RNA
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]

Ribonucleic acid or RNA is a nucleic acid, consisting of many nucleotides that form a polymer. Each nucleotide consists of a nitrogenous base, a ribose sugar, and a phosphate. RNA plays several important roles in the processes of translating genetic information from deoxyribonucleic acid (DNA) into proteins. One type of RNA acts as a messenger between DNA and the protein synthesis complexes known as ribosomes, others form vital portions of the structure of ribosomes, act as essential carrier molecules for amino acids to be used in protein synthesis, or change which genes are active.
RNA is very similar to DNA, but differs in a few important structural details: RNA is usually single stranded, while DNA is usually double stranded. RNA nucleotides contain ribose while DNA contains deoxyribose (a type of ribose that lacks one oxygen atom), and RNA uses the nucleotide uracil in its composition, instead of thymine which is present in DNA. RNA is transcribed from DNA by enzymes called RNA polymerases and is generally further processed by other enzymes, some of them guided by non-coding RNAs.
Chemical and stereochemical structure
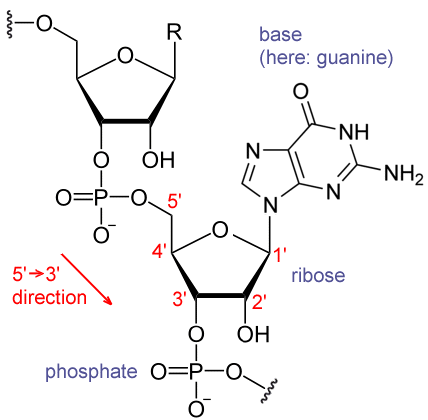
Each nucleotide in RNA contains a ribose, whose carbons are numbered 1' through 5'. The base – often adenine, cytosine, guanine or uracil – is attached to the 1' position. A phosphate group is attached to the 3' position of one ribose and the 5' position of the next. The phosphate groups have a negative charge each at physiological pH, making RNA a charged molecule.
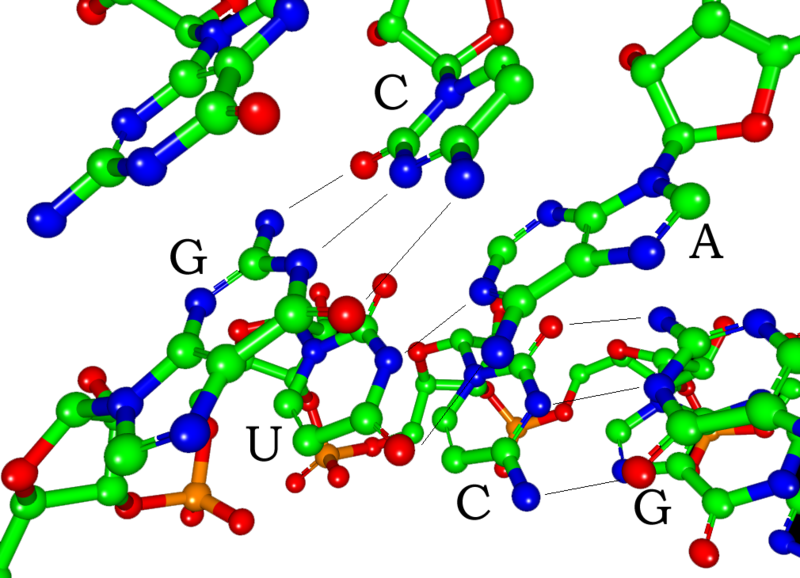
The bases often form hydrogen bonds between adenine and uracil and between cytosine and guanine, but other interactions are possible,[1] such as a group of adenine bases binding to each other in a bulge.[2]
There are also numerous modified bases and sugars found in RNA that serve many different roles. Pseudouridine (Ψ), in which the linkage between uracil and ribose is changed from a C–N bond to a C–C bond, and ribothymidine (T), are found in various places (most notably in the TΨC loop of tRNA).[3]
Another notable modified base is hypoxanthine, a deaminated guanine base whose nucleoside is called inosine. Inosine plays a key role in the Wobble Hypothesis of the genetic code.[4] There are nearly 100 other naturally occurring modified nucleosides,[5] of which pseudouridine and nucleosides with 2'-O-methylribose are the most common.[6]
The specific roles of many of these modifications in RNA are not fully understood. However, it is notable that in ribosomal RNA, many of the post-translational modifications occur in highly functional regions, such as the peptidyl transferase center and the subunit interface, implying that they are important for normal function.[7]
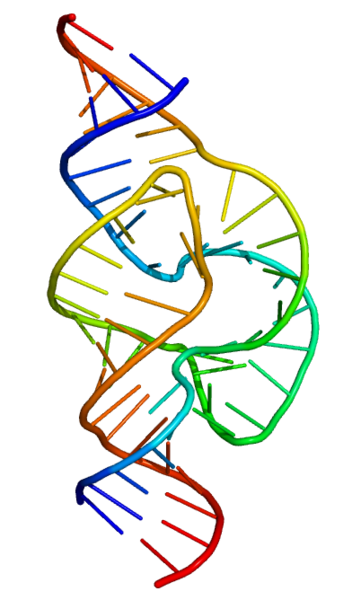
The most important structural feature of RNA, that distinguishes it from DNA is the presence of a hydroxyl group at the 2'-position of the ribose sugar. The presence of this functional group enforces the C3'-endo sugar conformation (as opposed to the C2'-endo conformation of the deoxyribose sugar in DNA) that causes the helix to adopt the A-form geometry rather than the B-form most commonly observed in DNA.[8] This results in a very deep and narrow major groove and a shallow and wide minor groove.[9]
A second consequence of the presence of the 2'-hydroxyl group is that in conformationally flexible regions of an RNA molecule (that is, not involved in formation of a double helix), it can chemically attack the adjacent phosphodiester bond to cleave the backbone.[10]
Comparison with DNA
RNA and DNA differ in three main ways. First, unlike DNA which is double-stranded, RNA is a single-stranded molecule in most of its biological roles and has a much shorter chain of nucleotides. Secondly, while DNA contains deoxyribose, RNA contains ribose, (there is no hydroxyl group attached to the pentose ring in the 2' position in DNA, whereas RNA has two hydroxyl groups). These hydroxyl groups make RNA less stable than DNA because it is more prone to hydrolysis. Thirdly, the complementary nucleotide to adenine is not thymine, as it is in DNA, but rather uracil, which is an unmethylated form of thymine.
Like DNA, most biologically active RNAs including tRNA, rRNA, snRNAs and other non-coding RNAs are extensively base paired to form double stranded helices. Structural analysis of these RNAs have revealed that they are highly structured. Unlike DNA, this structure is not just limited to long double-stranded helices but rather collections of short helices packed together into structures akin to proteins. In this fashion, RNAs can achieve chemical catalysis, like enzymes. For instance, determination of the structure of the ribosome – an enzyme that catalyzes peptide bond formation – revealed that its active site is composed entirely of RNA.
Synthesis
Synthesis of RNA is usually catalyzed by an enzyme - RNA polymerase, using DNA as a template. Initiation of synthesis begins with the binding of the enzyme to a promoter sequence in the DNA (usually found "upstream" of a gene). The DNA double helix is unwound by the helicase activity of the enzyme. The enzyme then progresses along the template strand in the 3’ -> 5’ direction, synthesizing a complementary RNA molecule with elongation occurring in the 5’ -> 3’ direction. The DNA sequence also dictates where termination of RNA synthesis will occur.[11]
There are also a number of RNA-dependent RNA polymerases as well that use RNA as their template for synthesis of a new strand of RNA. For instance, a number of RNA viruses (such as poliovirus) use this type of enzyme to replicate their genetic material.[12] Also, it is known that RNA-dependent RNA polymerases are required for the RNA interference pathway in many organisms.[13]
Biological roles
Messenger RNA (mRNA)
Messenger RNA is RNA that carries information from DNA to the ribosome sites of protein synthesis in the cell. In eukaryotic cells, once mRNA has been transcribed from DNA, it is "processed" before being exported from the nucleus into the cytoplasm, where it is bound to ribosomes and translated into its corresponding protein form with the help of tRNA. In prokaryotic cells, which do not have nucleus and cytoplasm compartments, mRNA can bind to ribosomes while it is being transcribed from DNA. After a certain amount of time the message degrades into its component nucleotides, usually with the assistance of ribonucleases.
Non-coding RNA
RNA genes (also known as non-coding RNA or small RNA) are genes that encode RNA which is not translated into a protein. The most prominent examples of RNA genes are transfer RNA (tRNA) and ribosomal RNA (rRNA), both of which are involved in the process of translation. Two other groups of non-coding RNA are microRNAs (miRNA) which regulate the gene expression and small nuclear RNAs (snRNA), a diverse class that includes for example the RNAs that form spliceosomes that excise introns from pre-mRNA.[14]
Certain RNAs are able to catalyse chemical reactions such as cutting and ligating other RNA molecules, and the catalysis of peptide bond formation in the ribosome; these are known as ribozymes.
Translation
In addition to mRNA, two types av RNA are involved in translation: ribosomal RNA (rRNA) and transfer RNA (tRNA).
Ribosomal RNA is the catalytic component of the ribosomes, the protein synthesis factories in the cell. Eukaryotic ribosomes contain four different rRNA molecules: 18S, 5.8S, 28S, and 5S rRNA. Three of the rRNA molecules are synthesized in the nucleolus, and one is synthesized elsewhere. rRNA molecules are extremely abundant and make up at least 80% of the RNA molecules found in a typical eukaryotic cell. In the cytoplasm, ribosomal RNA and protein combine to form a nucleoprotein called a ribosome. The ribosome binds mRNA and carries out protein synthesis. Several ribosomes may be attached to a single mRNA at any time.
Transfer RNA is a small RNA chain of about 74-95 nucleotides that transfers a specific amino acid to a growing polypeptide chain at the ribosomal site of protein synthesis, during translation. It has sites for amino-acid attachment and an anticodon region for codon recognition that binds to a specific sequence on the messenger RNA chain through hydrogen bonding.
Gene regulation
Several types of RNA can downregulate gene expression by being complementary to a part of a gene. MicroRNAs (miRNA; 21-22 nt) are found in eukaryotes and act through RNA interference (RNAi), where an effector complex of miRNA and enzymes can break down mRNA which the miRNA is complementary to, block the mRNA from being translated, or cause the promoter to be methylated which generally downregulates the gene.[15] Some miRNAs upregulate genes instead (RNA activation).[16] While small interfering RNAs (siRNA; 20-25 nt) are often produced by breakdown of viral RNA, there are also endogenous sources of siRNAs.[17] siRNAs act through RNA interference in a fashion similar to miRNAs. Animals have Piwi-interacting RNAs (piRNA; 29-30 nt) which are active in germline cells and are though to be a defense against transposons and play a role in gametogenesis.[18][19] X chromosome inactivation in female mammals is caused by Xist, an RNA which coats one X chromosome, inactiving it.[20] Antisense RNAs are widespread among bacteria; most downregulate a gene, but a few are activators of transcription.[21]
RNA modification
RNA can be modified after transcription not only by splicing, but also by having its bases modified to other bases than adenine, cytosine, guanine and uracil.
In eukaryotes, modifications of RNA bases are generally directed by small nucleolar RNAs (snoRNA), found in the nucleolus and cajal bodies. snoRNAs associate with enzymes and guide them to a spot on an RNA by basepairing to that RNA. These enzymes then perform the base modification. rRNA and tRNA are extensively modified, but snRNA and mRNA can also be the target of base modification.[22][23]
Double-stranded RNA
Double-stranded RNA (dsRNA) is RNA with two complementary strands, similar to the DNA found in all cells. dsRNA forms the genetic material of some viruses called double-stranded RNA viruses. In eukaryotes, long double-stranded RNA such as viral RNA can trigger RNA interference, where short dsRNA molecules called siRNAs (small interfering RNAs) can cause enzymes to break down specific mRNAs or silence the expression of genes. siRNA can also increase the transcription of a gene, a process called RNA activation.[24]
RNA world hypothesis
The RNA world hypothesis proposes that the earliest forms of life relied on RNA both to carry genetic information (like DNA does now) and to catalyze biochemical reactions like an enzyme. According to this hypothesis, descendants of these early lifeforms gradually integrated DNA and proteins into their metabolism.
RNA secondary structures
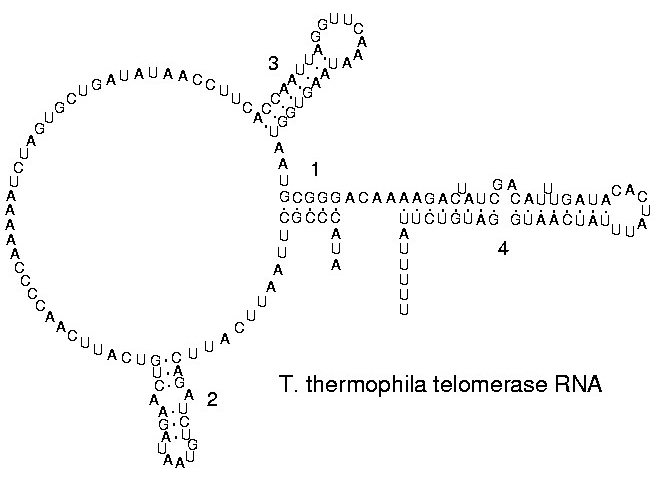
The functional form of single stranded RNA molecules, just like proteins, frequently requires a specific tertiary structure. The scaffold for this structure is provided by secondary structural elements which are hydrogen bonds within the molecule. This leads to several recognizable "domains" of secondary structure like hairpin loops, bulges and internal loops. The secondary structure of RNA molecules can be predicted computationally by calculating the minimum free energies (MFE) structure for all different combinations of hydrogen bondings and domains.[25] There has been a significant amount of research directed at the RNA structure prediction problem.
Online tools for MFE structure prediction from single sequences are provided by MFOLD and RNAfold.
Comparative studies of conserved RNA structures are significantly more accurate and provide evolutionary information. Computationally reasonable and accurate online tools for alignment folding are provided by KNetFold, RNAalifold and Pfold.
A package of RNA structure prediction programs is also available for Windows: RNAstructure.
A database of RNA sequences and secondary structures is available from Rfam, analyses and links to RNA analysis tools are available from Wikiomics.
List of RNA types
| Type | Abbr. | Function | Distribution |
|---|---|---|---|
| Messenger RNA | mRNA | Codes for protein | All cells |
| Ribosomal RNA | rRNA | Translation | All cells |
| Transfer RNA | tRNA | Translation | All cells |
| Small nuclear RNA | snRNA | Various | Eukaryotes and archaea [26] |
| Small nucleolar RNA | snoRNA | RNA editing | Eukaryotes and archaea [27] |
| MicroRNA | miRNA | Gene regulation | Eukaryotes |
| Piwi-interacting RNA | piRNA | Gene regulation | Animal germline cells |
| Small interfering RNA | siRNA | Gene regulation | Eukaryotes |
| Antisense RNA | aRNA | Gene regulation | Bacteria |
| Transfer-messenger RNA | tmRNA | Terminating translation | Bacteria |
| Signal recognition particle RNA | SRP RNA | mRNA tagging for export | All cells |
| Guide RNA | gRNA | RNA editing | Kinetoplastid mitochondria [28] |
| Ribozyme | Catalysis | All cells | |
| Transposon | Self-propagating | All cells | |
| Viroid | Self-propagating | Infected plants |
In addition, the genome of many types of viruses consists of RNA, namely double-stranded RNA viruses, positive-sense RNA viruses, negative-sense RNA viruses and most satellite viruses and reverse transcribing viruses.
History
Nucleic acids were discovered in 1868 by Friedrich Miescher, who called the material 'nuclein' since it was found in the nucleus.[29] It was later discovered that prokaryotic cells, which do not have a nucleus, also contain nucleic acids. The role of RNA in protein synthesis had been suspected since 1939, based on experiments carried out by Torbjörn Caspersson, Jean Brachet and Jack Schultz.[30] Gerard Marbaix isolated the first messenger RNA, for rabbit hemoglobin, and found it induced the synthesis of hemoglobin after injection into oocytes.[31] Finally, Severo Ochoa discovered the RNA, winning Ochoa the 1959 Nobel Prize for Medicine.[32] The sequence of the 77 nucleotides of a yeast RNA was found by Robert W. Holley in 1965,[33] winning Holley the 1968 Nobel Prize for Medicine. In 1976, Walter Fiers and his team at the University of Ghent determined the first complete nucleotide sequence of an RNA virus genome, that of bacteriophage MS2.[34]
While ribosomal RNA and transfer RNA were found early, since the late 1990s, many new RNA genes have been found, and thus RNA genes may play a much more significant role than previously thought. In the late 1990s and early 2000, there was persistent evidence of more complex transcription occurring in mammalian cells (and possibly others). This could point towards a more widespread use of RNA in biology, particularly in gene regulation. A particular class of non-coding RNA, microRNA, has been found in many eukaryotes and clearly plays an important role in regulating other genes, through RNA interference.
References
- ↑ Westhof E, Fritsch V (2000). "RNA folding: beyond Watson–Crick pairs". Structure. 8 (3): R55–R65.
- ↑ Barciszewski J, Frederic B, Clark C (1999). RNA biochemistry and biotechnology. Springer. pp. 73–87. ISBN 0792358627.
- ↑ Yu Q, Morrow CD (2001). "Identification of critical elements in the tRNA acceptor stem and TΨC loop necessary for human immunodeficiency virus type 1 infectivity". J Virol. 75 (10): 4902–06.
- ↑ Elliott MS, Trewyn RW (1983). "Inosine biosynthesis in transfer RNA by an enzymatic insertion of hypoxanthine". J. Biol. Chem. 259 (4): 2407–10.
- ↑ Söll D, RajBhandary U (1995). tRNA: Structure, biosynthesis, and function. ASM Press. p. 165. ISBN 155581073X.
- ↑ Kiss T (2001). "Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs". The EMBO Journal. 20: 3617–22.
- ↑ King TH, Liu B, McCully RR, Fournier MJ (2002). "Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center". Molecular Cell. 11 (2): 425–435.
- ↑ Salazar M, Fedoroff OY, Miller JM, Ribeiro NS, Reid BR (1992). "The DNA strand in DNAoRNA hybrid duplexes is neither B-form nor A-form in solution" (PDF). Biochemistry. 1993 (32): 4207–15.
- ↑ Hermann T, Patel DJ (2000). "RNA bulges as architectural and recognition motifs". Structure. 8 (3): R47–R54.
- ↑ Mikkola S, Nurmi K, Yousefi-Salakdeh E, Strömberg R, Lönnberg H (1999). "The mechanism of the metal ion promoted cleavage of RNA phosphodiester bonds involves a general acid catalysis by the metal aquo ion on the departure of the leaving group". Perkin transactions 2: 1619–1626.
- ↑ Nudler E, Gottesman ME (2002). "Transcription termination and anti-termination in E. coli". Genes to Cells. 7: 755–768.
- ↑ Jeffrey L Hansen, Alexander M Long, Steve C Schultz (1997). "Structure of the RNA-dependent RNA polymerase of poliovirus". Structure. 5 (8): 1109–1122.
- ↑ Ahlquist P (2002). "RNA-Dependent RNA Polymerases, Viruses, and RNA Silencing". Science. 296 (5571): 1270–1273.
- ↑ Berg JM, Tymoczko JL, Stryer L (2002). Biochemistry (5th edition ed.). WH Freeman and Company. pp. 781–808. ISBN 0-7167-4684-0.
- ↑ Matzke MA, Matzke AJM (2004). "Planting the seeds of a new paradigm". PLoS Biology. 2 (5): e133.
- ↑ Check E (2007). "RNA interference: hitting the on switch". Nature. 448 (7156): 855–858.
- ↑ Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert J, Bartel DP, Crété P (2004). "Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs". Molecular Cell. 16 (1): 69–79.
- ↑ Horwich MD, Li C Matranga C, Vagin V, Farley G, Wang P, Zamore PD (2007). "The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC". Current Biology. 17: 1265–72.
- ↑ Girard A, Sachidanandam R, Hannon GJ, Carmell MA (2006). "A germline-specific class of small RNAs binds mammalian Piwi proteins". Nature. 442: 199–202.
- ↑ Heard E, Mongelard F, Arnaud D, Chureau C, Vourc'h C, Avner P (1999). "Human XIST yeast artificial chromosome transgenes show partial X inactivation center function in mouse embryonic stem cells". Proc. Natl. Acad. Sci. USA. 96 (12): 6841–6846.
- ↑ Wagner EG, Altuvia S, Romby P (2002). "Antisense RNAs in bacteria and their genetic elements". Adv Genet. 46: 361–98.
- ↑ Covello PS, Gray MW (1989). "RNA editing in plant mitochondria". Nature. 341: 662–666.
- ↑ Omer AD, Ziesche S, Decatur WA, Fournier MJ, Dennis PP (2003). "RNA-modifying machines in archaea". Molecular Microbiology. 48 (3): 617–629.
- ↑ Doran G (2007). "RNAi – Is one suffix sufficient?". Journal of RNAi and Gene Silencing. 3 (1): 217–219.
- ↑ Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH (2004). "Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure" (PDF). Proc. Natl. Acad. Sci. U. S. A. 101 (19): 7287–7292.
- ↑ Thore S, Mayer C, Sauter C, Weeks S, Suck D (2003). "Crystal Structures of the Pyrococcus abyssi Sm Core and Its Complex with RNA". J. Biol. Chem. 278 (2): 1239–47.
- ↑ Kiss T (2001). "Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs". The EMBO Journal. 20: 3617–22.
- ↑ Alfonzo JD, Thiemann O, Simpson L (1997). "The mechanism of U insertion/deletion RNA editing in kinetoplastid mitochondria". Nucleic Acids Research. 25 (19): 3751–59.
- ↑ Dahm R (2005). "Friedrich Miescher and the discovery of DNA". Developmental Biology. 278 (2): 274–88. PMID 15680349.
- ↑ Nierhaus KH, Wilson DN (2004). Protein Synthesis and Ribosome Structure. Wiley-VCH. p. 3. ISBN 3527306382.
- ↑ Carlier M (2003). "L'ADN, cette "simple" molécule". Esprit libre.
- ↑ Ochoa S (1959). "Enzymatic synthesis of ribonucleic acid" (PDF). Nobel Lecture.
- ↑ Holley RW; et al. (1965). "Structure of a ribonucleic acid". Science. 147 (1664): 1462–1465.
- ↑ Fiers W; et al. (1976). "Complete nucleotide-sequence of bacteriophage MS2-RNA: primary and secondary structure of replicase gene". Nature. 260: 500–507.
External links
- RNA World website
- Nucleic Acid Database Images of DNA, RNA and complexes.
- RNAJunction Database: Extracted atomic models of RNA junction and kissing loop structures.
See also
ar:حمض ريبي نووي bn:আরএনএ zh-min-nan:RNA bs:Ribonukleinska kiselina br:Trenkenn ribonukleek bg:РНК ca:Àcid ribonucleic cs:RNA da:RNA de:Ribonukleinsäure et:Ribonukleiinhape el:RNA eo:RNA eu:Azido erribonukleiko fo:RNA gl:Ácido ribonucleico ko:RNA hr:Ribonukleinska kiselina id:Asam ribonukleat is:RKS it:RNA he:RNA la:Acidum ribonucleicum lv:Ribonukleīnskābe lb:RNS lt:Ribonukleino rūgštis hu:RNS mk:РНК mn:РНХ nl:RNA no:RNA nn:Ribonukleinsyre oc:Acid ribonucleïc simple:RNA sk:Ribonukleová kyselina sl:Ribonukleinska kislina sr:Рибонуклеинска киселина sh:Ribonukleinska kiselina fi:RNA sv:Ribonukleinsyra ta:ரைபோ கரு அமிலம் te:రైబో కేంద్రక ఆమ్లం th:อาร์เอ็นเอ uk:РНК ur:رائبو مرکزی ترشہ yo:RNA Template:Jb1