Human leukocyte antigen: Difference between revisions
m (Robot: Automated text replacement (-{{SIB}} + & -{{EH}} + & -{{EJ}} + & -{{Editor Help}} + & -{{Editor Join}} +)) |
m (Page link organ transplant rejections; noted T-helper cells are a.k.a. CD4+ (both these edits in the lead)) |
||
| Line 1: | Line 1: | ||
[[ | [[File:HLA.svg|frame|right|HLA region of Chromosome 6]] | ||
The '''human leukocyte antigen''' ('''HLA''') system or complex is a [[gene complex]] encoding the [[major histocompatibility complex]] (MHC) proteins in humans. These [[membrane protein|cell-surface protein]]s are responsible for the regulation of the [[immune system]] in humans. The HLA gene complex resides on a 3 [[base pair|Mbp]] stretch within [[Chromosome 6 (human)|chromosome 6]]p21. HLA genes are highly [[genetic polymorphism|polymorphic]], which means that they have many different [[alleles]], allowing them to fine-tune the [[adaptive immune system]]. The proteins encoded by certain genes are also known as ''[[Antigen|antigens]]'', as a result of their [[History and naming of human leukocyte antigens|historic discovery]] as factors in organ transplants. Different classes have different functions: | |||
HLAs corresponding to [[MHC class I]] ('''[[HLA-A|A]]''', '''[[HLA-B|B]]''', and '''[[HLA-C|C]]''') present peptides from inside the cell. For example, if the cell is infected by a virus, the HLA system brings fragments of the virus to the surface of the cell so that the cell can be destroyed by the immune system. These peptides are produced from digested proteins that are broken down in the [[proteasome]]s. In general, these particular peptides are small [[polymer]]s, about 9 [[amino acid]]s in length.{{Citation needed|date=March 2014}} Foreign antigens presented by MHC class I attract [[killer T-cell]]s (also called [[CD8]] positive- or cytotoxic T-cells) that destroy cells. MHC class I proteins associate with [[Beta-2 microglobulin|β2-microglobulin]], which unlike the HLA proteins is encoded by a gene on [[Chromosome 15 (human)|chromosome 15]]. | |||
HLAs corresponding to [[MHC class II]] ('''[[HLA-DP|DP]]''', '''[[HLA-DM|DM]]''', '''[[HLA-DOA|DOA]]''', '''[[HLA-DOB|DOB]]''', '''[[HLA-DQ|DQ]]''', and '''[[HLA-DR|DR]]''') present antigens from outside of the cell to T-lymphocytes. These particular antigens stimulate the multiplication of [[T-helper cell]]s (also called [[CD4]] positive T cells), which in turn stimulate [[antibody]]-producing [[B cell|B-cells]] to produce antibodies to that specific antigen. Self-antigens are suppressed by [[regulatory T cell]]s. | |||
HLAs corresponding to MHC class III encode components of the [[complement system]]. | |||
HLAs have other roles. They are important in disease defense. They are the major cause of [[transplant rejection|organ transplant rejections]]. They may protect against or fail to protect (if down-regulated by an infection) against cancers.<ref>{{cite journal | vauthors = Galbraith W, Wagner MC, Chao J, Abaza M, Ernst LA, Nederlof MA, Hartsock RJ, Taylor DL, Waggoner AS | title = Imaging cytometry by multiparameter fluorescence | journal = Cytometry | volume = 12 | issue = 7 | pages = 579–96 | year = 1991 | pmid = 1782829 | doi = 10.1002/cyto.990120702 | display-authors = 6 }}</ref> Mutations in HLA may be linked to [[autoimmune disease]] (examples: [[type I diabetes]], [[coeliac disease]]). HLA may also be related to people's perception of the odor of other people, and may be involved in mate selection, as at least one study found a lower-than-expected rate of HLA similarity between spouses in an isolated community.<ref>{{cite journal | vauthors = Brennan PA, Kendrick KM | title = Mammalian social odours: attraction and individual recognition | journal = Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences | volume = 361 | issue = 1476 | pages = 2061–78 | date = December 2006 | pmid = 17118924 | pmc = 1764843 | doi = 10.1098/rstb.2006.1931 }}</ref> | |||
Aside from the genes encoding the 6 major antigen-presenting proteins, there are a large number of other genes, many involved in immune function, located on the HLA complex. Diversity of HLAs in the human population is one aspect of disease defense, and, as a result, the chance of two unrelated individuals with identical HLA molecules on all [[locus (genetics)|loci]] is extremely low. HLA genes have historically been identified as a result of the ability to successfully transplant organs between HLA-similar individuals. | |||
==Functions== | |||
The [[protein]]s encoded by HLAs are those on the outer part of body cells that are (in effect) unique to that person. The [[immune system]] uses the HLAs to differentiate self cells and non-self cells. Any cell displaying that person's HLA type belongs to that person and, therefore, is not an invader. | |||
[[Image:DR beta 1 SEI topdown.JPG|frame|left|DR protein (DRA:DRB1*0101 gene products) with bound Staphylococcal enterotoxin ligand (subunit I-C), view is top down showing all DR amino acid residues within 5 Angstroms of the SEI peptide. {{PDB|2G9H}}]] | |||
== | ===In infectious disease=== | ||
When a foreign [[pathogen]] enters the body, specific [[cell (biology)|cells]] called [[antigen-presenting cell]]s (APCs) engulf the pathogen through a process called [[phagocytosis]]. Proteins from the pathogen are digested into small pieces ([[peptide]]s) and loaded onto HLA antigens (to be specific, [[MHC class II]]). They are then displayed by the [[antigen-presenting cell]]s to [[T helper cell|CD4+ helper T cells]],<ref name="pmid21727137">{{cite journal |vauthors=Taylor CJ, Bolton EM, Bradley JA | title=Immunological considerations for embryonic and induced pluripotent stem cell banking | journal=[[Philosophical Transactions of the Royal Society B]] | volume=366 | issue=1575 | year=2011 | pages=2312–2322 | doi=10.1098/rstb.2011.0030 | url= http://rstb.royalsocietypublishing.org/content/366/1575/2312.long | pmc=3130422 | pmid=21727137}}</ref> which then produce a variety of effects to eliminate the pathogen. | |||
Through a similar process, proteins (both native and foreign, such as the proteins of virus) produced inside most cells are displayed on HLAs (to be specific, [[MHC class I]]) on the cell surface. Infected cells can be recognized and destroyed by [[Cytotoxic T cell|CD8+ T cells]].<ref name="pmid21727137" /> | |||
The image off to the side shows a piece of a poisonous bacterial protein (SEI peptide) bound within the binding cleft portion of the HLA-DR1 molecule. In the illustration far below, a different view, one can see an entire DQ with a bound peptide in a similar cleft, as viewed from the side. Disease-related peptides fit into these "slots" much like a hand fits into a glove. When bound, peptides are presented to T cells. T cells require presentation via MHC molecules to recognize foreign antigens — a requirement known as [[MHC restriction]]. These cells have receptors that are similar to B cell receptors, and each cell recognizes only a few class II-peptide combinations. Once a T cell recognizes a peptide within an MHC class II molecule, it can stimulate B-cells that also recognize the same molecule in their B cell receptors. Thus, T cells help B cells make antibodies to the same foreign antigens. Each HLA can bind many peptides, and each person has 3 HLA types and can have 4 isoforms of DP, 4 isoforms of DQ and 4 Isoforms of DR (2 of DRB1, and 2 of DRB3, DRB4, or DRB5) for a total of 12 isoforms. In such heterozygotes, it is difficult for disease-related proteins to escape detection. | |||
===In graft rejection=== | |||
Any cell displaying some other HLA type is "non-self" and is seen as an invader by the body's immune system, resulting in the rejection of the tissue bearing those cells. This is particularly important in the case of transplanted tissue, because it could lead to [[transplant rejection]]. Because of the importance of HLA in transplantation, the HLA loci are some of the most frequently typed by serology and PCR. | |||
{| class="wikitable" style="float:right" | |||
|+ HLA and autoimmune diseases | |||
|-v | |||
! HLA allele !! Diseases with increased risk !! [[Relative risk]] | |||
|- | |||
|rowspan=3| [[HLA-B27]] | |||
| [[Ankylosing spondylitis]] || 12<ref name=Kumar5-7>Table 5-7 in: {{cite book |author1=Mitchell, Richard Sheppard |author2=Kumar, Vinay |author3=Abbas, Abul K. |author4=Fausto, Nelson |title=Robbins Basic Pathology|publisher=Saunders |location=Philadelphia |year= 2007|pages= |isbn=1-4160-2973-7 |oclc= |doi=}} 8th edition.</ref> | |||
|- | |||
| [[Postgonococcal arthritis]] || 14<ref name=Kumar5-7/> | |||
|-[[Post-salmonella arthritis]] | |||
| [[Acute anterior uveitis]] ||15<ref name=Kumar5-7/> | |||
|- | |||
| [[HLA-B47]] | |||
| [[Congenital adrenal hyperplasia due to 21-hydroxylase deficiency|21-hydroxylase deficiency]] || 15<ref name=Kumar5-7/> | |||
|- | |||
| [[HLA-DR2]] | |||
| [[Systemic lupus erythematosus]] || 2 to 3<ref name=Salmon>Values are given for Caucasians, according to [https://books.google.com/books?id=tj9JR3IITy8C&pg=PA61 Page 61] (right column) in: {{cite book |author1=Jane Salmon |author2=Wallace, Daniel J. |author3=Dubois, Edmund L. |author4=Kirou, Kyriakos A. |author5=Hahn, Bevra |author6=Lehman, Thomas A. |title=Dubois' lupus erythematosus |publisher=Lippincott Williams & Wilkins |location=Philadelphia |year=2007 |pages= |isbn=0-7817-9394-7 |oclc= |doi= |accessdate=}}</ref> | |||
|- | |||
|rowspan=4| [[HLA-DR3]] | |||
| [[Autoimmune hepatitis]] || 14<ref name=Kumar5-7/> | |||
|- | |||
| Primary [[Sjögren syndrome]] || 10<ref name=Kumar5-7/> | |||
|- | |||
| [[Diabetes mellitus type 1]] || 5<ref name=Kumar5-7/> | |||
|- | |||
| [[Systemic lupus erythematosus]] || 2 to 3<ref name=Salmon/> | |||
|- | |||
|rowspan=2| [[HLA-DR4]] | |||
| [[Rheumatoid arthritis]] || 4<ref name=Kumar5-7/> | |||
|- | |||
| [[Diabetes mellitus type 1]] || 6<ref name=Kumar5-7/> | |||
|- | |||
| HLA-DR3 and<br> -DR4 combined | |||
| [[Diabetes mellitus type 1]] || 15<ref name=Kumar5-7/> | |||
|- | |||
| [[HLA-DQ2]] and [[HLA-DQ8]] | |||
| [[Coeliac disease]] || 7<ref name="ref1595277555">{{cite journal | vauthors = Margaritte-Jeannin P, Babron MC, Bourgey M, Louka AS, Clot F, Percopo S, Coto I, Hugot JP, Ascher H, Sollid LM, Greco L, Clerget-Darpoux F | title = HLA-DQ relative risks for coeliac disease in European populations: a study of the European Genetics Cluster on Coeliac Disease | journal = Tissue Antigens | volume = 63 | issue = 6 | pages = 562–7 | date = June 2004 | pmid = 15140032 | doi = 10.1111/j.0001-2815.2004.00237.x }}</ref> | |||
|} | |||
===In autoimmunity=== | |||
HLA types are inherited, and some of them are connected with [[autoimmune disorder]]s and other diseases. People with certain HLA antigens are more likely to develop certain autoimmune diseases, such as [[type I diabetes]], [[ankylosing spondylitis]], [[coeliac disease|celiac disease]], [[systemic lupus erythematosus|SLE]] (systemic lupus erythematosus), [[myasthenia gravis]], [[inclusion body myositis]], [[Sjögren syndrome]], and [[narcolepsy]].<ref name="cell">{{cite journal|url=http://www.cell.com/AJHG/retrieve/pii/S0002929707631085|title=Complex HLA-DR and -DQ Interactions Confer Risk of Narcolepsy-Cataplexy in Three Ethnic Groups|accessdate=2014-01-25|doi=10.1086/318799|volume=68|journal=The American Journal of Human Genetics|pages=686–699}}</ref> HLA typing has led to some improvement and acceleration in the diagnosis of celiac disease and type 1 diabetes; however, for DQ2 typing to be useful, it requires either high-resolution B1*typing (resolving *02:01 from *02:02), DQA1*typing, or DR [[serotyping]]. Current serotyping can resolve, in one step, DQ8. HLA typing in autoimmunity is being increasingly used as a tool in diagnosis. In [[celiac disease]], it is the only effective means of discriminating between first-degree relatives that are at risk from those that are not at risk, prior to the appearance of sometimes-irreversible symptoms such as allergies and secondary autoimmune disease. | |||
===In cancer=== | |||
Some HLA-mediated diseases are directly involved in the promotion of cancer. Gluten-sensitive enteropathy is associated with increased prevalence of enteropathy-associated T-cell lymphoma, and DR3-DQ2 homozygotes are within the highest risk group, with close to 80% of gluten-sensitive enteropathy-associated T-cell lymphoma cases. More often, however, HLA molecules play a protective role, recognizing increases in antigens that are not tolerated because of low levels in the normal state. Abnormal cells might be targeted for apoptosis, which is thought to mediate many cancers before diagnosis. | |||
===In mate selection=== | |||
There is evidence for non-random mate choice with respect to certain genetic characteristics. This has led to a field known as [[Genetic matchmaking]]. | |||
''' | ==Classification== | ||
[[Image:MHC Class 1.svg|thumb|150px|right|Schematic representation of MHC class I]] | |||
'''MHC class I''' proteins form a functional receptor on most nucleated cells of the body. | |||
There are 3 major and 3 minor MHC class I genes in HLA. | |||
'''Major MHC class I''' | |||
'''MHC class I''' | *[[HLA-A]] | ||
*[[HLA-B]] | |||
*[[HLA-C]] | |||
Minor genes are [[HLA-E]], [[HLA-F]] and [[HLA-G]]. [[beta-2 microglobulin|β<sub>2</sub>-microglobulin]] binds with major and minor gene subunits to produce a heterodimer | |||
[[Image:DQ Illustration.PNG|frame|thumb|100px|right|Illustration of an HLA-DQ molecule (magenta and blue) | |||
with a bound ligand (yellow) floating on the plasma membrane of the cell.]] | |||
There are 3 major and 2 minor '''MHC class II''' proteins encoded by the HLA. | |||
There are 3 major and 2 minor MHC class II | |||
The genes of the class II combine to form heterodimeric (αβ) protein | The genes of the class II combine to form heterodimeric (αβ) protein | ||
receptors that are typically expressed on the surface of [[antigen presenting cells]]. | receptors that are typically expressed on the surface of [[antigen-presenting cells]]. | ||
'''Major MHC class II''' | |||
*[[HLA-DP]] | |||
**'''α'''-chain encoded by HLA-'''DPA1''' locus | |||
**'''β'''-chain encoded by [[HLA-DPB1|HLA-'''DPB1''']] locus | |||
*[[HLA-DQ]] | |||
**'''α'''-chain encoded by [[HLA-DQA1|HLA-'''DQA1''']] locus | |||
**'''β'''-chain encoded by [[HLA-DQB1|HLA-'''DQB1''']] locus | |||
*[[HLA-DR]] | |||
**'''α'''-chain encoded by HLA-'''DRA''' locus | |||
**'''4 β-chains''' (only 3 possible per person), encoded by HLA-'''DRB1''', '''DRB3''', '''DRB4''', '''DRB5''' loci | |||
The other MHC class II proteins, DM and DO, are used in the internal processing of antigens, loading the antigenic peptides generated from pathogens onto the HLA molecules of [[antigen-presenting cell]]. | |||
MHC | |||
===Nomenclature=== | |||
Modern HLA alleles are typically noted with a variety of levels of detail. Most designations begin with HLA- and the locus name, then * and some (even) number of digits specifying the allele. The first two digits specify a group of alleles. Older typing methodologies often could not completely distinguish alleles and so stopped at this level. The third through fourth digits specify a nonsynonymous allele. Digits five through six denote any synonymous mutations within the coding frame of the gene. The seventh and eighth digits distinguish mutations outside the coding region. Letters such as L, N, Q, or S may follow an allele's designation to specify an expression level or other non-genomic data known about it. Thus, a completely described allele may be up to 9 digits long, not including the HLA-prefix and locus notation.<ref>http://hla.alleles.org/nomenclature/naming.html</ref> | |||
{{further|History and naming of human leukocyte antigens}} | |||
=== | ===Variability=== | ||
[[Image:MHC expression.svg|thumb|Right|300px|Codominant expression of HLA genes.]] | |||
MHC loci are some of the most genetically variable coding loci in mammals, and the human HLA loci are no exceptions. Despite the fact that the human population went through a constriction more than 150,000 years ago that was capable of fixing many loci, the HLA loci appear to have survived such a constriction with a great deal of variation.<ref>{{cite book |title=Evolution and the Spiral of Technology |last=Shennan |first=Douglas H |year=2006 |publisher=Trafford Publishing |isbn=1-55212-518-1 }}</ref> Of the 9 loci mentioned above, most retained a dozen or more allele-groups for each locus, far more preserved variation than the vast majority of human loci. This is consistent with a heterozygous or [[balancing selection]] coefficient for these loci. In addition, some HLA loci are among the fastest-evolving coding regions in the human genome. One mechanism of diversification has been noted in the study of Amazonian tribes of South America that appear to have undergone intense [[gene conversion]] between variable | |||
alleles and loci within each HLA gene class.<ref name=Parham>{{cite journal | vauthors = Parham P, Ohta T | title = Population biology of antigen presentation by MHC class I molecules | journal = Science | volume = 272 | issue = 5258 | pages = 67–74 | date = April 1996 | pmid = 8600539 | doi = 10.1126/science.272.5258.67 }}.</ref> Less frequently, longer-range productive recombinations through HLA genes have been noted producing chimeric genes. | |||
Number of variant alleles at class I loci according to the IMGT-HLA database, last updated | Six loci have over 100 alleles that have been detected in the human population. Of these, the most variable are HLA B and HLA DRB1. As of 2012, the number of alleles that have been determined are listed in the table below. To interpret this table, it is necessary to consider that an allele is a variant of the nucleotide (DNA) sequence at a locus, such that each allele differs from all other alleles in at least one (single nucleotide polymorphism, SNP) position. Most of these changes result in a change in the amino acid sequences that result in slight to major functional differences in the protein. | ||
There are issues that limit this variation. Certain alleles like DQA1*05:01 and DQA1*05:05 encode proteins with identically processed products. Other alleles like DQB1*0201 and DQB1*0202 produce proteins that are functionally similar. For class II (DR, DP and DQ), amino acid variants within the receptor's peptide-binding cleft tend to produce molecules with different binding capability. | |||
====Tables of variant alleles==== | |||
Number of variant alleles at class I loci according to the IMGT-HLA database, last updated July 2014: | |||
{| border="0" cellspacing="0" cellpadding="0" align="center" style="text-align:center; margin-Left: 1em; border:1px #bbbbff solid; background:#f8f8ff;" | {| border="0" cellspacing="0" cellpadding="0" align="center" style="text-align:center; margin-Left: 1em; border:1px #bbbbff solid; background:#f8f8ff;" | ||
|+ align="left" style = "font-size:90%" | | |+ align="left" style = "font-size:90%" | | ||
|- style = "background:#fafaff; font-size:130%" | |- style = "background:#fafaff; font-size:130%" | ||
! style = "height:30px" colspan="2" align="center" valign = "center" | MHC class I | ! style = "height:30px" colspan="2" align="center" valign = "center" | MHC class I | ||
|- style = "background:#eeeeff;font-size:110%" | |- style = "background:#eeeeff;font-size:110%" | ||
! | ! width = "135" | locus | ||
| width = "85" | '''#'''<ref name=Marsh>{{cite journal | | | width = "85" | '''#'''<ref name=Marsh>{{cite journal | vauthors = Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Geraghty DE, Hansen JA, Hurley CK, Mach B, Mayr WR, Parham P, Petersdorf EW, Sasazuki T, Schreuder GM, Strominger JL, Svejgaard A, Terasaki PI, Trowsdale J | title = Nomenclature for factors of the HLA system, 2004 | journal = Tissue Antigens | volume = 65 | issue = 4 | pages = 301–69 | date = April 2005 | pmid = 15787720 | doi = 10.1111/j.1399-0039.2005.00379.x }}</ref><ref name=imgt>[http://www.ebi.ac.uk/imgt/hla/stats.html IMGT/HLA Database<!-- Bot generated title -->]</ref> | ||
|- style = "background:#f4f4ff" | |||
Erlich HA, Geraghty DE, Hansen JA, Hurley CK, Mach B, Mayr WR, Parham P, Petersdorf EW, Sasazuki T, Schreuder GM, Strominger JL, Svejgaard A, Terasaki PI, | ! style = "color:#aaaadd" | Major Antigens | ||
|- style = "background:#f4f4ff" | |||
|- style = "background:#f4f4ff" | ! HLA A | ||
! | | style="background:#f8f8ff" | 2,884 | ||
|- style = "background:#f4f4ff" | |- style = "background:#f4f4ff" | ||
! | ! HLA B | ||
| style="background:#f8f8ff" | | | style="background:#f8f8ff" | 3,589 | ||
|- style = "background:#f4f4ff" | |- style = "background:#f4f4ff" | ||
! | ! HLA C | ||
| style="background:#f8f8ff" | | | style="background:#f8f8ff" | 2,375 | ||
|- style = "background:#f4f4ff" | |||
! | |||
| style="background:#f8f8ff" | | |||
|- style = "background:#f4f4ff" | |- style = "background:#f4f4ff" | ||
| style = "color:#aaaadd" | Minor Antigens | | style = "color:#aaaadd" | Minor Antigens | ||
|-style = "background:#f4f4ff" | |-style = "background:#f4f4ff" | ||
| HLA E | | HLA E | ||
| style="background:#f8f8ff" | | | style="background:#f8f8ff" | 15 | ||
|- style = "background:#f4f4ff" | |- style = "background:#f4f4ff" | ||
| HLA F | | HLA F | ||
| style="background:#f8f8ff" | | | style="background:#f8f8ff" | 22 | ||
|- style = "background:#f4f4ff" | |- style = "background:#f4f4ff" | ||
| HLA G | | HLA G | ||
| style="background:#f8f8ff" | | | style="background:#f8f8ff" | 50 | ||
|- style = "background:#f4f4ff" | |- style = "background:#f4f4ff" | ||
|} | |} | ||
Number of variant alleles at class II loci (DP and | Number of variant alleles at class II loci (DM, DO, DP, DQ, and DR): | ||
{| border="0" cellspacing="0" cellpadding="0" align="center" style="text-align:center; margin-Left: 1em; border:1px #bbbbff solid; background:#f8f8ff;" | {| border="0" cellspacing="0" cellpadding="0" align="center" style="text-align:center; margin-Left: 1em; border:1px #bbbbff solid; background:#f8f8ff;" | ||
|+ align="left" style = "font-size:90%" | | |+ align="left" style = "font-size:90%" | | ||
|- style = "background:#fafaff; font-size:130%" | |- style = "background:#fafaff; font-size:130%" | ||
! style = "height:30px" colspan="5" align="center" valign = "center" | MHC class II | ! style = "height:30px" colspan="5" align="center" valign = "center" | MHC class II | ||
|- style = "background:#eeeeff;font-size:100%" | |- style = "background:#eeeeff;font-size:100%" | ||
! | ! width = "60" | HLA | ||
| width = "60" | -'''A1''' | | width = "60" | -'''A1''' | ||
| width = "60" | -'''B1''' | | width = "60" | -'''B1''' | ||
| width = "85" style = "font-size:90%" | '''-B3 to -B5'''<sup>1</sup> | | width = "85" style = "font-size:90%" | '''-B3 to -B5'''<sup>1</sup> | ||
| width = "60" style = "font-size:80%" | | | width = "60" style = "font-size:80%" | Theor. possible | ||
|- style = "background:#eeeeff;font-size:100%" | |- style = "background:#eeeeff;font-size:100%" | ||
! locus | ! locus | ||
| '''#'''<ref name = | | '''#'''<ref name=imgt/> || '''#'''<ref name=imgt/> ||'''#'''<ref name=imgt/> | ||
| width = "60" style = "font-size:80%" | | | width = "60" style = "font-size:80%" | combinations | ||
|- style = "background:#f8f8ff" | |- style = "background:#f8f8ff" | ||
! | ! style="background:#f4f4ff" | DM- | ||
| | | 7 || 13 || | ||
| style="background:#fefeff" | | | style="background:#fefeff" | 91 | ||
|- style = "background:#f8f8ff" | |- style = "background:#f8f8ff" | ||
! | ! style="background:#f4f4ff" | DO- | ||
| 12 || | | 12 || 13 || | ||
| style="background:#fefeff" | | | style="background:#fefeff" | 156 | ||
|- style = "background:#f8f8ff" | |- style = "background:#f8f8ff" | ||
! | ! style="background:#f4f4ff" | DP- | ||
| | | 38 || 422 || | ||
| style="background:#fefeff" | | | style="background:#fefeff" | 16,036 | ||
|- style = "background:#f8f8ff" | |- style = "background:#f8f8ff" | ||
! | ! style="background:#f4f4ff" | DQ- | ||
| | | 52 || 664 || | ||
| style="background:#fefeff" | | | style="background:#fefeff" | 34,528 | ||
|- style = "background:#f8f8ff" | |- style = "background:#f8f8ff" | ||
! | ! style="background:#f4f4ff" | DR- | ||
| | | 7 || 1540 || 93 | ||
| style="background:#fefeff" | | | style="background:#fefeff" | 11,431 | ||
|- style = "background:#f8f8ff" | |- style = "background:#f8f8ff" | ||
| colspan="5" style = "font-size:80%" | <sup>1</sup>DRB3, DRB4, DRB5 have variable presence in humans | | colspan="5" style = "font-size:80%" | <sup>1</sup>DRB3, DRB4, DRB5 have variable presence in humans | ||
|} | |||
====Sequence feature variant type (SFVT)==== | |||
The large extent of variability in HLA genes poses significant challenges in investigating the role of HLA genetic variations in diseases. Disease association studies typically treat each HLA allele as a single complete unit, which does not illuminate the parts of the molecule associated with disease. Karp D. R. et al. describes a novel [[sequence feature variant type]] (SFVT) approach for HLA genetic analysis that categorizes HLA proteins into biologically relevant smaller sequence features (SFs), and their variant types (VTs).<ref name=Karp>{{cite journal | vauthors = Karp DR, Marthandan N, Marsh SG, Ahn C, Arnett FC, Deluca DS, Diehl AD, Dunivin R, Eilbeck K, Feolo M, Guidry PA, Helmberg W, Lewis S, Mayes MD, Mungall C, Natale DA, Peters B, Petersdorf E, Reveille JD, Smith B, Thomson G, Waller MJ, Scheuermann RH | title = Novel sequence feature variant type analysis of the HLA genetic association in systemic sclerosis | journal = Human Molecular Genetics | volume = 19 | issue = 4 | pages = 707–19 | date = February 2010 | pmid = 19933168 | pmc = 2807365 | doi = 10.1093/hmg/ddp521 }}</ref> Sequence features are combinations of amino acid sites defined based on structural information (e.g., beta-sheet 1), functional information (e.g., peptide antigen-binding), and polymorphism. These sequence features can be overlapping and continuous or discontinuous in the linear sequence. Variant types for each sequence feature are defined based upon all known polymorphisms in the HLA locus being described. SFVT categorization of HLA is applied in genetic association analysis so that the effects and roles of the epitopes shared by several HLA alleles can be identified. Sequence features and their variant types have been described for all classical HLA proteins; the international repository of HLA SFVTs will be maintained at IMGT/HLA database.<ref>{{cite web |url= http://www.ebi.ac.uk/imgt/hla/|title = IMGT/HLA Database}}</ref> A tool to convert HLA alleles into their component SFVTs can be found on the Immunology Database and Analysis Portal (ImmPort) website.<ref>{{cite web|url=https://www.immport.org/immportWeb/home/home.do?loginType=full |title=Immunology Database and Analysis Portal (ImmPort) |deadurl=yes |archiveurl=https://web.archive.org/web/20110726174823/https://www.immport.org/immportWeb/home/home.do?loginType=full |archivedate=2011-07-26 |df= }}</ref> | |||
====Common, well-documented and rare alleles==== | |||
Although the number of individual HLA alleles that have been identified is large, approximately 40% of these alleles appear to be unique, having only been identified in single individuals.<ref name="middleton">{{cite journal | vauthors = Middleton D, Gonzalez F, Fernandez-Vina M, Tiercy JM, Marsh SG, Aubrey M, Bicalho MG, Canossi A, Carter V, Cate S, Guerini FR, Loiseau P, Martinetti M, Moraes ME, Morales V, Perasaari J, Setterholm M, Sprague M, Tavoularis S, Torres M, Vidal S, Witt C, Wohlwend G, Yang KL | title = A bioinformatics approach to ascertaining the rarity of HLA alleles | journal = Tissue Antigens | volume = 74 | issue = 6 | pages = 480–5 | date = December 2009 | pmid = 19793314 | doi = 10.1111/j.1399-0039.2009.01361.x }}</ref><ref name="gonzalez">{{cite journal | vauthors = Gonzalez-Galarza FF, Mack SJ, Hollenbach J, Fernandez-Vina M, Setterholm M, Kempenich J, Marsh SG, Jones AR, Middleton D | title = 16(th) IHIW: extending the number of resources and bioinformatics analysis for the investigation of HLA rare alleles | journal = International Journal of Immunogenetics | volume = 40 | issue = 1 | pages = 60–5 | date = February 2013 | pmid = 23198982 | doi = 10.1111/iji.12030 }}</ref> Roughly a third of alleles have been reported more than three times in unrelated individuals.<ref name="gonzalez" /><ref name="cano">{{cite journal | vauthors = Cano P, Klitz W, Mack SJ, Maiers M, Marsh SG, Noreen H, Reed EF, Senitzer D, Setterholm M, Smith A, Fernández-Viña M | title = Common and well-documented HLA alleles: report of the Ad-Hoc committee of the american society for histocompatiblity and immunogenetics | journal = Human Immunology | volume = 68 | issue = 5 | pages = 392–417 | date = May 2007 | pmid = 17462507 | doi = 10.1016/j.humimm.2007.01.014 }}</ref> Because of this variation in the rate at which of individual HLA alleles are detected, attempts have been made to categorize alleles at each expressed HLA locus in terms of their prevalence. The result is a catalog of common and well-documented (CWD) HLA alleles,<ref name="cano" /><ref name="mack">{{cite journal | vauthors = Mack SJ, Cano P, Hollenbach JA, He J, Hurley CK, Middleton D, Moraes ME, Pereira SE, Kempenich JH, Reed EF, Setterholm M, Smith AG, Tilanus MG, Torres M, Varney MD, Voorter CE, Fischer GF, Fleischhauer K, Goodridge D, Klitz W, Little AM, Maiers M, Marsh SG, Müller CR, Noreen H, Rozemuller EH, Sanchez-Mazas A, Senitzer D, Trachtenberg E, Fernandez-Vina M | title = Common and well-documented HLA alleles: 2012 update to the CWD catalogue | journal = Tissue Antigens | volume = 81 | issue = 4 | pages = 194–203 | date = April 2013 | pmid = 23510415 | doi = 10.1111/tan.12093 | pmc=3634360}}</ref> and a catalogue of rare and very rare HLA alleles.<ref name="middleton" /><ref name="gonzalez" /> | |||
Common HLA alleles are defined as having been observed with a frequency of at least 0.001 in reference populations of at least 1500 individuals.<ref name="cano" /><ref name="mack" /> Well-documented HLA alleles were originally defined as having been reported at least three times in unrelated individuals,<ref name="cano" /> and are now defined as having been detected at least five times in unrelated individuals via the application of a sequence-based typing (SBT) method, or at least three times via a SBT method and in a specific haplotype in unrelated individuals.<ref name="mack" /> Rare alleles are defined as those that have been reported one to four times, and very rare alleles as those reported only once.<ref name="middleton" /><ref name="gonzalez" /> | |||
=====Table of HLA alleles in each prevalence category===== | |||
While the current CWD and rare or very rare designations were developed using different datasets and different versions of the [[HLA Informatics Group|IMGT/HLA Database]],<ref name="gonzalez" /><ref name="mack" /> the approximate fraction of alleles at each HLA locus in each category is shown below. | |||
{| class="wikitable" style="text-align:center" | |||
|- | |||
! HLA<br>Locus !! No. common<br>alleles<ref name="mack" /> !! % common<br>alleles<ref name="mack" /> !! No. well-documented<br>alleles<ref name="mack" /> !! % well-documented<br>alleles<ref name="mack" /> !! No. rare<br>alleles<ref name="gonzalez" /> !! % rare<br>alleles<ref name="gonzalez" /> !! No. very rare<br>alleles<ref name="gonzalez" /> !! % very rare<br>alleles<ref name="gonzalez" /> !! % alleles<br>categorized | |||
|- | |||
| [[HLA-A|A]] || 68 || 3.4% || 178 || 8.8% || 145 || 21.5% || 280 || 41.6% || ~75% | |||
|- | |||
| [[HLA-B|B]] || 125 || 4.8% || 242 || 9.3% || 190 || 17.6% || 468 || 43.5% || ~75% | |||
|- | |||
| [[HLA-C|C]] || 44 || 2.8% || 102 || 6.6% || 77 || 21.4% || 154 || 42.8% || ~74% | |||
|- | |||
| [[HLA-DRB1|DRB1]] || 79 || 6.8% || 147 || 12.7% || 133 || 22.7% || 206 || 35.2% || ~77% | |||
|- | |||
| [[HLA-DRB3|DRB3]] || 5 || 8.6% || 7 || 12.1% || || || || || ~21% | |||
|- | |||
| [[HLA-DRB4|DRB4]] || 6 || 40.0% || 2 || 13.3% || || || || || ~53% | |||
|- | |||
| [[HLA-DRB5|DRB5]] || 5 || 25.0% || 3 || 15.0% || || || || || ~40% | |||
|- | |||
| [[HLA-DQA1|DQA1]] || 15 || 31.9% || 4 || 8.5% || 9 || 26.5% || 7 || 20.6% || ~88% | |||
|- | |||
| [[HLA-DQB1|DQB1]] || 22 || 12.5% || 8 || 4.5% || 26 || 28.9% || 42 || 45.2% || ~91% | |||
|- | |||
| [[HLA-DPA1|DPA1]] || 6 || 17.6% || 0 || 0.0% || 4 || 14.8% || 15 || 55.6% || ~88% | |||
|- | |||
| [[HLA-DPB1|DPB1]] || 40 || 28.8% || 14 || 9.0% || 29 || 22.7% || 29 || 32.8% || ~90% | |||
|- | |||
| All Loci || 415 || 5.3% || 707 || 9.0% || 613 || 20.6% || 1214 || 40.8% || ~76% | |||
|- | |||
|} | |} | ||
===Examining HLA types=== | ===Examining HLA types=== | ||
====Serotype and allele names==== | |||
There are two parallel systems of nomenclature that are applied to HLA. The first, and oldest, system is based on serological (antibody based) recognition. In this system, antigens were eventually assigned letters and numbers (e.g., [[HLA-B27]] or, shortened, B27). A parallel system that allowed more refined definition of alleles was developed. In this system, an | |||
"HLA" is used in conjunction with a letter, *, and a four-or-more-digit number (e.g., HLA-B*08:01, A*68:01, A*24:02:01N N=Null) to designate a specific [[allele]] at a given HLA [[locus (genetics)|locus]]. HLA loci can be further classified into [[MHC class I]] and [[MHC class II]] (or rarely, D locus). Every two years, a nomenclature is put forth to aid researchers in interpreting | |||
serotypes to alleles.<ref name=Marsh /> | |||
====Serotyping==== | |||
{{Further|Serotype}} | |||
In order to create a typing reagent, blood from animals or humans would be taken, the blood cells allowed to separate from the serum, and the serum diluted to its optimal sensitivity and used to type cells from other individuals or animals. Thus, serotyping became a way of crudely identifying HLA receptors and receptor isoforms. Over the years, serotyping antibodies became more refined as techniques for increasing sensitivity improved and new serotyping antibodies continue to appear. One of the goals of serotype analysis is to fill gaps in the analysis. It is possible to predict based on 'square root','maximum-likelihood' method, or analysis of familial haplotypes to account for adequately typed alleles. These studies using serotyping techniques frequently revealed, in particular for non-European or north East Asian populations a large number of null or blank serotypes. This was particularly problematic for the Cw locus until recently, and almost half of the Cw serotypes went untyped in the 1991 survey of the human population. | |||
There are several types of serotypes. A broad antigen serotype is a crude measure of identity of cells. For example, HLA A9 serotype recognizes cells of A23- and A24-bearing individuals. It may also recognize cells that A23 and A24 miss because of small variations. A23 and A24 are split antigens, but antibodies specific to either are typically used more often than antibodies to broad antigens. | |||
==== | ====Cellular typing==== | ||
A representative cellular assay is the mixed lymphocyte culture (MLC) and used to determine the HLA class II types.<ref name=Hurley>Hurley CK (1997). "DNA-based typing of HLA for transplantation." ''In'' Leffell MS, Donnenberg AD, Rose NR, eds. (1997) ''Handbook of Human Immunology.'' pp. 521–55, Boca Raton: [[CRC Press]], {{ISBN|0-8493-0134-3}}.</ref> The cellular assay is more sensitive in detecting HLA differences than serotyping. This is because minor differences unrecognized by alloantisera can stimulate T cells. This typing is designated as Dw types. Serotyped DR1 has cellularly defined as either of Dw1 or of Dw20 and so on for other serotyped DRs. Table<ref name=Bodmer>{{cite journal | vauthors = Bodmer JG, Marsh SG, Albert ED, Bodmer WF, Dupont B, Erlich HA, Mach B, Mayr WR, Parham P, Sasazuki T | title = Nomenclature for factors of the HLA system, 1991 | journal = Human Immunology | volume = 34 | issue = 1 | pages = 4–18 | date = May 1992 | pmid = 1399721 | doi = 10.1016/0198-8859(92)90079-3 | display-authors = 6 }}</ref> shows associated cellular specificities for DR alleles. However, cellular typing has inconsistency in the reaction between cellular-type individuals, sometimes resulting differently from predicted. Together with difficulty of cellular assay in generating and maintaining cellular typing reagents, cellular assay is being replaced by DNA-based typing method.<ref name=Hurley/> | |||
====Gene sequencing==== | |||
Minor reactions to subregions that show similarity to other types can be observed to the gene products of alleles of a serotype group. The sequence of the antigens determines the antibody reactivities, and so having a good sequencing capability (or sequence-based typing) obviates the need for serological reactions. Therefore, different serotype reactions may indicate the need to sequence a person's HLA to determine a new '''gene sequence'''. | |||
Broad antigen types are still useful, such as typing very diverse populations with many unidentified HLA alleles (Africa, Arabia,<ref name=Valluri>{{cite journal | vauthors = Valluri V, Valluei V, Mustafa M, Santhosh A, Middleton D, Alvares M, Alvales M, El Haj E, Gumama O, Abdel-Wareth L, Abdel-Waieth L | title = Frequencies of HLA-A, HLA-B, HLA-DR, and HLA-DQ phenotypes in the United Arab Emirates population | journal = Tissue Antigens | volume = 66 | issue = 2 | pages = 107–13 | date = August 2005 | pmid = 16029430 | doi = 10.1111/j.1399-0039.2005.00441.x }}</ref> Southeastern Iran<ref name=Farjadin>{{cite journal | vauthors = Farjadian S, Naruse T, Kawata H, Ghaderi A, Bahram S, Inoko H | title = Molecular analysis of HLA allele frequencies and haplotypes in Baloch of Iran compared with related populations of Pakistan | journal = Tissue Antigens | volume = 64 | issue = 5 | pages = 581–7 | date = November 2004 | pmid = 15496201 | doi = 10.1111/j.1399-0039.2004.00302.x }}</ref> and Pakistan, India<ref name=Shankarkumar>{{cite journal | vauthors = Shankarkumar U, Prasanavar D, Ghosh K, Mohanty D | title = HLA A*02 allele frequencies and B haplotype associations in Western Indians | journal = Human Immunology | volume = 64 | issue = 5 | pages = 562–6 | date = May 2003 | pmid = 12691707 | doi = 10.1016/S0198-8859(03)00032-6 }}</ref>). Africa, Southern Iran, and Arabia show the difficulty in typing areas that were settled earlier. Allelic diversity makes it necessary to use broad antigen typing followed by '''gene sequencing''' because there is an increased risk of misidentifying by serotyping techniques. | |||
Broad antigen types are still useful, such as typing very diverse populations with many unidentified HLA alleles (Africa, Arabia,<ref name=Valluri>{{cite journal | | |||
In the end, a workshop, based on sequence, decides which new allele goes into which serogroup either by sequence or reactivity. Once the sequence is verified it is assigned a number. For example, a new allele of B44 may get a serotype B* | In the end, a workshop, based on sequence, decides which new allele goes into which serogroup either by sequence or by reactivity. Once the sequence is verified, it is assigned a number. For example, a new allele of B44 may get a serotype (i.e. B44) and allele ID i.e. B*44:65, as it is the 65th B44 allele discovered. Marsh et al. (2005)<ref name=Marsh/> can be considered a code book for HLA serotypes and genotypes, and a new book biannually with monthly updates in ''Tissue Antigens''. | ||
==== | ====Phenotyping==== | ||
Gene typing is different from gene sequencing and serotyping. | Gene typing is different from gene sequencing and serotyping. | ||
With this strategy PCR primers specific to a variant region of DNA are used (called | With this strategy, PCR primers specific to a variant region of DNA are used (called [[Single Specific Primer-PCR|SSP-PCR]]). If a product of the right size is found, the assumption is that the HLA allele has been identified. New gene sequences often result in an increasing appearance of ambiguity. Because gene typing is based on SSP-PCR, it is possible that new variants, in particular in the class I and DRB1 loci, may be missed. | ||
For SSP-PCR within the clinical situation is often used for identifying HLA '''phenotypes'''. An example of an extended phenotype for a person might be: | For example, SSP-PCR within the clinical situation is often used for identifying HLA '''phenotypes'''. An example of an extended phenotype for a person might be: | ||
A< | A<span style="color:Purple;">*01:01</span>/<span style="color:Purple;">*03:01</span>, C<span style="color:Green;">*07:01</span>/<span style="color:Green;">*07:02</span>, B<span style="color:Magenta;">*07:02</span>/<span style="color:Magenta;">*08:01</span>, DRB1<span style="color:Brown;">*03:01</span>/<span style="color:Brown;">*15:01</span>, | ||
DQA1< | DQA1<span style="color:DarkOrchid;">*05:01</span>/<span style="color:DarkOrchid;">*01:02</span>, DQB1<span style="color:DarkGreen;">*02:01</span>/<span style="color:DarkGreen;">*06:02</span> | ||
In general, this is identical to the extended serotype: | |||
A1,A3,B7,B8,DR3,DR15(2), DQ2,DQ6(1) | A1,A3,B7,B8,DR3,DR15(2), DQ2,DQ6(1) | ||
For many populations such as the Japanese or European populations so many patients have been typed that new alleles are relatively rare, and thus SSP-PCR is more than adequate for allele resolution. Haplotypes can be obtained by typing family members | For many populations, such as the Japanese or European populations, so many patients have been typed that new alleles are relatively rare, and thus SSP-PCR is more than adequate for allele resolution. Haplotypes can be obtained by typing family members in areas of the world where SSP-PCR is unable to recognize alleles and typing requires the sequencing of new alleles. Areas of the world where SSP-PCR or serotyping may be inadequate include Central Africa, Eastern Africa, parts of southern Africa, Arabia, S. Iran, Pakistan, and India. | ||
====Haplotypes==== | |||
An HLA haplotype is a series of HLA "genes" (loci-alleles) by chromosome, one passed from the mother and one from the father. | |||
The phenotype exampled above is one of the more common in Ireland and is the result of two common genetic ''haplotypes'': | |||
The phenotype exampled above is one of the more common in Ireland and is | |||
the result of two common genetic | |||
A< | A<span style="color:Purple;">*01:01</span> ; C<span style="color:Green;">*07:01</span> ; B<span style="color:Magenta;">*08:01</span> ; DRB1<span style="color:Brown;">*03:01</span> ; DQA1<span style="color:DarkOrchid;">*05:01</span> ; DQB1<span style="color:DarkGreen;">*02:01</span> | ||
(By serotyping A1-Cw7-B8-DR3-DQ2) | (By serotyping [[HLA A1-B8-DR3-DQ2|A1-Cw7-B8-DR3-DQ2]]) | ||
which is called ' 'super B8' ' or ' 'ancestral haplotype' ' and | which is called ' 'super B8' ' or ' 'ancestral haplotype' ' and | ||
A< | A<span style="color:Purple;">*03:01</span> ; C<span style="color:Green;">*07:02</span> ; B<span style="color:Magenta;">*07:02</span> ; DRB1<span style="color:Brown;">*15:01</span> ; DQA1<span style="color:DarkOrchid;">*01:02</span> ; DQB1<span style="color:DarkGreen;">*06:02</span> | ||
(By serotyping A3-Cw7-B7-DR15-DQ6 or the older version "A3-B7-DR2-DQ1") | (By serotyping A3-Cw7-B7-DR15-DQ6 or the older version "A3-B7-DR2-DQ1") | ||
These haplotypes can be used to trace migrations in the human population | These haplotypes can be used to trace migrations in the human population | ||
because they are often much like a fingerprint of an event that has occurred | because they are often much like a fingerprint of an event that has occurred | ||
in evolution. The Super-B8 haplotype is enriched in the Western Irish, declines along gradients away from that region and is only | in evolution. The Super-B8 haplotype is enriched in the Western Irish, declines along gradients away from that region, and is found only in areas of the world where Western Europeans have migrated. The "A3-B7-DR2-DQ1" is more widely spread, from Eastern Asia to Iberia. The Super-B8 haplotype is associated with a number of diet-associated autoimmune diseases. There are 100,000s of extended haplotypes, but only a few show a visible and nodal character in the human population. | ||
==Role of allelic variation== | |||
Studies of humans and animals imply a heterozygous selection mechanism operating on these loci as an explanation for this variability.<ref name=Apanius>{{cite journal | vauthors = Apanius V, Penn D, Slev PR, Ruff LR, Potts WK | title = The nature of selection on the major histocompatibility complex | journal = Critical Reviews in Immunology | volume = 17 | issue = 2 | pages = 179–224 | year = 1997 | pmid = 9094452 | doi = 10.1615/critrevimmunol.v17.i2.40 }}.</ref> One credible mechanism is sexual selection in which females are able to detect males with different HLA relative to their own type.<ref name=Wedekind>{{cite journal | vauthors = Wedekind C, Seebeck T, Bettens F, Paepke AJ | title = MHC-dependent mate preferences in humans | journal = Proceedings of the Royal Society B: Biological Sciences | volume = 260 | issue = 1359 | pages = 245–9 | date = June 1995 | pmid = 7630893 | doi = 10.1098/rspb.1995.0087 }}</ref> While the DQ and DP encoding loci have fewer alleles, combinations of A1:B1 can produce a theoretical potential of 7,755 DQ and 5,270 DP αβ heterodimers, respectively. While nowhere near this number of isoforms exist in the human population, each individual can carry 4 variable DQ and DP isoforms, increasing the potential number of antigens that these receptors can present to the immune system. | |||
Studies of the variable positions of DP, DR, and DQ reveal that peptide antigen contact residues on class II molecules are most frequently the site of variation in the protein primary structure. Therefore, through a combination of intense allelic variation and/or subunit pairing, the class II 'peptide' receptors are capable of binding an almost endless variation of peptides of 9 amino acids or longer in length, protecting interbreeding subpopulations from nascent or epidemic diseases. Individuals in a population frequently have different haplotypes, and this results in many combinations, even in small groups. This diversity enhances the survival of such groups, and thwarts evolution of epitopes in pathogens, which would otherwise be able to be shielded from the immune system. | |||
==Antibodies== | |||
HLA antibodies are typically not naturally occurring, and with few exceptions are formed as a result of an immunologic challenge to a foreign material containing non-self HLAs via blood transfusion, pregnancy (paternally inherited antigens), or organ or tissue transplant. | |||
Antibodies against disease-associated HLA haplotypes have been proposed as a treatment for severe autoimmune diseases.<ref name = "MHCpeptide_immunize">{{cite journal | vauthors = Oshima M, Deitiker P, Ashizawa T, Atassi MZ | title = Vaccination with a MHC class II peptide attenuates cellular and humoral responses against tAChR and suppresses clinical EAMG | journal = Autoimmunity | volume = 35 | issue = 3 | pages = 183–90 | date = May 2002 | pmid = 12389643 | doi = 10.1080/08916930290022270 }}</ref> | |||
Donor-specific HLA antibodies have been found to be associated with graft failure in kidney, heart, lung, and liver transplantation. | |||
== | ==HLA matching for sick siblings== | ||
{{Main|Savior sibling}} | |||
In some diseases requiring [[hematopoietic stem cell transplantation]], [[preimplantation genetic diagnosis]] may be used to give rise to a sibling with matching HLA, although there are ethical considerations.<ref>{{cite journal | vauthors = Verlinsky Y, Rechitsky S, Schoolcraft W, Strom C, Kuliev A | title = Preimplantation diagnosis for Fanconi anemia combined with HLA matching | journal = JAMA | volume = 285 | issue = 24 | pages = 3130–3 | date = June 2001 | pmid = 11427142 | doi = 10.1001/jama.285.24.3130 | url = http://jama.jamanetwork.com/article.aspx?articleid=193944 }}</ref> | |||
== | == See also == | ||
*[[HCP5]] | |||
*[[List of human leukocyte antigen alleles associated with cutaneous conditions]] | |||
== References == | |||
{{reflist|30em}} | |||
== Bibliography == | |||
* Daniel M. Davis, ''The Compatibility Gene'', London, [[Penguin Books]], 2014 ({{ISBN|978-0-241-95675-5}}). | |||
* Zsolt Harsanyi and Richard Hutton, Richard, ''Genetic Prophecy: Beyond the Double Helix'', London: Granada, 1982 ({{ISBN|0-246-11760-5}}). | |||
== External links == | == External links == | ||
*[http://www.ebi.ac.uk/imgt/hla/ IMGT/HLA Sequence Database] at [[European Bioinformatics Institute]] | *[http://www.ebi.ac.uk/imgt/hla/ IMGT/HLA Sequence Database] at [[European Bioinformatics Institute]] | ||
*[http://hla.alleles.org/ hla.alleles.org] | |||
*[https://web.archive.org/web/20031203131321/http://www.anthonynolan.org.uk/HIG/ HLA Informatics Group] at [[The Anthony Nolan Trust]] | |||
*[http://www.bshi.org.uk British Society for Histocompatibility and Immunogenetics] | |||
*[http://ashi-hla.org American Society for Histocompatibility and Immunogenetics] | *[http://ashi-hla.org American Society for Histocompatibility and Immunogenetics] | ||
*[http://www.efiweb. | *[http://www.efiweb.eu European Federation for Immunogenetics] | ||
*[http://www.histocheck.org HistoCheck HLA matching tool for organ and stem cell transplantation] | *[http://www.histocheck.org HistoCheck HLA matching tool for organ and stem cell transplantation] | ||
*[http://www.allelefrequencies.net Allele Frequencies at Variable Immune related loci] | *[http://www.allelefrequencies.net Allele Frequencies at Variable Immune related loci] | ||
*{{MeshName|Human+leukocyte+antigens}} | *{{MeshName|Human+leukocyte+antigens}} | ||
*[https://www.ncbi.nlm.nih.gov/gv/mhc/ dbMHC Home, NCBI's database of the Major Histocompatibility Complex] | |||
*[http://www.allelefrequencies.net/hla6001a.asp Rare Alleles Project at the AlleleFrequencies Net Database (AFND)] | |||
*[http://cwd.immunogenomics.org Common and Well-Documented (CWD) alleles catalogue] | |||
{{Surface antigens}} | {{Surface antigens}} | ||
{{Use dmy dates|date=April 2017}} | |||
[[Category:Gene complexes]] | |||
[[Category:Immune system]] | [[Category:Immune system]] | ||
[[Category:Human MHC haplogroups|0]] | [[Category:Human MHC haplogroups|0]] | ||
[[Category:Genes on chromosome 6 | [[Category:Genes on human chromosome 6]] | ||
Revision as of 23:52, 28 November 2017
The human leukocyte antigen (HLA) system or complex is a gene complex encoding the major histocompatibility complex (MHC) proteins in humans. These cell-surface proteins are responsible for the regulation of the immune system in humans. The HLA gene complex resides on a 3 Mbp stretch within chromosome 6p21. HLA genes are highly polymorphic, which means that they have many different alleles, allowing them to fine-tune the adaptive immune system. The proteins encoded by certain genes are also known as antigens, as a result of their historic discovery as factors in organ transplants. Different classes have different functions:
HLAs corresponding to MHC class I (A, B, and C) present peptides from inside the cell. For example, if the cell is infected by a virus, the HLA system brings fragments of the virus to the surface of the cell so that the cell can be destroyed by the immune system. These peptides are produced from digested proteins that are broken down in the proteasomes. In general, these particular peptides are small polymers, about 9 amino acids in length.[citation needed] Foreign antigens presented by MHC class I attract killer T-cells (also called CD8 positive- or cytotoxic T-cells) that destroy cells. MHC class I proteins associate with β2-microglobulin, which unlike the HLA proteins is encoded by a gene on chromosome 15.
HLAs corresponding to MHC class II (DP, DM, DOA, DOB, DQ, and DR) present antigens from outside of the cell to T-lymphocytes. These particular antigens stimulate the multiplication of T-helper cells (also called CD4 positive T cells), which in turn stimulate antibody-producing B-cells to produce antibodies to that specific antigen. Self-antigens are suppressed by regulatory T cells.
HLAs corresponding to MHC class III encode components of the complement system.
HLAs have other roles. They are important in disease defense. They are the major cause of organ transplant rejections. They may protect against or fail to protect (if down-regulated by an infection) against cancers.[1] Mutations in HLA may be linked to autoimmune disease (examples: type I diabetes, coeliac disease). HLA may also be related to people's perception of the odor of other people, and may be involved in mate selection, as at least one study found a lower-than-expected rate of HLA similarity between spouses in an isolated community.[2]
Aside from the genes encoding the 6 major antigen-presenting proteins, there are a large number of other genes, many involved in immune function, located on the HLA complex. Diversity of HLAs in the human population is one aspect of disease defense, and, as a result, the chance of two unrelated individuals with identical HLA molecules on all loci is extremely low. HLA genes have historically been identified as a result of the ability to successfully transplant organs between HLA-similar individuals.
Functions
The proteins encoded by HLAs are those on the outer part of body cells that are (in effect) unique to that person. The immune system uses the HLAs to differentiate self cells and non-self cells. Any cell displaying that person's HLA type belongs to that person and, therefore, is not an invader.

In infectious disease
When a foreign pathogen enters the body, specific cells called antigen-presenting cells (APCs) engulf the pathogen through a process called phagocytosis. Proteins from the pathogen are digested into small pieces (peptides) and loaded onto HLA antigens (to be specific, MHC class II). They are then displayed by the antigen-presenting cells to CD4+ helper T cells,[3] which then produce a variety of effects to eliminate the pathogen.
Through a similar process, proteins (both native and foreign, such as the proteins of virus) produced inside most cells are displayed on HLAs (to be specific, MHC class I) on the cell surface. Infected cells can be recognized and destroyed by CD8+ T cells.[3]
The image off to the side shows a piece of a poisonous bacterial protein (SEI peptide) bound within the binding cleft portion of the HLA-DR1 molecule. In the illustration far below, a different view, one can see an entire DQ with a bound peptide in a similar cleft, as viewed from the side. Disease-related peptides fit into these "slots" much like a hand fits into a glove. When bound, peptides are presented to T cells. T cells require presentation via MHC molecules to recognize foreign antigens — a requirement known as MHC restriction. These cells have receptors that are similar to B cell receptors, and each cell recognizes only a few class II-peptide combinations. Once a T cell recognizes a peptide within an MHC class II molecule, it can stimulate B-cells that also recognize the same molecule in their B cell receptors. Thus, T cells help B cells make antibodies to the same foreign antigens. Each HLA can bind many peptides, and each person has 3 HLA types and can have 4 isoforms of DP, 4 isoforms of DQ and 4 Isoforms of DR (2 of DRB1, and 2 of DRB3, DRB4, or DRB5) for a total of 12 isoforms. In such heterozygotes, it is difficult for disease-related proteins to escape detection.
In graft rejection
Any cell displaying some other HLA type is "non-self" and is seen as an invader by the body's immune system, resulting in the rejection of the tissue bearing those cells. This is particularly important in the case of transplanted tissue, because it could lead to transplant rejection. Because of the importance of HLA in transplantation, the HLA loci are some of the most frequently typed by serology and PCR.
| HLA allele | Diseases with increased risk | Relative risk |
|---|---|---|
| HLA-B27 | Ankylosing spondylitis | 12[4] |
| Postgonococcal arthritis | 14[4] | |
| Acute anterior uveitis | 15[4] | |
| HLA-B47 | 21-hydroxylase deficiency | 15[4] |
| HLA-DR2 | Systemic lupus erythematosus | 2 to 3[5] |
| HLA-DR3 | Autoimmune hepatitis | 14[4] |
| Primary Sjögren syndrome | 10[4] | |
| Diabetes mellitus type 1 | 5[4] | |
| Systemic lupus erythematosus | 2 to 3[5] | |
| HLA-DR4 | Rheumatoid arthritis | 4[4] |
| Diabetes mellitus type 1 | 6[4] | |
| HLA-DR3 and -DR4 combined |
Diabetes mellitus type 1 | 15[4] |
| HLA-DQ2 and HLA-DQ8 | Coeliac disease | 7[6] |
In autoimmunity
HLA types are inherited, and some of them are connected with autoimmune disorders and other diseases. People with certain HLA antigens are more likely to develop certain autoimmune diseases, such as type I diabetes, ankylosing spondylitis, celiac disease, SLE (systemic lupus erythematosus), myasthenia gravis, inclusion body myositis, Sjögren syndrome, and narcolepsy.[7] HLA typing has led to some improvement and acceleration in the diagnosis of celiac disease and type 1 diabetes; however, for DQ2 typing to be useful, it requires either high-resolution B1*typing (resolving *02:01 from *02:02), DQA1*typing, or DR serotyping. Current serotyping can resolve, in one step, DQ8. HLA typing in autoimmunity is being increasingly used as a tool in diagnosis. In celiac disease, it is the only effective means of discriminating between first-degree relatives that are at risk from those that are not at risk, prior to the appearance of sometimes-irreversible symptoms such as allergies and secondary autoimmune disease.
In cancer
Some HLA-mediated diseases are directly involved in the promotion of cancer. Gluten-sensitive enteropathy is associated with increased prevalence of enteropathy-associated T-cell lymphoma, and DR3-DQ2 homozygotes are within the highest risk group, with close to 80% of gluten-sensitive enteropathy-associated T-cell lymphoma cases. More often, however, HLA molecules play a protective role, recognizing increases in antigens that are not tolerated because of low levels in the normal state. Abnormal cells might be targeted for apoptosis, which is thought to mediate many cancers before diagnosis.
In mate selection
There is evidence for non-random mate choice with respect to certain genetic characteristics. This has led to a field known as Genetic matchmaking.
Classification
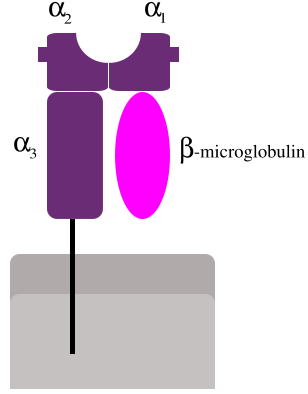
MHC class I proteins form a functional receptor on most nucleated cells of the body.
There are 3 major and 3 minor MHC class I genes in HLA.
Major MHC class I
Minor genes are HLA-E, HLA-F and HLA-G. β2-microglobulin binds with major and minor gene subunits to produce a heterodimer
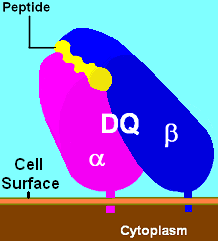
There are 3 major and 2 minor MHC class II proteins encoded by the HLA. The genes of the class II combine to form heterodimeric (αβ) protein receptors that are typically expressed on the surface of antigen-presenting cells.
Major MHC class II
- HLA-DP
- α-chain encoded by HLA-DPA1 locus
- β-chain encoded by HLA-DPB1 locus
- HLA-DQ
- HLA-DR
- α-chain encoded by HLA-DRA locus
- 4 β-chains (only 3 possible per person), encoded by HLA-DRB1, DRB3, DRB4, DRB5 loci
The other MHC class II proteins, DM and DO, are used in the internal processing of antigens, loading the antigenic peptides generated from pathogens onto the HLA molecules of antigen-presenting cell.
Nomenclature
Modern HLA alleles are typically noted with a variety of levels of detail. Most designations begin with HLA- and the locus name, then * and some (even) number of digits specifying the allele. The first two digits specify a group of alleles. Older typing methodologies often could not completely distinguish alleles and so stopped at this level. The third through fourth digits specify a nonsynonymous allele. Digits five through six denote any synonymous mutations within the coding frame of the gene. The seventh and eighth digits distinguish mutations outside the coding region. Letters such as L, N, Q, or S may follow an allele's designation to specify an expression level or other non-genomic data known about it. Thus, a completely described allele may be up to 9 digits long, not including the HLA-prefix and locus notation.[8]
Variability
MHC loci are some of the most genetically variable coding loci in mammals, and the human HLA loci are no exceptions. Despite the fact that the human population went through a constriction more than 150,000 years ago that was capable of fixing many loci, the HLA loci appear to have survived such a constriction with a great deal of variation.[9] Of the 9 loci mentioned above, most retained a dozen or more allele-groups for each locus, far more preserved variation than the vast majority of human loci. This is consistent with a heterozygous or balancing selection coefficient for these loci. In addition, some HLA loci are among the fastest-evolving coding regions in the human genome. One mechanism of diversification has been noted in the study of Amazonian tribes of South America that appear to have undergone intense gene conversion between variable alleles and loci within each HLA gene class.[10] Less frequently, longer-range productive recombinations through HLA genes have been noted producing chimeric genes.
Six loci have over 100 alleles that have been detected in the human population. Of these, the most variable are HLA B and HLA DRB1. As of 2012, the number of alleles that have been determined are listed in the table below. To interpret this table, it is necessary to consider that an allele is a variant of the nucleotide (DNA) sequence at a locus, such that each allele differs from all other alleles in at least one (single nucleotide polymorphism, SNP) position. Most of these changes result in a change in the amino acid sequences that result in slight to major functional differences in the protein.
There are issues that limit this variation. Certain alleles like DQA1*05:01 and DQA1*05:05 encode proteins with identically processed products. Other alleles like DQB1*0201 and DQB1*0202 produce proteins that are functionally similar. For class II (DR, DP and DQ), amino acid variants within the receptor's peptide-binding cleft tend to produce molecules with different binding capability.
Tables of variant alleles
Number of variant alleles at class I loci according to the IMGT-HLA database, last updated July 2014:
| MHC class I | |
|---|---|
| locus | #[11][12] |
| Major Antigens | |
| HLA A | 2,884 |
| HLA B | 3,589 |
| HLA C | 2,375 |
| Minor Antigens | |
| HLA E | 15 |
| HLA F | 22 |
| HLA G | 50 |
Number of variant alleles at class II loci (DM, DO, DP, DQ, and DR):
| MHC class II | ||||
|---|---|---|---|---|
| HLA | -A1 | -B1 | -B3 to -B51 | Theor. possible |
| locus | #[12] | #[12] | #[12] | combinations |
| DM- | 7 | 13 | 91 | |
| DO- | 12 | 13 | 156 | |
| DP- | 38 | 422 | 16,036 | |
| DQ- | 52 | 664 | 34,528 | |
| DR- | 7 | 1540 | 93 | 11,431 |
| 1DRB3, DRB4, DRB5 have variable presence in humans | ||||
Sequence feature variant type (SFVT)
The large extent of variability in HLA genes poses significant challenges in investigating the role of HLA genetic variations in diseases. Disease association studies typically treat each HLA allele as a single complete unit, which does not illuminate the parts of the molecule associated with disease. Karp D. R. et al. describes a novel sequence feature variant type (SFVT) approach for HLA genetic analysis that categorizes HLA proteins into biologically relevant smaller sequence features (SFs), and their variant types (VTs).[13] Sequence features are combinations of amino acid sites defined based on structural information (e.g., beta-sheet 1), functional information (e.g., peptide antigen-binding), and polymorphism. These sequence features can be overlapping and continuous or discontinuous in the linear sequence. Variant types for each sequence feature are defined based upon all known polymorphisms in the HLA locus being described. SFVT categorization of HLA is applied in genetic association analysis so that the effects and roles of the epitopes shared by several HLA alleles can be identified. Sequence features and their variant types have been described for all classical HLA proteins; the international repository of HLA SFVTs will be maintained at IMGT/HLA database.[14] A tool to convert HLA alleles into their component SFVTs can be found on the Immunology Database and Analysis Portal (ImmPort) website.[15]
Common, well-documented and rare alleles
Although the number of individual HLA alleles that have been identified is large, approximately 40% of these alleles appear to be unique, having only been identified in single individuals.[16][17] Roughly a third of alleles have been reported more than three times in unrelated individuals.[17][18] Because of this variation in the rate at which of individual HLA alleles are detected, attempts have been made to categorize alleles at each expressed HLA locus in terms of their prevalence. The result is a catalog of common and well-documented (CWD) HLA alleles,[18][19] and a catalogue of rare and very rare HLA alleles.[16][17]
Common HLA alleles are defined as having been observed with a frequency of at least 0.001 in reference populations of at least 1500 individuals.[18][19] Well-documented HLA alleles were originally defined as having been reported at least three times in unrelated individuals,[18] and are now defined as having been detected at least five times in unrelated individuals via the application of a sequence-based typing (SBT) method, or at least three times via a SBT method and in a specific haplotype in unrelated individuals.[19] Rare alleles are defined as those that have been reported one to four times, and very rare alleles as those reported only once.[16][17]
Table of HLA alleles in each prevalence category
While the current CWD and rare or very rare designations were developed using different datasets and different versions of the IMGT/HLA Database,[17][19] the approximate fraction of alleles at each HLA locus in each category is shown below.
| HLA Locus |
No. common alleles[19] |
% common alleles[19] |
No. well-documented alleles[19] |
% well-documented alleles[19] |
No. rare alleles[17] |
% rare alleles[17] |
No. very rare alleles[17] |
% very rare alleles[17] |
% alleles categorized |
|---|---|---|---|---|---|---|---|---|---|
| A | 68 | 3.4% | 178 | 8.8% | 145 | 21.5% | 280 | 41.6% | ~75% |
| B | 125 | 4.8% | 242 | 9.3% | 190 | 17.6% | 468 | 43.5% | ~75% |
| C | 44 | 2.8% | 102 | 6.6% | 77 | 21.4% | 154 | 42.8% | ~74% |
| DRB1 | 79 | 6.8% | 147 | 12.7% | 133 | 22.7% | 206 | 35.2% | ~77% |
| DRB3 | 5 | 8.6% | 7 | 12.1% | ~21% | ||||
| DRB4 | 6 | 40.0% | 2 | 13.3% | ~53% | ||||
| DRB5 | 5 | 25.0% | 3 | 15.0% | ~40% | ||||
| DQA1 | 15 | 31.9% | 4 | 8.5% | 9 | 26.5% | 7 | 20.6% | ~88% |
| DQB1 | 22 | 12.5% | 8 | 4.5% | 26 | 28.9% | 42 | 45.2% | ~91% |
| DPA1 | 6 | 17.6% | 0 | 0.0% | 4 | 14.8% | 15 | 55.6% | ~88% |
| DPB1 | 40 | 28.8% | 14 | 9.0% | 29 | 22.7% | 29 | 32.8% | ~90% |
| All Loci | 415 | 5.3% | 707 | 9.0% | 613 | 20.6% | 1214 | 40.8% | ~76% |
Examining HLA types
Serotype and allele names
There are two parallel systems of nomenclature that are applied to HLA. The first, and oldest, system is based on serological (antibody based) recognition. In this system, antigens were eventually assigned letters and numbers (e.g., HLA-B27 or, shortened, B27). A parallel system that allowed more refined definition of alleles was developed. In this system, an "HLA" is used in conjunction with a letter, *, and a four-or-more-digit number (e.g., HLA-B*08:01, A*68:01, A*24:02:01N N=Null) to designate a specific allele at a given HLA locus. HLA loci can be further classified into MHC class I and MHC class II (or rarely, D locus). Every two years, a nomenclature is put forth to aid researchers in interpreting serotypes to alleles.[11]
Serotyping
In order to create a typing reagent, blood from animals or humans would be taken, the blood cells allowed to separate from the serum, and the serum diluted to its optimal sensitivity and used to type cells from other individuals or animals. Thus, serotyping became a way of crudely identifying HLA receptors and receptor isoforms. Over the years, serotyping antibodies became more refined as techniques for increasing sensitivity improved and new serotyping antibodies continue to appear. One of the goals of serotype analysis is to fill gaps in the analysis. It is possible to predict based on 'square root','maximum-likelihood' method, or analysis of familial haplotypes to account for adequately typed alleles. These studies using serotyping techniques frequently revealed, in particular for non-European or north East Asian populations a large number of null or blank serotypes. This was particularly problematic for the Cw locus until recently, and almost half of the Cw serotypes went untyped in the 1991 survey of the human population.
There are several types of serotypes. A broad antigen serotype is a crude measure of identity of cells. For example, HLA A9 serotype recognizes cells of A23- and A24-bearing individuals. It may also recognize cells that A23 and A24 miss because of small variations. A23 and A24 are split antigens, but antibodies specific to either are typically used more often than antibodies to broad antigens.
Cellular typing
A representative cellular assay is the mixed lymphocyte culture (MLC) and used to determine the HLA class II types.[20] The cellular assay is more sensitive in detecting HLA differences than serotyping. This is because minor differences unrecognized by alloantisera can stimulate T cells. This typing is designated as Dw types. Serotyped DR1 has cellularly defined as either of Dw1 or of Dw20 and so on for other serotyped DRs. Table[21] shows associated cellular specificities for DR alleles. However, cellular typing has inconsistency in the reaction between cellular-type individuals, sometimes resulting differently from predicted. Together with difficulty of cellular assay in generating and maintaining cellular typing reagents, cellular assay is being replaced by DNA-based typing method.[20]
Gene sequencing
Minor reactions to subregions that show similarity to other types can be observed to the gene products of alleles of a serotype group. The sequence of the antigens determines the antibody reactivities, and so having a good sequencing capability (or sequence-based typing) obviates the need for serological reactions. Therefore, different serotype reactions may indicate the need to sequence a person's HLA to determine a new gene sequence.
Broad antigen types are still useful, such as typing very diverse populations with many unidentified HLA alleles (Africa, Arabia,[22] Southeastern Iran[23] and Pakistan, India[24]). Africa, Southern Iran, and Arabia show the difficulty in typing areas that were settled earlier. Allelic diversity makes it necessary to use broad antigen typing followed by gene sequencing because there is an increased risk of misidentifying by serotyping techniques.
In the end, a workshop, based on sequence, decides which new allele goes into which serogroup either by sequence or by reactivity. Once the sequence is verified, it is assigned a number. For example, a new allele of B44 may get a serotype (i.e. B44) and allele ID i.e. B*44:65, as it is the 65th B44 allele discovered. Marsh et al. (2005)[11] can be considered a code book for HLA serotypes and genotypes, and a new book biannually with monthly updates in Tissue Antigens.
Phenotyping
Gene typing is different from gene sequencing and serotyping. With this strategy, PCR primers specific to a variant region of DNA are used (called SSP-PCR). If a product of the right size is found, the assumption is that the HLA allele has been identified. New gene sequences often result in an increasing appearance of ambiguity. Because gene typing is based on SSP-PCR, it is possible that new variants, in particular in the class I and DRB1 loci, may be missed.
For example, SSP-PCR within the clinical situation is often used for identifying HLA phenotypes. An example of an extended phenotype for a person might be:
A*01:01/*03:01, C*07:01/*07:02, B*07:02/*08:01, DRB1*03:01/*15:01, DQA1*05:01/*01:02, DQB1*02:01/*06:02
In general, this is identical to the extended serotype: A1,A3,B7,B8,DR3,DR15(2), DQ2,DQ6(1)
For many populations, such as the Japanese or European populations, so many patients have been typed that new alleles are relatively rare, and thus SSP-PCR is more than adequate for allele resolution. Haplotypes can be obtained by typing family members in areas of the world where SSP-PCR is unable to recognize alleles and typing requires the sequencing of new alleles. Areas of the world where SSP-PCR or serotyping may be inadequate include Central Africa, Eastern Africa, parts of southern Africa, Arabia, S. Iran, Pakistan, and India.
Haplotypes
An HLA haplotype is a series of HLA "genes" (loci-alleles) by chromosome, one passed from the mother and one from the father.
The phenotype exampled above is one of the more common in Ireland and is the result of two common genetic haplotypes:
A*01:01 ; C*07:01 ; B*08:01 ; DRB1*03:01 ; DQA1*05:01 ; DQB1*02:01 (By serotyping A1-Cw7-B8-DR3-DQ2)
which is called ' 'super B8' ' or ' 'ancestral haplotype' ' and
A*03:01 ; C*07:02 ; B*07:02 ; DRB1*15:01 ; DQA1*01:02 ; DQB1*06:02 (By serotyping A3-Cw7-B7-DR15-DQ6 or the older version "A3-B7-DR2-DQ1")
These haplotypes can be used to trace migrations in the human population because they are often much like a fingerprint of an event that has occurred in evolution. The Super-B8 haplotype is enriched in the Western Irish, declines along gradients away from that region, and is found only in areas of the world where Western Europeans have migrated. The "A3-B7-DR2-DQ1" is more widely spread, from Eastern Asia to Iberia. The Super-B8 haplotype is associated with a number of diet-associated autoimmune diseases. There are 100,000s of extended haplotypes, but only a few show a visible and nodal character in the human population.
Role of allelic variation
Studies of humans and animals imply a heterozygous selection mechanism operating on these loci as an explanation for this variability.[25] One credible mechanism is sexual selection in which females are able to detect males with different HLA relative to their own type.[26] While the DQ and DP encoding loci have fewer alleles, combinations of A1:B1 can produce a theoretical potential of 7,755 DQ and 5,270 DP αβ heterodimers, respectively. While nowhere near this number of isoforms exist in the human population, each individual can carry 4 variable DQ and DP isoforms, increasing the potential number of antigens that these receptors can present to the immune system.
Studies of the variable positions of DP, DR, and DQ reveal that peptide antigen contact residues on class II molecules are most frequently the site of variation in the protein primary structure. Therefore, through a combination of intense allelic variation and/or subunit pairing, the class II 'peptide' receptors are capable of binding an almost endless variation of peptides of 9 amino acids or longer in length, protecting interbreeding subpopulations from nascent or epidemic diseases. Individuals in a population frequently have different haplotypes, and this results in many combinations, even in small groups. This diversity enhances the survival of such groups, and thwarts evolution of epitopes in pathogens, which would otherwise be able to be shielded from the immune system.
Antibodies
HLA antibodies are typically not naturally occurring, and with few exceptions are formed as a result of an immunologic challenge to a foreign material containing non-self HLAs via blood transfusion, pregnancy (paternally inherited antigens), or organ or tissue transplant.
Antibodies against disease-associated HLA haplotypes have been proposed as a treatment for severe autoimmune diseases.[27]
Donor-specific HLA antibodies have been found to be associated with graft failure in kidney, heart, lung, and liver transplantation.
HLA matching for sick siblings
In some diseases requiring hematopoietic stem cell transplantation, preimplantation genetic diagnosis may be used to give rise to a sibling with matching HLA, although there are ethical considerations.[28]
See also
References
- ↑ Galbraith W, Wagner MC, Chao J, Abaza M, Ernst LA, Nederlof MA, et al. (1991). "Imaging cytometry by multiparameter fluorescence". Cytometry. 12 (7): 579–96. doi:10.1002/cyto.990120702. PMID 1782829.
- ↑ Brennan PA, Kendrick KM (December 2006). "Mammalian social odours: attraction and individual recognition". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 361 (1476): 2061–78. doi:10.1098/rstb.2006.1931. PMC 1764843. PMID 17118924.
- ↑ 3.0 3.1 Taylor CJ, Bolton EM, Bradley JA (2011). "Immunological considerations for embryonic and induced pluripotent stem cell banking". Philosophical Transactions of the Royal Society B. 366 (1575): 2312–2322. doi:10.1098/rstb.2011.0030. PMC 3130422. PMID 21727137.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 4.8 4.9 Table 5-7 in: Mitchell, Richard Sheppard; Kumar, Vinay; Abbas, Abul K.; Fausto, Nelson (2007). Robbins Basic Pathology. Philadelphia: Saunders. ISBN 1-4160-2973-7. 8th edition.
- ↑ 5.0 5.1 Values are given for Caucasians, according to Page 61 (right column) in: Jane Salmon; Wallace, Daniel J.; Dubois, Edmund L.; Kirou, Kyriakos A.; Hahn, Bevra; Lehman, Thomas A. (2007). Dubois' lupus erythematosus. Philadelphia: Lippincott Williams & Wilkins. ISBN 0-7817-9394-7.
- ↑ Margaritte-Jeannin P, Babron MC, Bourgey M, Louka AS, Clot F, Percopo S, Coto I, Hugot JP, Ascher H, Sollid LM, Greco L, Clerget-Darpoux F (June 2004). "HLA-DQ relative risks for coeliac disease in European populations: a study of the European Genetics Cluster on Coeliac Disease". Tissue Antigens. 63 (6): 562–7. doi:10.1111/j.0001-2815.2004.00237.x. PMID 15140032.
- ↑ "Complex HLA-DR and -DQ Interactions Confer Risk of Narcolepsy-Cataplexy in Three Ethnic Groups". The American Journal of Human Genetics. 68: 686–699. doi:10.1086/318799. Retrieved 2014-01-25.
- ↑ http://hla.alleles.org/nomenclature/naming.html
- ↑ Shennan, Douglas H (2006). Evolution and the Spiral of Technology. Trafford Publishing. ISBN 1-55212-518-1.
- ↑ Parham P, Ohta T (April 1996). "Population biology of antigen presentation by MHC class I molecules". Science. 272 (5258): 67–74. doi:10.1126/science.272.5258.67. PMID 8600539..
- ↑ 11.0 11.1 11.2 Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Geraghty DE, Hansen JA, Hurley CK, Mach B, Mayr WR, Parham P, Petersdorf EW, Sasazuki T, Schreuder GM, Strominger JL, Svejgaard A, Terasaki PI, Trowsdale J (April 2005). "Nomenclature for factors of the HLA system, 2004". Tissue Antigens. 65 (4): 301–69. doi:10.1111/j.1399-0039.2005.00379.x. PMID 15787720.
- ↑ 12.0 12.1 12.2 12.3 IMGT/HLA Database
- ↑ Karp DR, Marthandan N, Marsh SG, Ahn C, Arnett FC, Deluca DS, Diehl AD, Dunivin R, Eilbeck K, Feolo M, Guidry PA, Helmberg W, Lewis S, Mayes MD, Mungall C, Natale DA, Peters B, Petersdorf E, Reveille JD, Smith B, Thomson G, Waller MJ, Scheuermann RH (February 2010). "Novel sequence feature variant type analysis of the HLA genetic association in systemic sclerosis". Human Molecular Genetics. 19 (4): 707–19. doi:10.1093/hmg/ddp521. PMC 2807365. PMID 19933168.
- ↑ "IMGT/HLA Database".
- ↑ "Immunology Database and Analysis Portal (ImmPort)". Archived from the original on 2011-07-26.
- ↑ 16.0 16.1 16.2 Middleton D, Gonzalez F, Fernandez-Vina M, Tiercy JM, Marsh SG, Aubrey M, Bicalho MG, Canossi A, Carter V, Cate S, Guerini FR, Loiseau P, Martinetti M, Moraes ME, Morales V, Perasaari J, Setterholm M, Sprague M, Tavoularis S, Torres M, Vidal S, Witt C, Wohlwend G, Yang KL (December 2009). "A bioinformatics approach to ascertaining the rarity of HLA alleles". Tissue Antigens. 74 (6): 480–5. doi:10.1111/j.1399-0039.2009.01361.x. PMID 19793314.
- ↑ 17.0 17.1 17.2 17.3 17.4 17.5 17.6 17.7 17.8 Gonzalez-Galarza FF, Mack SJ, Hollenbach J, Fernandez-Vina M, Setterholm M, Kempenich J, Marsh SG, Jones AR, Middleton D (February 2013). "16(th) IHIW: extending the number of resources and bioinformatics analysis for the investigation of HLA rare alleles". International Journal of Immunogenetics. 40 (1): 60–5. doi:10.1111/iji.12030. PMID 23198982.
- ↑ 18.0 18.1 18.2 18.3 Cano P, Klitz W, Mack SJ, Maiers M, Marsh SG, Noreen H, Reed EF, Senitzer D, Setterholm M, Smith A, Fernández-Viña M (May 2007). "Common and well-documented HLA alleles: report of the Ad-Hoc committee of the american society for histocompatiblity and immunogenetics". Human Immunology. 68 (5): 392–417. doi:10.1016/j.humimm.2007.01.014. PMID 17462507.
- ↑ 19.0 19.1 19.2 19.3 19.4 19.5 19.6 19.7 Mack SJ, Cano P, Hollenbach JA, He J, Hurley CK, Middleton D, Moraes ME, Pereira SE, Kempenich JH, Reed EF, Setterholm M, Smith AG, Tilanus MG, Torres M, Varney MD, Voorter CE, Fischer GF, Fleischhauer K, Goodridge D, Klitz W, Little AM, Maiers M, Marsh SG, Müller CR, Noreen H, Rozemuller EH, Sanchez-Mazas A, Senitzer D, Trachtenberg E, Fernandez-Vina M (April 2013). "Common and well-documented HLA alleles: 2012 update to the CWD catalogue". Tissue Antigens. 81 (4): 194–203. doi:10.1111/tan.12093. PMC 3634360. PMID 23510415.
- ↑ 20.0 20.1 Hurley CK (1997). "DNA-based typing of HLA for transplantation." In Leffell MS, Donnenberg AD, Rose NR, eds. (1997) Handbook of Human Immunology. pp. 521–55, Boca Raton: CRC Press, ISBN 0-8493-0134-3.
- ↑ Bodmer JG, Marsh SG, Albert ED, Bodmer WF, Dupont B, Erlich HA, et al. (May 1992). "Nomenclature for factors of the HLA system, 1991". Human Immunology. 34 (1): 4–18. doi:10.1016/0198-8859(92)90079-3. PMID 1399721.
- ↑ Valluri V, Valluei V, Mustafa M, Santhosh A, Middleton D, Alvares M, Alvales M, El Haj E, Gumama O, Abdel-Wareth L, Abdel-Waieth L (August 2005). "Frequencies of HLA-A, HLA-B, HLA-DR, and HLA-DQ phenotypes in the United Arab Emirates population". Tissue Antigens. 66 (2): 107–13. doi:10.1111/j.1399-0039.2005.00441.x. PMID 16029430.
- ↑ Farjadian S, Naruse T, Kawata H, Ghaderi A, Bahram S, Inoko H (November 2004). "Molecular analysis of HLA allele frequencies and haplotypes in Baloch of Iran compared with related populations of Pakistan". Tissue Antigens. 64 (5): 581–7. doi:10.1111/j.1399-0039.2004.00302.x. PMID 15496201.
- ↑ Shankarkumar U, Prasanavar D, Ghosh K, Mohanty D (May 2003). "HLA A*02 allele frequencies and B haplotype associations in Western Indians". Human Immunology. 64 (5): 562–6. doi:10.1016/S0198-8859(03)00032-6. PMID 12691707.
- ↑ Apanius V, Penn D, Slev PR, Ruff LR, Potts WK (1997). "The nature of selection on the major histocompatibility complex". Critical Reviews in Immunology. 17 (2): 179–224. doi:10.1615/critrevimmunol.v17.i2.40. PMID 9094452..
- ↑ Wedekind C, Seebeck T, Bettens F, Paepke AJ (June 1995). "MHC-dependent mate preferences in humans". Proceedings of the Royal Society B: Biological Sciences. 260 (1359): 245–9. doi:10.1098/rspb.1995.0087. PMID 7630893.
- ↑ Oshima M, Deitiker P, Ashizawa T, Atassi MZ (May 2002). "Vaccination with a MHC class II peptide attenuates cellular and humoral responses against tAChR and suppresses clinical EAMG". Autoimmunity. 35 (3): 183–90. doi:10.1080/08916930290022270. PMID 12389643.
- ↑ Verlinsky Y, Rechitsky S, Schoolcraft W, Strom C, Kuliev A (June 2001). "Preimplantation diagnosis for Fanconi anemia combined with HLA matching". JAMA. 285 (24): 3130–3. doi:10.1001/jama.285.24.3130. PMID 11427142.
Bibliography
- Daniel M. Davis, The Compatibility Gene, London, Penguin Books, 2014 (ISBN 978-0-241-95675-5).
- Zsolt Harsanyi and Richard Hutton, Richard, Genetic Prophecy: Beyond the Double Helix, London: Granada, 1982 (ISBN 0-246-11760-5).
External links
- IMGT/HLA Sequence Database at European Bioinformatics Institute
- hla.alleles.org
- HLA Informatics Group at The Anthony Nolan Trust
- British Society for Histocompatibility and Immunogenetics
- American Society for Histocompatibility and Immunogenetics
- European Federation for Immunogenetics
- HistoCheck HLA matching tool for organ and stem cell transplantation
- Allele Frequencies at Variable Immune related loci
- Human+leukocyte+antigens at the US National Library of Medicine Medical Subject Headings (MeSH)
- dbMHC Home, NCBI's database of the Major Histocompatibility Complex
- Rare Alleles Project at the AlleleFrequencies Net Database (AFND)
- Common and Well-Documented (CWD) alleles catalogue