Hepatitis B virus
| Hepatitis B virus | ||||||||
|---|---|---|---|---|---|---|---|---|
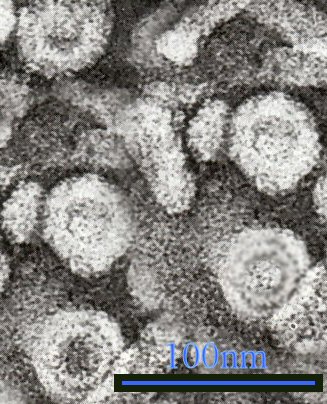 Micrograph showing hepatitis B virions - Author: GrahamColm na projektu Wikipedie v jazyce angličtina, CC BY 3.0, https://commons.wikimedia.org/w/index.php?curid=6032684
| ||||||||
| Virus classification | ||||||||
|
|
Hepatitis B |
|
Diagnosis |
|
Treatment |
|
Case Studies |
|
Hepatitis B virus On the Web |
|
American Roentgen Ray Society Images of Hepatitis B virus |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]; Associate Editor(s)-in-Chief: João André Alves Silva, M.D. [2]
Overview
The hepatitis B virus is responsible for causing hepatitis B. HBV is a double stranded DNA virus belonging to the family Hepadnaviridae. The viral particle consists of an outer lipid envelope and an icosahedral nucleocapsid core composed of protein. The nucleocapsid encloses the viral DNA. HBV DNA polymerase has reverse transcriptase activity. It shows tropism for hepatocytes. Humans are the only natural reservoir.[1] The virus is divided into four major serotypes (adr, adw, ayr, ayw) based on antigenic epitopes presented on its envelope proteins and ten genotypes (A-J) according to overall nucleotide sequence variation of the genome.[2][3]
History
The earliest record of an epidemic caused by Hepatitis B virus was made by Lurman in 1885.[4] An outbreak of smallpox occurred in Bremen in 1883 and 1,289 shipyard employees were vaccinated with lymph from other people. After several weeks, and up to eight months later, 191 of the vaccinated workers became ill with jaundice and were diagnosed as suffering from serum hepatitis. Other employees who had been inoculated with different batches of lymph remained healthy. Lurman's paper, now regarded as a classical example of an epidemiological study, proved that contaminated lymph was the source of the outbreak. Later, numerous similar outbreaks were reported following the introduction, in 1909, of hypodermic needles that were used, and more importantly reused, for administering Salvarsan for the treatment of syphilis. The virus was not discovered until 1965 when Baruch Blumberg, then working at the National Institutes of Health (NIH), discovered the Australia antigen (later known to be Hepatitis B surface antigen, or HBsAg) in the blood of Australian aboriginal people.[5] Although a virus had been suspected since the research published by MacCallum in 1947.[6] In 1970, D.S. Dane and others discovered the virus particle by electron microscopy.[7] By the early 1980s the genome of the virus had been sequenced,[8] and the first vaccines were being tested.[9]
Microbiology
Structure

Hepatitis B virus (HBV) is a member of the Hepadnavirus family.[1]
The viral particle (virion) consists of an outer lipid envelope and an icosahedral nucleocapsid core composed of protein. The nucleocapsid encloses the viral DNA and a DNA polymerase, that has reverse transcriptase activity.[10]
The outer envelope contains embedded proteins that are involved in viral binding of, and entry into, susceptible cells. The virus is one of the smallest enveloped animal viruses, with a virion diameter of 42nm, but pleomorphic forms exist, including filamentous and spherical bodies lacking a core. These particles are not infectious and are composed of the lipid and protein that form part of the surface of the virion, which is called the surface antigen (HBsAg), and is produced in excess during the life cycle of the virus.[11]
The protein of the virion coat is termed "surface antigen" or HBsAg. It is sometimes extended as a tubular tail on one side of the virus particle. The surface antigen is generally produced in vast excess, and is found in the blood of infected individuals in the form of filamentous and spherical particles. Filamentous particles are identical to the virion "tails" - they vary in length and have a mean diameter of about 22nm. They sometimes display regular, non-helical transverse striations.[12]
Genome
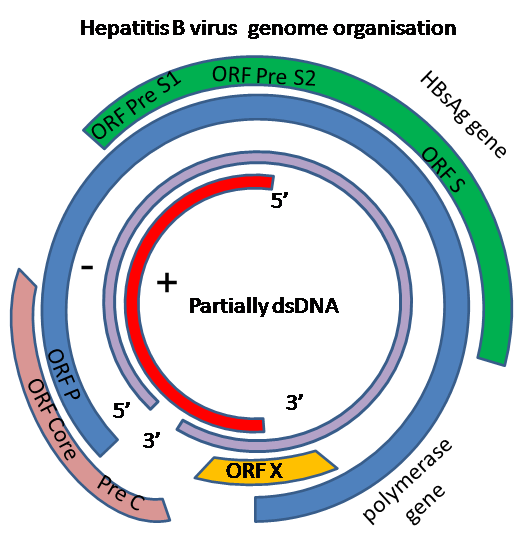
- HBV virion DNA is a relaxed circular, partially duplex molecule of 3.2 kb, whose circularity is maintained by 5' cohesive ends.[12]
- A virion-associated polymerase can repair gaps and generate a fully duplex genome. Negative strand DNA is the template for the synthesis of the viral mRNA transcripts. HBV DNA has a very compact coding organization with four partially overlapping open reading frames (ORFs) that are translated into seven known proteins.[12]
- Noncoding regions are not present.[12]
- Genomic, P, and pre-C and C RNAs
- L protein mRNA
- M and S protein mRNAs
- X protein mRNA
They are referred to as the genomic, pre-S1, S, and X promoters, respectively.
- Two major classes of transcripts exist:
- ORF P encodes the viral polymerase and the terminal protein found on minus strand DNA. ORF C encodes the structural protein of the nucleocapsid and the HBeAg, and ORF S/pre-S encodes the viral surface glycoproteins. The product of ORF X is a poorly understood regulatory protein that enhances the expression of heterologous and homologous cellular genes in trans.
- The three envelope glycoproteins are not distributed uniformly among the various HBV particle types. Subviral 22 nm particles are composed predominantly of S proteins, with variable amounts of M proteins and few or no L proteins. Virus particles are enriched for L proteins. L proteins carry the receptor recognition domain, which allows efficient binding to cell surface receptors.
- HBcAg is the most conserved polypeptide among the mammalian hepadnaviruses with 68% homology between HBV and GSHV and 92% between GSHV and WHV. Furthermore, it plays important roles in the encapsidation of the viral pregenomic RNA. The polymerase protein is quite immunogenic during both acute and chronic infection.[12]
Replication
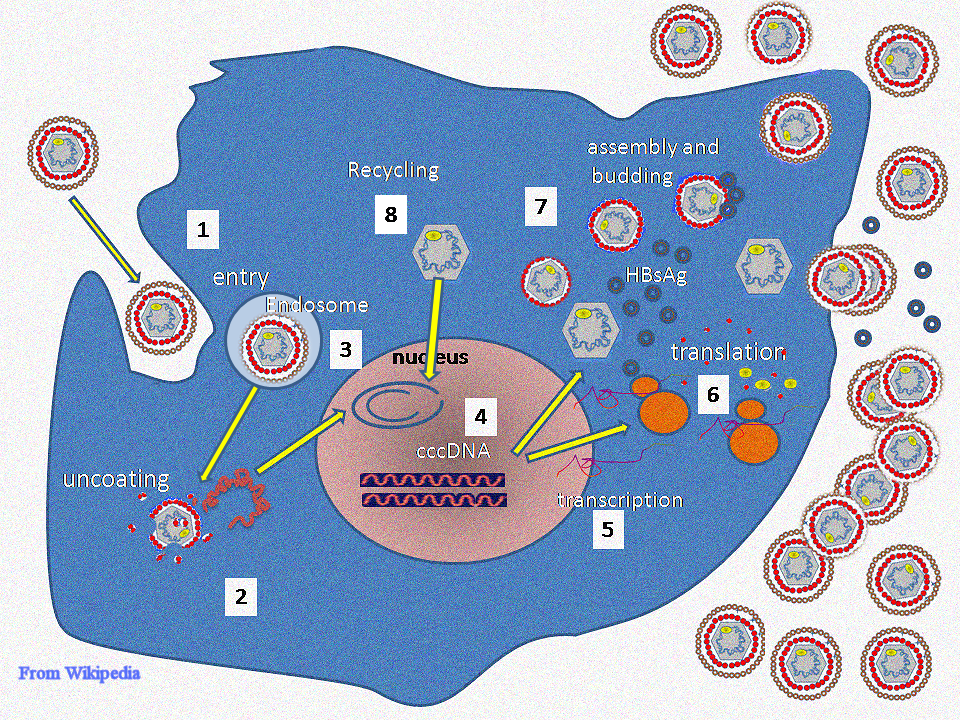
The life cycle of Hepatitis B virus is complex. Hepatitis B is one of a few known non-retroviral viruses which use reverse transcription as a part of its replication process. The virus gains entry into the cell by binding to a receptor on the surface of the cell and enters it by endocytosis. Because the virus multiplies via RNA made by a host enzyme, the viral genomic DNA has to be transferred to the cell nucleus by host proteins called chaperones. The partially double stranded viral DNA is then made fully double stranded and transformed into closed circular supercoiled DNA (cccDNA) that serves as a template for transcription of four viral mRNAs. The largest mRNA, (which is longer than the viral genome), is used to make the new copies of the genome and to make the capsid core protein and the viral DNA polymerase. These four viral transcripts undergo additional processing and go on to form progeny virions which are released from the cell or returned to the nucleus and re-cycled to produce even more copies.[14][15] The long mRNA is then transported back to the cytoplasm where the virion P protein synthesizes DNA via its reverse transcriptase activity.
Serotypes and Genotypes
The virus is divided into:
- Four major serotypes (adr, adw, ayr, ayw) based on antigenic epitopes presented on its envelope proteins
- Ten genotypes (A-J) according to overall nucleotide sequence variation of the genome
- The genotypes have a distinct geographical distribution and are used in tracing the evolution and transmission of the virus.
- Differences between genotypes affect:[16][2][3]
- Outcome (worst in genotype C)
- Seroconversion (lower in genotype C)
- Tendency to chronicity (higher in genotype A)
- Likelihood of complications (higher in genotype C)
- Response to treatment and possibly vaccination (better response to IFN therapy in genotypes A and B. No association was found between genotypes and response to nucleosides or nucleotides)
Transmission
Transmission results from exposure to infectious blood or body fluids containing blood. Possible forms of transmission include (but are not limited to) unprotected sexual contact, blood transfusions, re-use of contaminated needles & syringes, and vertical transmission from mother to child during childbirth. Without intervention, a mother who is positive for the hepatitis B surface antigen confers a 20% risk of passing the infection to her offspring at the time of birth. This risk is as high as 90% if the mother is also positive for the hepatitis B e antigen. HBV can be transmitted between family members within households, possibly by contact of nonintact skin or mucous membrane with secretions or saliva containing HBV.[17] However, at least 30% of reported hepatitis B among adults cannot be associated with an identifiable risk factor.[18]
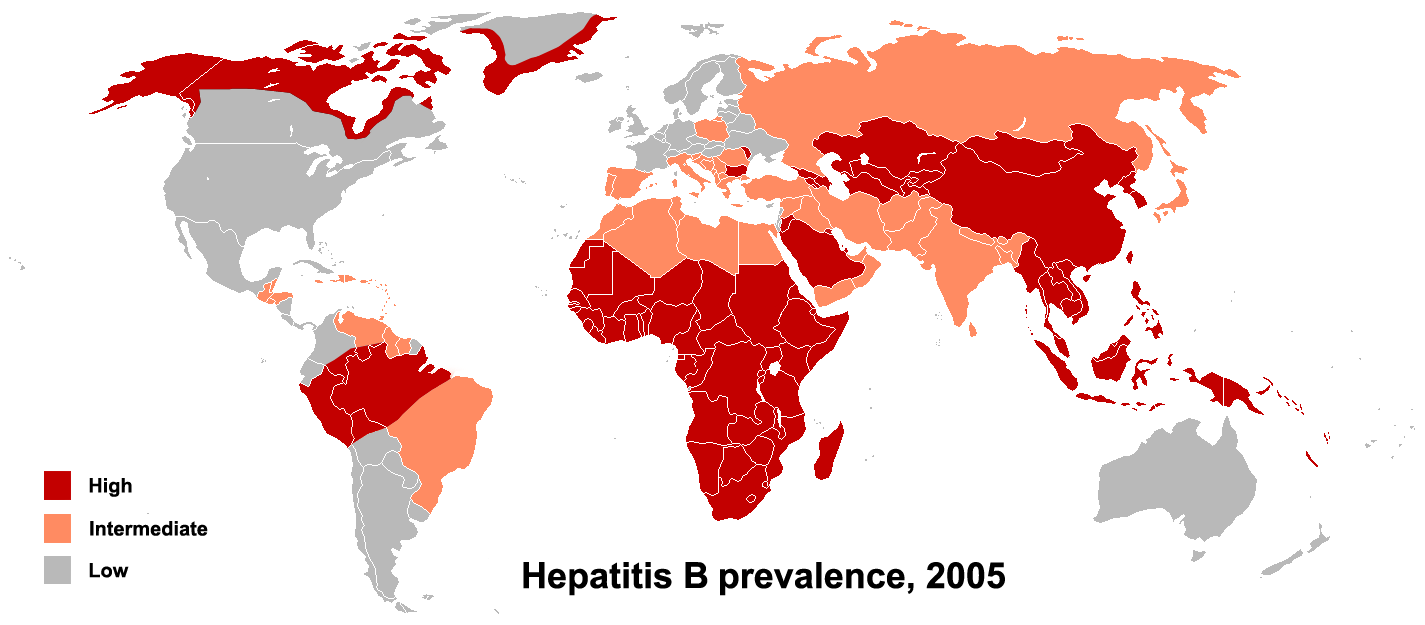
The primary method of transmission reflects the prevalence of chronic HBV infection in a given area. In low prevalence areas such as the continental United States and Western Europe, where less than 2% of the population is chronically infected, injection drug abuse and unprotected sex are the primary methods, although other factors may be important.[19] In moderate prevalence areas, which include Eastern Europe, Russia, and Japan, where 2-7% of the population is chronically infected, the disease is predominantly spread among children. In high prevalence areas such as China and South East Asia, transmission during childbirth is most common, although in other areas of high endemicity such as Africa, transmission during childhood is a significant factor.[20] The prevalence of chronic HBV infection in areas of high endemicity is at least 8%.
Tropism
Hepatitis B virus shows tropism for hepatocytes.[12]
Natural Reservoir
The natural reservoir for hepatitis B virus is man. Closely related hepadnaviruses have been found in woodchucks and ducks, but they are not infectious for humans.[21]
Pathogenesis
The hepatitis B virus primarily interferes with the functions of the liver by replicating in liver cells, known as hepatocytes. During HBV infection, the host immune response causes both hepatocellular damage and viral clearance. Although the innate immune response does not play a significant role in these processes, the adaptive immune response, particularly virus-specific cytotoxic T lymphocytes (CTLs), contributes to most of the liver injury associated with HBV infection. By killing infected cells and by producing antiviral cytokines capable of purging HBV from viable hepatocytes, CTLs eliminate the virus.[22] Although liver damage is initiated and mediated by the CTLs, antigen-nonspecific inflammatory cells can worsen CTL-induced immunopathology, and platelets activated at the site of infection may facilitate the accumulation of CTLs into the liver.[23]
See also
- Hepatitis A
- Hepatitis B in China
- Hepatitis C
- Hepatitis D
- Hepatitis E
- Hepatitis F
- Hepatitis G
- Jade Ribbon Campaign
- Maurice Hilleman
- Baruch S. Blumberg
References
- ↑ 1.0 1.1 Zuckerman AJ (1996). "Hepatitis Viruses". In Baron S; et al. Baron's Medical Microbiology (4th ed.). University of Texas Medical Branch. ISBN 0-9631172-1-1.
- ↑ 2.0 2.1 Magnius LO, Norder H (1995). "Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene". Intervirology. 38 (1–2): 24–34. PMID 8666521.
|access-date=requires|url=(help) - ↑ 3.0 3.1 Lin CL, Kao JH (2011). "The clinical implications of hepatitis B virus genotype: Recent advances". J Gastroenterol Hepatol. 26 Suppl 1: 123–30. doi:10.1111/j.1440-1746.2010.06541.x. PMID 21199523.
- ↑ Lurman A. (1885) Eine icterus epidemic. (In German). Berl Klin Woschenschr 22:20–3.
- ↑ Alter HJ, Blumberg BS (1966). "Further studies on a "new" human isoprecipitin system (Australia antigen)". Blood. 27 (3): 297–309. PMID 5930797.
- ↑ MacCallum, F.O., Homologous serum hepatitis. Lancet 2, 691, (1947)
- ↑ Dane DS, Cameron CH, Briggs M (1970). "Virus-like particles in serum of patients with Australia-antigen-associated hepatitis". Lancet. 1 (7649): 695–8. PMID 4190997.
- ↑ Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P (1979). "Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli". Nature. 281 (5733): 646–50. PMID 399327.
- ↑ "Hepatitis B vaccine". Lancet. 2 (8206): 1229–30. 1980. PMID 6108398.
- ↑ Locarnini S (2004). "Molecular virology of hepatitis B virus". Seminars in Liver Disease. 24 Suppl 1: 3–10. doi:10.1055/s-2004-828672. PMID 15192795. Retrieved 2012-02-08.
- ↑ Howard CR (1986). "The biology of hepadnaviruses". The Journal of General Virology. 67 ( Pt 7): 1215–35. PMID 3014045. Retrieved 2012-02-08. Unknown parameter
|month=ignored (help) - ↑ 12.0 12.1 12.2 12.3 12.4 12.5 12.6 12.7 "Hepatitis B" (PDF).
- ↑ Plotkin, Stanley (1999). Vaccines. Philadelphia: W.B. Saunders Co. ISBN 0721674437.
- ↑ Beck J, Nassal M (2007). "Hepatitis B virus replication". World J. Gastroenterol. 13 (1): 48–64. PMID 17206754.
- ↑ Bruss V (2007). "Hepatitis B virus morphogenesis". World J. Gastroenterol. 13 (1): 65–73. PMID 17206755.
- ↑ Kramvis A, Kew M, François G (2005). "Hepatitis B virus genotypes". Vaccine. 23 (19): 2409–23. doi:10.1016/j.vaccine.2004.10.045. PMID 15752827. Retrieved 2012-02-08. Unknown parameter
|month=ignored (help) - ↑ Petersen NJ, Barrett DH, Bond WW, Berquist KR, Favero MS, Bender TR, Maynard JE (1976). "Hepatitis B surface antigen in saliva, impetiginous lesions, and the environment in two remote Alaskan villages". Appl. Environ. Microbiol. 32 (4): 572–574. PMID 791124.
- ↑ Shapiro CN (1993). "Epidemiology of hepatitis B". Pediatr. Infect. Dis. J. 12 (5): 433–7. PMID 8392167.
- ↑ Redd JT, Baumbach J, Kohn W; et al. (2007). "Patient-to-patient transmission of hepatitis B virus associated with oral surgery" (PDF). J Infect Dis. 195 (9): 1311&ndash, 4. Retrieved 2007-12-12.
- ↑ Alter MJ (2003). "Epidemiology and prevention of hepatitis B". Semin. Liver Dis. 23 (1): 39–46. doi:10.1055/s-2003-37583. PMID 12616449.
- ↑ "Hepatitis B".
- ↑ Iannacone M, Sitia G, Ruggeri ZM, Guidotti LG (2007). "HBV pathogenesis in animal models: recent advances on the role of platelets". J. Hepatol. 46 (4): 719–26. doi:10.1016/j.jhep.2007.01.007. PMID 17316876.
- ↑ Iannacone M, Sitia G, Isogawa M, Marchese P, Castro M, Lowenstein P, Chisari F, Ruggeri Z, Guidotti L (2005). "Platelets mediate cytotoxic T lymphocyte-induced liver damage". Nature Medicine. 11 (11): 1167–1169. PMID 16258538.
External links
- NIH collection of links to relevant articles on Hepatitis B
- Hepatitis B Foundation, non-profit organization dedicated to the global problem of hepatitis B
- CDC webpage on Hepatitis B
- ThinkB: Hepatitis B information for Asians. Multiple language content available.
- Pediatric Hepatitis Report compiled by Parents of Kids with Infectious Diseases
- Advances in Hepatitis B Research: From Virology to Clinical Management
- An illustration of subunit vaccine development for Hepatitis B - Nova Online
- American Liver Foundation: Comprehensive information about Hepatitis B, including links to chapters for finding local resources
- Children's Liver Disease Foundation