Tipranavir microbiology
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]; Associate Editor(s)-in-Chief: Ahmed Zaghw, M.D. [2]
Microbiology
Mechanism of Action
Tipranavir (TPV) is an HIV-1 protease inhibitor that inhibits the virus-specific processing of the viral Gag and Gag-Pol polyproteins in HIV-1 infected cells, thus preventing formation of mature virions.
Antiviral Activity
Tipranavir inhibits the replication of laboratory strains of HIV-1 and clinical isolates in acute models of T-cell infection, with 50% effective concentrations (EC50) ranging from 0.03 to 0.07 μM (18-42 ng/mL). Tipranavir demonstrates antiviral activity in cell culture against a broad panel of HIV-1 group M non-clade B isolates (A, C, D, F, G, H, CRF01 AE, CRF02 AG, CRF12 BF). Group O and HIV-2 isolates have reduced susceptibility in cell culture to tipranavir with EC50 values ranging from 0.164 -1 μM and 0.233-0.522 μM, respectively. When used with other antiretroviral agents in cell culture, the combination of tipranavir was additive to antagonistic with other protease inhibitors (amprenavir, atazanavir, indinavir, lopinavir, nelfinavir, ritonavir, and saquinavir) and generally additive with the NNRTIs (delavirdine, efavirenz, and nevirapine) and the NRTIs (abacavir, didanosine, emtricitabine, lamivudine, stavudine, tenofovir, and zidovudine). Tipranavir was synergistic with the HIV-1 fusion inhibitor enfuvirtide. There was no antagonism of the cell culture combinations of tipranavir with either adefovir or ribavirin, used in the treatment of viral hepatitis.
Resistance
In cell culture:
HIV-1 isolates with a decreased susceptibility to tipranavir have been selected in cell culture and obtained from patients treated with APTIVUS/ritonavir (TPV/ritonavir). After 9 months of culture in TPV-containing media, HIV-1 isolates with 87-fold reduced susceptibility to tipranavir were selected in cell culture; these contained 10 protease substitutions that developed in the following order: L33F, I84V, K45I, I13V, V32I, V82L, M36I, A71V, L10F, and I54V/T. Changes in the Gag polyprotein CA/P2 cleavage site were also observed following drug selection. Experiments with site-directed mutants of HIV-1 showed that the presence of 6 substitutions in the protease coding sequence (I13V, V32I, L33F, K45I, V82L, I84V) conferred >10-fold reduced susceptibility to tipranavir.
Clinical Studies of Treatment-Experienced Patients:
In controlled clinical trials 1182.12 and 1182.48, multiple protease inhibitor-resistant HIV-1 isolates from 59 treatment-experienced adult patients who received APTIVUS/ritonavir and experienced virologic rebound developed amino acid substitutions that were associated with resistance to tipranavir. The most common amino acid substitutions that developed on 500/200 mg APTIVUS/ritonavir in greater than 20% of APTIVUS/ritonavir virologic failure isolates were L33V/I/F, V82T, and I84V. Other substitutions that developed in 10 to 20% of APTIVUS/ritonavir virologic failure isolates included L10V/I/S, I13V, E35D/G/N, I47V, I54A/M/V, K55R, V82L, and L89V/M. Evolution at protease gag polyprotein cleavage sites was also observed. Among 28 pediatric patients in clinical trial 1182.14 who experienced virologic failure or non-response, the emergent protease amino acid codon substitutions were similar to those observed in adult virologic failure isolates.
In clinical trials 1182.12 and 1182.48 tipranavir resistance was detected at virologic rebound after an average of 38 weeks of APTIVUS/ritonavir treatment with a median 14-fold decrease in tipranavir susceptibility. Similarly, reduced tipranavir susceptibility was associated with emergent mutations in pediatric patient isolates.
Cross-resistance
Cross-resistance among protease inhibitors has been observed. Tipranavir had <4-fold decreased susceptibility against 90% (94/105) of HIV-1 clinical isolates resistant to amprenavir, atazanavir, indinavir, lopinavir, nelfinavir, ritonavir, or saquinavir. Tipranavir-resistant viruses which emerged in cell culture from wild-type HIV-1 had decreased susceptibility to the protease inhibitors amprenavir, atazanavir, indinavir, lopinavir, nelfinavir and ritonavir but remained sensitive to saquinavir.
Baseline Genotype and Virologic Outcome Analyses
Genotypic and/or phenotypic analysis of baseline virus may aid in determining tipranavir susceptibility before initiation of APTIVUS/ritonavir therapy. Several analyses were conducted to evaluate the impact of specific substitutions and combination of substitutions on virologic outcome. Both the type and number of baseline protease inhibitor substitutions as well as use of additional active agents (e.g., enfuvirtide) affected APTIVUS/ritonavir response rates in controlled clinical trials 1182.12 and 1182.48 through Week 48 of treatment.
Regression analyses of baseline and/or on-treatment HIV-1 genotypes from 860 treatment-experienced patients in Phase 2 and 3 trials demonstrated that amino acid substitutions at 16 codons in the HIV-1 protease coding sequence were associated with reduced virologic responses and/or reduced tipranavir susceptibility: L10V, I13V, K20M/R/V, L33F, E35G, M36I, K43T, M46L, I47V, I54A/M/V, Q58E, H69K, T74P, V82L/T, N83D or I84V.
As-treated analyses were also conducted to assess virologic outcome by the number of primary protease inhibitor substitutions present at baseline. Response rates were reduced if five or more protease inhibitor-associated substitutions were present at baseline and subjects did not receive concomitant new enfuvirtide with APTIVUS/ritonavir. See Table 9
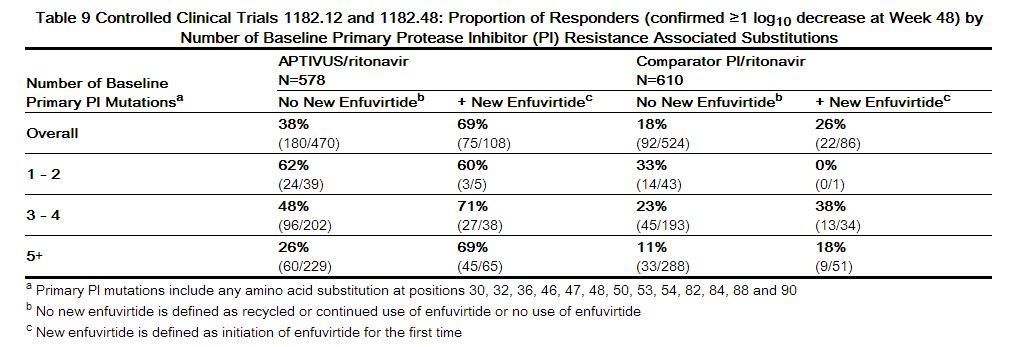
|
The median change from baseline in plasma HIV-1 RNA at weeks 2, 4, 8, 16, 24 and 48 was evaluated by the number of baseline primary protease inhibitor resistance associated substitutions (1-4 or ≥5) in subjects who received APTIVUS/ritonavir with or without new enfuvirtide. The following observations were made:
Approximately 1.5 log10 decrease in HIV-1 RNA at early time points (Week 2) regardless of the number of baseline primary protease inhibitor resistance associated substitutions (1-4 or 5+). Subjects with 5 or more primary protease inhibitor resistance associated substitutions in their HIV-1 at baseline who received APTIVUS/ritonavir without new enfuvirtide (n=303) began to lose their antiviral response after Week 4. Early HIV-1 RNA decreases (1.5-2 log10) were sustained through Week 48 in subjects with 5 or more primary protease inhibitor resistance associated substitutions at baseline who received new enfuvirtide with APTIVUS/ritonavir (n=74).
Baseline Phenotype and Virologic Outcome Analyses APTIVUS/ritonavir response rates were also assessed by baseline tipranavir phenotype. Relationships between baseline phenotypic susceptibility to tipranavir, mutations at protease amino acid codons 33, 82, 84 and 90, tipranavir resistance-associated mutations, and response to APTIVUS/ritonavir therapy at Week-48 are summarized in Tables 10 and 11. These baseline phenotype groups are not meant to represent clinical susceptibility breakpoints for APTIVUS/ritonavir because the data are based on the select 1182.12 and 1182.48 patient population. The data are provided to give clinicians information on the likelihood of virologic success based on pre-treatment susceptibility to APTIVUS/ritonavir in protease inhibitor-experienced patients.
 |
Analyses of pediatric clinical trial 1182.14 also demonstrated that response to therapy was influenced by the number of baseline protease inhibitor mutations present.[1]
References
Adapted from the FDA Package Insert.