and proliferation signals, which lead to branching morphogenesis.[21] Of note, HGF, differently from most mitogens, induces sustained RAS activation, and thus prolonged MAPK activity.[22]
- The PI3K pathway is activated in two ways: PI3K can be either downstream of RAS, or it can be recruited directly through the multifunctional docking site.[23] Activation of the PI3K pathway is currently associated with cell motility through remodeling of adhesion to the extracellular matrix as well as localized recruitment of transducers involved in cytoskeletal reorganization, such as RAC1 and PAK. PI3K activation also triggers a survival signal due to activation of the AKT pathway.[8]
- The STAT pathway, together with the sustained MAPK activation, is necessary for the HGF-induced branching morphogenesis. MET activates the STAT3 transcription factor directly, through an SH2 domain.[24]
- The beta-catenin pathway, a key component of the Wnt signaling pathway, translocates into the nucleus following MET activation and participates in transcriptional regulation of numerous genes.[25]
- The Notch pathway, through transcriptional activation of Delta ligand (see DLL3).[14][26]

Role in development
MET mediates a complex program known as invasive growth.[8] Activation of MET triggers mitogenesis, and morphogenesis.[27][28]
During embryonic development, transformation of the flat, two-layer germinal disc into a three-dimensional body depends on transition of some cells from an epithelial phenotype to spindle-shaped cells with motile behaviour, a mesenchymal phenotype. This process is referred to as epithelial-mesenchymal transition (EMT).[29] Later in embryonic development, MET is crucial for gastrulation, angiogenesis, myoblast migration, bone remodeling, and nerve sprouting among others.[30] MET is essential for embryogenesis, because MET −/− mice die in utero due to severe defects in placental development.[31] Along with Ectodysplasin A, it has been shown to be involved in the differentiation of anatomical placodes, precursors of scales, feathers and hair follicles in vertebrates.[32] Furthermore, MET is required for such critical processes as liver regeneration and wound healing during adulthood.[8]
HGF/MET axis is also involved in myocardial development. Both HGF and MET receptor mRNAs are co-expressed in cardiomyocytes from E7.5, soon after the heart has been determined, to E9.5. Transcripts for HGF ligand and receptor are first detected before the occurrence of cardiac beating and looping, and persist throughout the looping stage, when heart morphology begins to elaborate.[33] In avian studies, HGF was found in the myocardial layer of the atrioventricular canal, in a developmental stage in which the epithelial to mesenchymal transformation (EMT) of the endocardial cushion occurs.[34] However, MET is not essential for heart development, since α-MHCMet-KO mice show normal heart development.[35]
Expression
Tissue distribution
MET is normally expressed by epithelial cells.[8] However, MET is also found on endothelial cells, neurons, hepatocytes, hematopoietic cells, melanocytes and neonatal cardiomyocytes.[28][36] HGF expression is restricted to cells of mesenchymal origin.[29]
Transcriptional control
MET transcription is activated by HGF and several growth factors.[37] MET promoter has four putative binding sites for Ets, a family of transcription factors that control several invasive growth genes.[37] ETS1 activates MET transcription in vitro.[38] MET transcription is activated by hypoxia-inducible factor 1 (HIF1), which is activated by low concentration of intracellular oxygen.[39] HIF1 can bind to one of the several hypoxia response elements (HREs) in the MET promoter.[29] Hypoxia also activates transcription factor AP-1, which is involved in MET transcription.[29]
Clinical significance
Role in cancer
MET pathway plays an important role in the development of cancer through:
- activation of key oncogenic pathways (RAS, PI3K, STAT3, beta-catenin);
- angiogenesis (sprouting of new blood vessels from pre-existing ones to supply a tumor with nutrients);
- scatter (cells dissociation due to metalloprotease production), which often leads to metastasis.[40]
Coordinated down-regulation of both MET and its downstream effector extracellular signal-regulated kinase 2 (ERK2) by miR-199a* may be effective in inhibiting not only cell proliferation but also motility and invasive capabilities of tumor cells.[41]
MET amplification has emerged as a potential biomarker of the clear cell tumor subtype.[42]
The amplification of the cell surface receptor MET often drives resistance to anti-EGFR therapies in colorectal cancer.[43]
Role in autism
The SFARIgene database lists MET with an autism score of 2.0, which indicates that it is a strong candidate for playing a role in cases of autism. The database also identifies at least one study that found a role for MET in cases of schizophrenia. The gene was first implicated in autism in a study that identified a polymorphism in the promoter of the MET gene.[44] The polymorphism reduces transcription by 50%. Further, the variant as an autism risk polymorphism has been replicated, and shown to be enriched in children with autism and gastrointestinal disturbances.[45] A rare mutation that appears in two family members, one with autism and the other with a social and communication disorder.[46] The role of the receptor in brain development is distinct from its role in other developmental processes. Activation of the MET receptor regulates synapse formation[47][48][49][50][51] and can impact the development and function of circuits involved in social and emotional behavior.[52]
Role in heart function
In adult mice, MET is required to protect cardiomyocytes by preventing age-related oxidative stress, apoptosis, fibrosis and cardiac dysfunction.[35] Moreover, MET inhibitors, such as Crizotinib or PF-04254644, have been tested by short-term treatments in cellular and preclinical models, and have been shown to induce cardiomyocytes death through ROS production, activation of caspases, metabolism alteration and blockage of ion channels.[53][54]
In the injured heart, HGF/MET axis plays important roles in cardioprotection by promoting pro-survival (anti-apoptotic and anti-autophagic) effects in cardiomyocytes, angiogenesis, inhibition of fibrosis, anti-inflammatory and immunomodulatory signals, and regeneration through activation of cardiac stem cells.[55][56]
Interaction with tumour suppressor genes
PTEN
PTEN (phosphatase and tensin homolog) is a tumor suppressor gene encoding a protein PTEN, which possesses lipid and protein phosphatase-dependent as well as phosphatase-independent activities.[57] PTEN protein phosphatase is able to interfere with MET signaling by dephosphorylating either PIP3 generated by PI3K, or the p52 isoform of SHC. SHC dephosphorylation inhibits recruitment of the GRB2 adapter to activated MET.[14]
VHL
There is evidence of correlation between inactivation of VHL tumor suppressor gene and increased MET signaling in renal cell carcinoma (RCC) and also in malignant transformations of the heart.[58][59]
Cancer therapies targeting HGF/MET
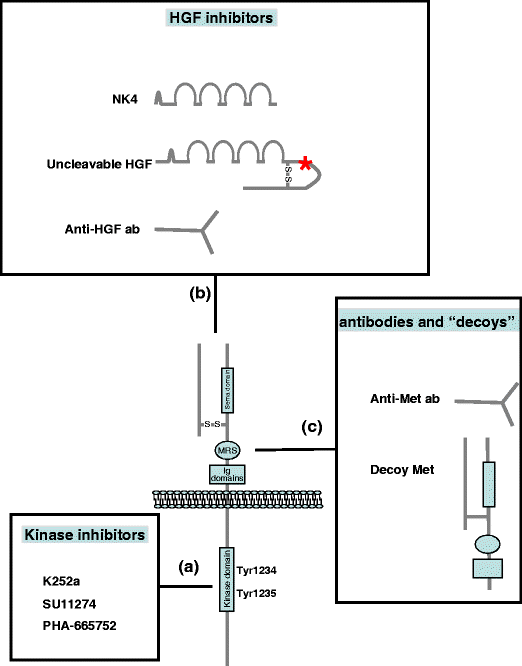
Since tumor invasion and metastasis are the main cause of death in cancer patients, interfering with MET signaling appears to be a promising therapeutic approach. A comprehensive list of HGF and MET targeted experimental therapeutics for oncology now in human clinical trials can be found here.
MET kinase inhibitors
Kinase inhibitors are low molecular weight molecules that prevent ATP binding to MET, thus inhibiting receptor transphosphorylation and recruitment of the downstream effectors. The limitations of kinase inhibitors include the facts that they only inhibit kinase-dependent MET activation, and that none of them is fully specific for MET.
- K252a (Fermentek Biotechnology) is a staurosporine analogue isolated from Nocardiopsis sp. soil fungi, and it is a potent inhibitor of all receptor tyrosine kinases (RTKs). At nanomolar concentrations, K252a inhibits both the wild type and the mutant (M1268T) MET function.[60]
- SU11274 (SUGEN) specifically inhibits MET kinase activity and its subsequent signaling. SU11274 is also an effective inhibitor of the M1268T and H1112Y MET mutants, but not the L1213V and Y1248H mutants.[61] SU11274 has been demonstrated to inhibit HGF-induced motility and invasion of epithelial and carcinoma cells.[62]
- PHA-665752 (Pfizer) specifically inhibits MET kinase activity, and it has been demonstrated to represses both HGF-dependent and constitutive MET phosphorylation.[63] Furthermore, some tumors harboring MET amplifications are highly sensitive to treatment with PHA-665752.[64]
- ARQ197 (ArQule) is a promising selective inhibitor of MET, which entered a phase 2 clinical trial in 2008. (Failed a phase 3 in 2017)
- Foretinib (XL880, Exelixis) targets multiple receptor tyrosine kinases (RTKs) with growth-promoting and angiogenic properties. The primary targets of foretinib are MET, VEGFR2, and KDR. Foretinib has completed a phase 2 clinical trials with indications for papillary renal cell carcinoma, gastric cancer, and head and neck cancer.[65]
- SGX523 (SGX Pharmaceuticals) specifically inhibits MET at low nanomolar concentrations.
- MP470 (SuperGen) is a novel inhibitor of c-KIT, MET, PDGFR, Flt3, and AXL. Phase I clinical trial of MP470 had been announced in 2007.
HGF inhibitors
Since HGF is the only known ligand of MET, formation of a HGF:MET complex blocks MET biological activity. For this purpose, truncated HGF, anti-HGF neutralizing antibodies, and an uncleavable form of HGF have been utilized so far. The major limitation of HGF inhibitors is that they block only HGF-dependent MET activation.
- NK4 competes with HGF as it binds MET without inducing receptor activation, thus behaving as a full antagonist. NK4 is a molecule bearing the N-terminal hairpin and the four kringle domains of HGF. Moreover, NK4 is structurally similar to angiostatins, which is why it possesses anti-angiogenic activity.[66]
- Neutralizing anti-HGF antibodies were initially tested in combination, and it was shown that at least three antibodies, acting on different HGF epitopes, are necessary to prevent MET tyrosine kinase activation.[67] More recently, it has been demonstrated that fully human monoclonal antibodies can individually bind and neutralize human HGF, leading to regression of tumors in mouse models.[68] Two anti-HGF antibodies are currently available: the humanized AV299 (AVEO), and the fully human AMG102 (Amgen).
- Uncleavable HGF is an engineered form of pro-HGF carrying a single amino-acid substitution, which prevents the maturation of the molecule. Uncleavable HGF is capable of blocking MET-induced biological responses by binding MET with high affinity and displacing mature HGF. Moreover, uncleavable HGF competes with the wild-type endogenous pro-HGF for the catalytic domain of proteases that cleave HGF precursors. Local and systemic expression of uncleavable HGF inhibits tumor growth and, more importantly, prevents metastasis.[69]
Decoy MET
Decoy MET refers to a soluble truncated MET receptor. Decoys are able to inhibit MET activation mediated by both HGF-dependent and independent mechanisms, as decoys prevent both the ligand binding and the MET receptor homodimerization. CGEN241 (Compugen) is a decoy MET that is highly efficient in inhibiting tumor growth and preventing metastasis in animal models.[70]
Immunotherapy targeting MET
Drugs used for immunotherapy can act either passively by enhancing the immunologic response to MET-expressing tumor cells, or actively by stimulating immune cells and altering differentiation/growth of tumor cells.[71]
Passive immunotherapy
Administering monoclonal antibodies (mAbs) is a form of passive immunotherapy. MAbs facilitate destruction of tumor cells by complement-dependent cytotoxicity (CDC) and cell-mediated cytotoxicity (ADCC). In CDC, mAbs bind to specific antigen, leading to activation of the complement cascade, which in turn leads to formation of pores in tumor cells. In ADCC, the Fab domain of a mAb binds to a tumor antigen, and Fc domain binds to Fc receptors present on effector cells (phagocytes and NK cells), thus forming a bridge between an effector and a target cells. This induces the effector cell activation, leading to phagocytosis of the tumor cell by neutrophils and macrophages. Furthermore, NK cells release cytotoxic molecules, which lyse tumor cells.[71]
- DN30 is monoclonal anti-MET antibody that recognizes the extracellular portion of MET. DN30 induces both shedding of the MET ectodomain as well as cleavage of the intracellular domain, which is successively degraded by proteasome machinery. As a consequence, on one side MET is inactivated, and on the other side the shed portion of extracellular MET hampers activation of other MET receptors, acting as a decoy. DN30 inhibits tumour growth and prevents metastasis in animal models.[72]
- OA-5D5 is one-armed monoclonal anti-MET antibody that was demonstrated to inhibit orthotopic pancreatic[73] and glioblastoma[74] tumor growth and to improve survival in tumor xenograft models. OA-5D5 is produced as a recombinant protein in Escherichia coli. It is composed of murine variable domains for the heavy and light chains with human IgG1 constant domains. The antibody blocks HGF binding to MET in a competitive fashion.
Active immunotherapy
Active immunotherapy to MET-expressing tumors can be achieved by administering cytokines, such as interferons (IFNs) and interleukins (IL-2), which triggers non-specific stimulation of numerous immune cells. IFNs have been tested as therapies for many types of cancers and have demonstrated therapeutic benefits. IL-2 has been approved by the U.S. Food and Drug Administration (FDA) for the treatment of renal cell carcinoma and metastatic melanoma, which often have deregulated MET activity.[71]
Interactions
Met has been shown to interact with:
See also
References
- ↑ Bottaro DP, Rubin JS, Faletto DL, Chan AM, Kmiecik TE, Vande Woude GF, Aaronson SA (February 1991). "Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product". Science. 251 (4995): 802–4. doi:10.1126/science.1846706. PMID 1846706.
- ↑ Galland F, Stefanova M, Lafage M, Birnbaum D (1992). "Localization of the 5' end of the MCF2 oncogene to human chromosome 15q15----q23". Cytogenet. Cell Genet. 60 (2): 114–6. doi:10.1159/000133316. PMID 1611909.
- ↑ Cooper CS (January 1992). "The met oncogene: from detection by transfection to transmembrane receptor for hepatocyte growth factor". Oncogene. 7 (1): 3–7. PMID 1531516.
- ↑ Johnson M, Koukoulis G, Kochhar K, Kubo C, Nakamura T, Iyer A (Sep 1995). "Selective tumorigenesis in non-parenchymal liver epithelial cell lines by hepatocyte growth factor transfection". Cancer Letters. 96 (1): 37–48. doi:10.1016/0304-3835(95)03915-j. PMID 7553606.
- ↑ Kochhar KS, Johnson ME, Volpert O, Iyer AP (1995). "Evidence for autocrine basis of transformation in NIH-3T3 cells transfected with met/HGF receptor gene". Growth Factors. 12 (4): 303–13. doi:10.3109/08977199509028968. PMID 8930021.
- ↑ "Entrez Gene: MET met proto-oncogene (hepatocyte growth factor receptor)".
- ↑ Dean M, Park M, Le Beau MM, Robins TS, Diaz MO, Rowley JD, Blair DG, Vande Woude GF (1985). "The human met oncogene is related to the tyrosine kinase oncogenes". Nature. 318 (6044): 385–8. doi:10.1038/318385a0. PMID 4069211.
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 Gentile A, Trusolino L, Comoglio PM (March 2008). "The Met tyrosine kinase receptor in development and cancer". Cancer Metastasis Rev. 27 (1): 85–94. doi:10.1007/s10555-007-9107-6. PMID 18175071.
- ↑ 9.0 9.1 Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF (December 2003). "Met, metastasis, motility and more". Nat. Rev. Mol. Cell Biol. 4 (12): 915–25. doi:10.1038/nrm1261. PMID 14685170.
- ↑ Gandino L, Longati P, Medico E, Prat M, Comoglio PM (January 1994). "Phosphorylation of serine 985 negatively regulates the hepatocyte growth factor receptor kinase". J. Biol. Chem. 269 (3): 1815–20. PMID 8294430.
- ↑ Peschard P, Fournier TM, Lamorte L, Naujokas MA, Band H, Langdon WY, Park M (November 2001). "Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein". Mol. Cell. 8 (5): 995–1004. doi:10.1016/S1097-2765(01)00378-1. PMID 11741535.
- ↑ 12.0 12.1 Ponzetto C, Bardelli A, Zhen Z, Maina F, dalla Zonca P, Giordano S, Graziani A, Panayotou G, Comoglio PM (April 1994). "A multifunctional docking site mediates signaling and transformation by the hepatocyte growth factor/scatter factor receptor family". Cell. 77 (2): 261–71. doi:10.1016/0092-8674(94)90318-2. PMID 7513258.
- ↑ Maina F, Casagranda F, Audero E, Simeone A, Comoglio PM, Klein R, Ponzetto C (November 1996). "Uncoupling of Grb2 from the Met receptor in vivo reveals complex roles in muscle development". Cell. 87 (3): 531–42. doi:10.1016/S0092-8674(00)81372-0. PMID 8898205.
- ↑ 14.0 14.1 14.2 14.3 Abounader R, Reznik T, Colantuoni C, Martinez-Murillo F, Rosen EM, Laterra J (December 2004). "Regulation of c-Met-dependent gene expression by PTEN". Oncogene. 23 (57): 9173–82. doi:10.1038/sj.onc.1208146. PMID 15516982.
- ↑ Johnson M, Kochhar K, Nakamura T, Iyer A (Jul 1995). "Hepatocyte growth factor-induced signal transduction in two normal mouse epithelial cell lines". Biochemistry and Molecular Biology International. 36 (3): 465–74. PMID 7549943.
- ↑ 16.0 16.1 Pelicci G, Giordano S, Zhen Z, Salcini AE, Lanfrancone L, Bardelli A, Panayotou G, Waterfield MD, Ponzetto C, Pelicci PG (April 1995). "The motogenic and mitogenic responses to HGF are amplified by the Shc adaptor protein". Oncogene. 10 (8): 1631–8. PMID 7731718.
- ↑ Weidner KM, Di Cesare S, Sachs M, Brinkmann V, Behrens J, Birchmeier W (November 1996). "Interaction between Gab1 and the c-Met receptor tyrosine kinase is responsible for epithelial morphogenesis". Nature. 384 (6605): 173–6. doi:10.1038/384173a0. PMID 8906793.
- ↑ Furge KA, Zhang YW, Vande Woude GF (November 2000). "Met receptor tyrosine kinase: enhanced signaling through adapter proteins". Oncogene. 19 (49): 5582–9. doi:10.1038/sj.onc.1203859. PMID 11114738.
- ↑ Gual P, Giordano S, Anguissola S, Parker PJ, Comoglio PM (January 2001). "Gab1 phosphorylation: a novel mechanism for negative regulation of HGF receptor signaling". Oncogene. 20 (2): 156–66. doi:10.1038/sj.onc.1204047. PMID 11313945.
- ↑ Gual P, Giordano S, Williams TA, Rocchi S, Van Obberghen E, Comoglio PM (March 2000). "Sustained recruitment of phospholipase C-gamma to Gab1 is required for HGF-induced branching tubulogenesis". Oncogene. 19 (12): 1509–18. doi:10.1038/sj.onc.1203514. PMID 10734310.
- ↑ O'Brien LE, Tang K, Kats ES, Schutz-Geschwender A, Lipschutz JH, Mostov KE (July 2004). "ERK and MMPs sequentially regulate distinct stages of epithelial tubule development". Dev. Cell. 7 (1): 21–32. doi:10.1016/j.devcel.2004.06.001. PMID 15239951.
- ↑ Marshall CJ (January 1995). "Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation". Cell. 80 (2): 179–85. doi:10.1016/0092-8674(95)90401-8. PMID 7834738.
- ↑ Graziani A, Gramaglia D, Cantley LC, Comoglio PM (November 1991). "The tyrosine-phosphorylated hepatocyte growth factor/scatter factor receptor associates with phosphatidylinositol 3-kinase". J. Biol. Chem. 266 (33): 22087–90. PMID 1718989.
- ↑ Boccaccio C, Andò M, Tamagnone L, Bardelli A, Michieli P, Battistini C, Comoglio PM (January 1998). "Induction of epithelial tubules by growth factor HGF depends on the STAT pathway". Nature. 391 (6664): 285–8. doi:10.1038/34657. PMID 9440692.
- ↑ Monga SP, Mars WM, Pediaditakis P, Bell A, Mulé K, Bowen WC, Wang X, Zarnegar R, Michalopoulos GK (April 2002). "Hepatocyte growth factor induces Wnt-independent nuclear translocation of beta-catenin after Met-beta-catenin dissociation in hepatocytes". Cancer Res. 62 (7): 2064–71. PMID 11929826.
- ↑ Gude NA, Emmanuel G, Wu W, Cottage CT, Fischer K, Quijada P, Muraski JA, Alvarez R, Rubio M, Schaefer E, Sussman MA (May 2008). "Activation of Notch-mediated protective signaling in the myocardium". Circ. Res. 102 (9): 1025–35. doi:10.1161/CIRCRESAHA.107.164749. PMC 3760732. PMID 18369158.
- ↑ Johnson M, Koukoulis G, Matsumoto K, Nakamura T, Iyer A (Jun 1993). "Hepatocyte growth factor induces proliferation and morphogenesis in nonparenchymal epithelial liver cells". Hepatology. 17 (6): 1052–61. doi:10.1016/0270-9139(93)90122-4. PMID 8514254.
- ↑ 28.0 28.1 "he fields of HGF/c-Met involvement". HealthValue. Retrieved 2009-06-13.
- ↑ 29.0 29.1 29.2 29.3 Boccaccio C, Comoglio PM (August 2006). "Invasive growth: a MET-driven genetic programme for cancer and stem cells". Nat. Rev. Cancer. 6 (8): 637–45. doi:10.1038/nrc1912. PMID 16862193.
- ↑ Birchmeier C, Gherardi E (October 1998). "Developmental roles of HGF/SF and its receptor, the c-Met tyrosine kinase". Trends Cell Biol. 8 (10): 404–10. doi:10.1016/S0962-8924(98)01359-2. PMID 9789329.
- ↑ Uehara Y, Minowa O, Mori C, Shiota K, Kuno J, Noda T, Kitamura N (February 1995). "Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor". Nature. 373 (6516): 702–5. doi:10.1038/373702a0. PMID 7854453.
- ↑ Barrow-McGee R, Kishi N, Joffre C, Ménard L, Hervieu A, Bakhouche BA, et al. (2016). "Beta 1-integrin-c-Met cooperation reveals an inside-in survival signalling on autophagy-related endomembranes". Nature Communications. 7: 11942. doi:10.1038/ncomms11942. PMC 4931016. PMID 27336951.
- ↑ Rappolee DA, Iyer A, Patel Y (Jun 1996). "Hepatocyte growth factor and its receptor are expressed in cardiac myocytes during early cardiogenesis". Circulation Research. 78 (6): 1028–36. doi:10.1161/01.RES.78.6.1028. PMID 8635233.
- ↑ Song W, Majka SM, McGuire PG (1999). "Hepatocyte growth factor expression in the developing myocardium: evidence for a role in the regulation of the mesenchymal cell phenotype and urokinase expression". Developmental Dynamics. 214 (1): 92–100. doi:10.1002/(SICI)1097-0177(199901)214:1<92::AID-DVDY9>3.0.CO;2-X. PMID 9915579.
- ↑ 35.0 35.1 Arechederra M, Carmona R, González-Nuñez M, Gutiérrez-Uzquiza A, Bragado P, Cruz-González I, Cano E, Guerrero C, Sánchez A, López-Novoa JM, Schneider MD, Maina F, Muñoz-Chápuli R, Porras A (Dec 2013). "Met signaling in cardiomyocytes is required for normal cardiac function in adult mice". Biochimica et Biophysica Acta. 1832 (12): 2204–15. doi:10.1016/j.bbadis.2013.08.008. PMID 23994610.
- ↑ Leo C, Sala V, Morello M, Chiribiri A, Riess I, Mancardi D, Schiaffino S, Ponzetto C, Crepaldi T (2011-02-09). "Activated Met signalling in the developing mouse heart leads to cardiac disease". PLOS ONE. 6 (2): e14675. doi:10.1371/journal.pone.0014675. PMC 3036588. PMID 21347410.
- ↑ 37.0 37.1 Shirasaki F, Makhluf HA, LeRoy C, Watson DK, Trojanowska M (December 1999). "Ets transcription factors cooperate with Sp1 to activate the human tenascin-C promoter". Oncogene. 18 (54): 7755–64. doi:10.1038/sj.onc.1203360. PMID 10618716.
- ↑ Gambarotta G, Boccaccio C, Giordano S, Andŏ M, Stella MC, Comoglio PM (November 1996). "Ets up-regulates MET transcription". Oncogene. 13 (9): 1911–7. PMID 8934537.
- ↑ Pennacchietti S, Michieli P, Galluzzo M, Mazzone M, Giordano S, Comoglio PM (April 2003). "Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene". Cancer Cell. 3 (4): 347–61. doi:10.1016/S1535-6108(03)00085-0. PMID 12726861.
- ↑ "HGF/c-Met and cancer". HealthValue. Retrieved 2009-06-13.
- ↑ Kim S, Lee UJ, Kim MN, Lee EJ, Kim JY, Lee MY, Choung S, Kim YJ, Choi YC (June 2008). "MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2)". J. Biol. Chem. 283 (26): 18158–66. doi:10.1074/jbc.M800186200. PMID 18456660.
- ↑ del Carmen MG, Birrer M, Schorge JO (September 2012). "Clear cell carcinoma of the ovary: a review of the literature". Gynecol. Oncol. 126 (3): 481–90. doi:10.1016/j.ygyno.2012.04.021. PMID 22525820.
- ↑ Bardelli A, Corso S, Bertotti A, Hobor S, Valtorta E, Siravegna G, Sartore-Bianchi A, Scala E, Cassingena A, Zecchin D, Apicella M, Migliardi G, Galimi F, Lauricella C, Zanon C, Perera T, Veronese S, Corti G, Amatu A, Gambacorta M, Diaz LA, Sausen M, Velculescu VE, Comoglio P, Trusolino L, Di Nicolantonio F, Giordano S, Siena S (June 2013). "Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer". Cancer Discov. 3 (6): 658–73. doi:10.1158/2159-8290.CD-12-0558. PMC 4078408. PMID 23729478.
- ↑ Campbell DB, Sutcliffe JS, Ebert PJ, Militerni R, Bravaccio C, Trillo S, Elia M, Schneider C, Melmed R, Sacco R, Persico AM, Levitt P (2006). "A genetic variant that disrupts MET transcription is associated with autism". Proc. Natl. Acad. Sci. U.S.A. 103 (45): 16834–9. doi:10.1073/pnas.0605296103. PMC 1838551. PMID 17053076.
- ↑ Campbell DB, Buie TM, Winter H, Bauman M, Sutcliffe JS, Perrin JM, Levitt P (2009). "Distinct genetic risk based on association of MET in families with co-occurring autism and gastrointestinal conditions". Pediatrics. 123 (3): 1018–24. doi:10.1542/peds.2008-0819. PMID 19255034.
- ↑ Lambert N, Wermenbol V, Pichon B, Acosta S, van den Ameele J, Perazzolo C, Messina D, Musumeci MF, Dessars B, De Leener A, Abramowicz M, Vilain C (2014). "A familial heterozygous null mutation of MET in autism spectrum disorder". Autism Res. 7 (5): 617–22. doi:10.1002/aur.1396. PMID 24909855.
- ↑ Qiu S, Lu Z, Levitt P (2014). "MET receptor tyrosine kinase controls dendritic complexity, spine morphogenesis, and glutamatergic synapse maturation in the hippocampus". J. Neurosci. 34 (49): 16166–79. doi:10.1523/JNEUROSCI.2580-14.2014. PMC 4252539. PMID 25471559.
- ↑ Eagleson KL, Milner TA, Xie Z, Levitt P (2013). "Synaptic and extrasynaptic location of the receptor tyrosine kinase met during postnatal development in the mouse neocortex and hippocampus". J. Comp. Neurol. 521 (14): 3241–59. doi:10.1002/cne.23343. PMC 3942873. PMID 23787772.
- ↑ Judson MC, Eagleson KL, Levitt P (2011). "A new synaptic player leading to autism risk: Met receptor tyrosine kinase". J Neurodev Disord. 3 (3): 282–92. doi:10.1007/s11689-011-9081-8. PMC 3261279. PMID 21509596.
- ↑ Qiu S, Anderson CT, Levitt P, Shepherd GM (2011). "Circuit-specific intracortical hyperconnectivity in mice with deletion of the autism-associated Met receptor tyrosine kinase". J. Neurosci. 31 (15): 5855–64. doi:10.1523/JNEUROSCI.6569-10.2011. PMC 3086026. PMID 21490227.
- ↑ Judson MC, Eagleson KL, Wang L, Levitt P (2010). "Evidence of cell-nonautonomous changes in dendrite and dendritic spine morphology in the met-signaling-deficient mouse forebrain". J. Comp. Neurol. 518 (21): 4463–78. doi:10.1002/cne.22467. PMC 2952412. PMID 20853516.
- ↑ Rudie JD, Hernandez LM, Brown JA, Beck-Pancer D, Colich NL, Gorrindo P, Thompson PM, Geschwind DH, Bookheimer SY, Levitt P, Dapretto M. "Autism-associated promoter variant in MET impacts functional and structural brain networks". Neuron. 75: 904–15. doi:10.1016/j.neuron.2012.07.010. PMC 3454529. PMID 22958829.
- ↑ Doherty KR, Wappel RL, Talbert DR, Trusk PB, Moran DM, Kramer JW, Brown AM, Shell SA, Bacus S (October 2013). "Multi-parameter in vitro toxicity testing of crizotinib, sunitinib, erlotinib, and nilotinib in human cardiomyocytes". Toxicology and Applied Pharmacology. 272 (1): 245–55. doi:10.1016/j.taap.2013.04.027. PMID 23707608.
- ↑ Aguirre SA, Heyen JR, Collette W, Bobrowski W, Blasi ER (April 2010). "Cardiovascular effects in rats following exposure to a receptor tyrosine kinase inhibitor". Toxicologic Pathology. 38 (3): 416–28. doi:10.1177/0192623310364027. PMID 20231546.
- ↑ Schmoldt A, Benthe HF, Haberland G, Scott WA, Mahoney E, Pounds JG, Long GJ, Rosen JF (February 1991). "Cellular and molecular toxicity of lead in bone". Environmental Health Perspectives. 91 (17): 17–32. doi:10.3390/biomedicines2040247. PMC 1519349. PMID 2040247.
- ↑ Sala V, Crepaldi T (May 2011). "Novel therapy for myocardial infarction: can HGF/Met be beneficial?". Cellular and Molecular Life Sciences. 68 (10): 1703–17. doi:10.1007/s00018-011-0633-6. PMID 21327916.
- ↑ Maehama T, Dixon JE (May 1998). "The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate". J. Biol. Chem. 273 (22): 13375–8. doi:10.1074/jbc.273.22.13375. PMID 9593664.
- ↑ Morris MR, Gentle D, Abdulrahman M, Maina EN, Gupta K, Banks RE, Wiesener MS, Kishida T, Yao M, Teh B, Latif F, Maher ER (June 2005). "Tumor suppressor activity and epigenetic inactivation of hepatocyte growth factor activator inhibitor type 2/SPINT2 in papillary and clear cell renal cell carcinoma". Cancer Res. 65 (11): 4598–606. doi:10.1158/0008-5472.CAN-04-3371. PMID 15930277.
- ↑ Lei L, Mason S, Liu D, Huang Y, Marks C, Hickey R, Jovin IS, Pypaert M, Johnson RS, Giordano FJ (June 2008). "Hypoxia-inducible factor-dependent degeneration, failure, and malignant transformation of the heart in the absence of the von Hippel-Lindau protein". Molecular and Cellular Biology. 28 (11): 3790–803. doi:10.1128/MCB.01580-07. PMC 2423296. PMID 18285456.
- ↑ Morotti A, Mila S, Accornero P, Tagliabue E, Ponzetto C (July 2002). "K252a inhibits the oncogenic properties of Met, the HGF receptor". Oncogene. 21 (32): 4885–93. doi:10.1038/sj.onc.1205622. PMID 12118367.
- ↑ Berthou S, Aebersold DM, Schmidt LS, Stroka D, Heigl C, Streit B, Stalder D, Gruber G, Liang C, Howlett AR, Candinas D, Greiner RH, Lipson KE, Zimmer Y (July 2004). "The Met kinase inhibitor SU11274 exhibits a selective inhibition pattern toward different receptor mutated variants". Oncogene. 23 (31): 5387–93. doi:10.1038/sj.onc.1207691. PMID 15064724.
- ↑ Wang X, Le P, Liang C, Chan J, Kiewlich D, Miller T, Harris D, Sun L, Rice A, Vasile S, Blake RA, Howlett AR, Patel N, McMahon G, Lipson KE (November 2003). "Potent and selective inhibitors of the Met [hepatocyte growth factor/scatter factor (HGF/SF) receptor] tyrosine kinase block HGF/SF-induced tumor cell growth and invasion". Mol. Cancer Ther. 2 (11): 1085–92. PMID 14617781.
- ↑ Christensen JG, Schreck R, Burrows J, Kuruganti P, Chan E, Le P, Chen J, Wang X, Ruslim L, Blake R, Lipson KE, Ramphal J, Do S, Cui JJ, Cherrington JM, Mendel DB (November 2003). "A selective small molecule inhibitor of c-Met kinase inhibits c-Met-dependent phenotypes in vitro and exhibits cytoreductive antitumor activity in vivo". Cancer Res. 63 (21): 7345–55. PMID 14612533.
- ↑ Smolen GA, Sordella R, Muir B, Mohapatra G, Barmettler A, Archibald H, Kim WJ, Okimoto RA, Bell DW, Sgroi DC, Christensen JG, Settleman J, Haber DA (February 2006). "Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752". Proc. Natl. Acad. Sci. U.S.A. 103 (7): 2316–21. doi:10.1073/pnas.0508776103. PMC 1413705. PMID 16461907.
- ↑ Poster
- ↑ Matsumoto K, Nakamura T (April 2003). "NK4 (HGF-antagonist/angiogenesis inhibitor) in cancer biology and therapeutics". Cancer Sci. 94 (4): 321–7. doi:10.1111/j.1349-7006.2003.tb01440.x. PMID 12824898.
- ↑ Cao B, Su Y, Oskarsson M, Zhao P, Kort EJ, Fisher RJ, Wang LM, Vande Woude GF (June 2001). "Neutralizing monoclonal antibodies to hepatocyte growth factor/scatter factor (HGF/SF) display antitumor activity in animal models". Proc. Natl. Acad. Sci. U.S.A. 98 (13): 7443–8. doi:10.1073/pnas.131200498. PMC 34688. PMID 11416216.
- ↑ Burgess T, Coxon A, Meyer S, Sun J, Rex K, Tsuruda T, Chen Q, Ho SY, Li L, Kaufman S, McDorman K, Cattley RC, Sun J, Elliott G, Zhang K, Feng X, Jia XC, Green L, Radinsky R, Kendall R (February 2006). "Fully human monoclonal antibodies to hepatocyte growth factor with therapeutic potential against hepatocyte growth factor/c-Met-dependent human tumors". Cancer Res. 66 (3): 1721–9. doi:10.1158/0008-5472.CAN-05-3329. PMID 16452232.
- ↑ Mazzone M, Basilico C, Cavassa S, Pennacchietti S, Risio M, Naldini L, Comoglio PM, Michieli P (November 2004). "An uncleavable form of pro-scatter factor suppresses tumor growth and dissemination in mice". J. Clin. Invest. 114 (10): 1418–32. doi:10.1172/JCI22235. PMC 525743. PMID 15545993.
- ↑ Michieli P, Mazzone M, Basilico C, Cavassa S, Sottile A, Naldini L, Comoglio PM (July 2004). "Targeting the tumor and its microenvironment by a dual-function decoy Met receptor". Cancer Cell. 6 (1): 61–73. doi:10.1016/j.ccr.2004.05.032. PMID 15261142.
- ↑ 71.0 71.1 71.2 Reang P, Gupta M, Kohli K (2006). "Biological response modifiers in cancer". MedGenMed. 8 (4): 33. PMC 1868326. PMID 17415315.
- ↑ Petrelli A, Circosta P, Granziero L, Mazzone M, Pisacane A, Fenoglio S, Comoglio PM, Giordano S (March 2006). "Ab-induced ectodomain shedding mediates hepatocyte growth factor receptor down-regulation and hampers biological activity". Proc. Natl. Acad. Sci. U.S.A. 103 (13): 5090–5. doi:10.1073/pnas.0508156103. PMC 1458799. PMID 16547140.
- ↑ Jin H, Yang R, Zheng Z, Romero M, Ross J, Bou-Reslan H, Carano RA, Kasman I, Mai E, Young J, Zha J, Zhang Z, Ross S, Schwall R, Colbern G, Merchant M (June 2008). "MetMAb, the one-armed 5D5 anti-c-Met antibody, inhibits orthotopic pancreatic tumor growth and improves survival". Cancer Res. 68 (11): 4360–8. doi:10.1158/0008-5472.CAN-07-5960. PMID 18519697.
- ↑ Martens T, Schmidt NO, Eckerich C, Fillbrandt R, Merchant M, Schwall R, Westphal M, Lamszus K (October 2006). "A novel one-armed anti-c-Met antibody inhibits glioblastoma growth in vivo". Clin. Cancer Res. 12 (20 Pt 1): 6144–52. doi:10.1158/1078-0432.CCR-05-1418. PMID 17062691.
- ↑ Davies G, Jiang WG, Mason MD (2001). "HGF/SF modifies the interaction between its receptor c-Met, and the E-cadherin/catenin complex in prostate cancer cells". Int. J. Mol. Med. 7 (4): 385–8. doi:10.3892/ijmm.7.4.385. PMID 11254878.
- ↑ Petrelli A, Gilestro GF, Lanzardo S, Comoglio PM, Migone N, Giordano S (2002). "The endophilin-CIN85-Cbl complex mediates ligand-dependent downregulation of c-Met". Nature. 416 (6877): 187–90. doi:10.1038/416187a. PMID 11894096.
- ↑ Ng C, Jackson RA, Buschdorf JP, Sun Q, Guy GR, Sivaraman J (2008). "Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates". EMBO J. 27 (5): 804–16. doi:10.1038/emboj.2008.18. PMC 2265755. PMID 18273061.
- ↑ Grisendi S, Chambraud B, Gout I, Comoglio PM, Crepaldi T (2001). "Ligand-regulated binding of FAP68 to the hepatocyte growth factor receptor". J. Biol. Chem. 276 (49): 46632–8. doi:10.1074/jbc.M104323200. PMID 11571281.
- ↑ Ponzetto C, Zhen Z, Audero E, Maina F, Bardelli A, Basile ML, Giordano S, Narsimhan R, Comoglio P (1996). "Specific uncoupling of GRB2 from the Met receptor. Differential effects on transformation and motility". J. Biol. Chem. 271 (24): 14119–23. doi:10.1074/jbc.271.24.14119. PMID 8662889.
- ↑ Liang Q, Mohan RR, Chen L, Wilson SE (1998). "Signaling by HGF and KGF in corneal epithelial cells: Ras/MAP kinase and Jak-STAT pathways". Invest. Ophthalmol. Vis. Sci. 39 (8): 1329–38. PMID 9660480.
- ↑ Comoglio PM (1993). "Structure, biosynthesis and biochemical properties of the HGF receptor in normal and malignant cells". EXS. 65: 131–65. PMID 8380735.
- ↑ Naldini L, Weidner KM, Vigna E, Gaudino G, Bardelli A, Ponzetto C, Narsimhan RP, Hartmann G, Zarnegar R, Michalopoulos GK (1991). "Scatter factor and hepatocyte growth factor are indistinguishable ligands for the MET receptor". EMBO J. 10 (10): 2867–78. PMC 452997. PMID 1655405.
- ↑ Hiscox S, Jiang WG (1999). "Association of the HGF/SF receptor, c-met, with the cell-surface adhesion molecule, E-cadherin, and catenins in human tumor cells". Biochem. Biophys. Res. Commun. 261 (2): 406–11. doi:10.1006/bbrc.1999.1002. PMID 10425198.
- ↑ Wang D, Li Z, Messing EM, Wu G (2002). "Activation of Ras/Erk pathway by a novel MET-interacting protein RanBPM". J. Biol. Chem. 277 (39): 36216–22. doi:10.1074/jbc.M205111200. PMID 12147692.
Further reading
- Peruzzi B, Bottaro DP (2006). "Targeting the c-Met signaling pathway in cancer". Clin. Cancer Res. 12 (12): 3657–60. doi:10.1158/1078-0432.CCR-06-0818. PMID 16778093.
- Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF (December 2003). "Met, metastasis, motility and more". Nat. Rev. Mol. Cell Biol. 4 (12): 915–25. doi:10.1038/nrm1261. PMID 14685170.
- Zhang YW, Vande Woude GF (February 2003). "HGF/SF-met signaling in the control of branching morphogenesis and invasion". J. Cell. Biochem. 88 (2): 408–17. doi:10.1002/jcb.10358. PMID 12520544.
- Paumelle R, Tulasne D, Kherrouche Z, Plaza S, Leroy C, Reveneau S, Vandenbunder B, Fafeur V, Tulashe D, Reveneau S (April 2002). "Hepatocyte growth factor/scatter factor activates the ETS1 transcription factor by a RAS-RAF-MEK-ERK signaling pathway". Oncogene. 21 (15): 2309–19. doi:10.1038/sj.onc.1205297. PMID 11948414.
- Comoglio PM (1993). "Structure, biosynthesis and biochemical properties of the HGF receptor in normal and malignant cells". EXS. 65: 131–65. PMID 8380735.
- Maulik G, Shrikhande A, Kijima T, Ma PC, Morrison PT, Salgia R (2002). "Role of the hepatocyte growth factor receptor, c-Met, in oncogenesis and potential for therapeutic inhibition". Cytokine Growth Factor Rev. 13 (1): 41–59. doi:10.1016/S1359-6101(01)00029-6. PMID 11750879.
- Ma PC, Maulik G, Christensen J, Salgia R (2003). "c-Met: structure, functions and potential for therapeutic inhibition". Cancer Metastasis Rev. 22 (4): 309–25. doi:10.1023/A:1023768811842. PMID 12884908.
- Knudsen BS, Edlund M (2004). "Prostate cancer and the met hepatocyte growth factor receptor". Adv. Cancer Res. Advances in Cancer Research. 91: 31–67. doi:10.1016/S0065-230X(04)91002-0. ISBN 978-0-12-006691-9. PMID 15327888.
- Dharmawardana PG, Giubellino A, Bottaro DP (2004). "Hereditary papillary renal carcinoma type I". Curr. Mol. Med. 4 (8): 855–68. doi:10.2174/1566524043359674. PMID 15579033.
- Kemp LE, Mulloy B, Gherardi E (2006). "Signalling by HGF/SF and Met: the role of heparan sulphate co-receptors". Biochem. Soc. Trans. 34 (Pt 3): 414–7. doi:10.1042/BST0340414. PMID 16709175.
External links
- Proto-Oncogene+Proteins+c-met at the US National Library of Medicine Medical Subject Headings (MeSH)
- UniProtKB/Swiss-Prot entry P08581: MET_HUMAN, ExPASy (Expert Protein Analysis System) proteomics server of the Swiss Institute of Bioinformatics (SIB)
- A table with references to significant roles of MET in cancer
- Human MET genome location and MET gene details page in the UCSC Genome Browser.