Diabetes mellitus type 1 pathophysiology: Difference between revisions
No edit summary |
No edit summary |
||
| Line 18: | Line 18: | ||
*[[Genes]] associated with [[Diabetes mellitus]] include the following: <ref name="pmid27302272">{{cite journal| author=Pociot F, Lernmark Å| title=Genetic risk factors for type 1 diabetes. | journal=Lancet | year= 2016 | volume= 387 | issue= 10035 | pages= 2331-9 | pmid=27302272 | doi=10.1016/S0140-6736(16)30582-7 | pmc= | url=https://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=27302272 }}</ref><ref name="pmid27625010">{{cite journal| author=Safari-Alighiarloo N, Taghizadeh M, Tabatabaei SM, Shahsavari S, Namaki S, Khodakarim S et al.| title=Identification of new key genes for type 1 diabetes through construction and analysis of protein-protein interaction networks based on blood and pancreatic islet transcriptomes. | journal=J Diabetes | year= 2016 | volume= | issue= | pages= | pmid=27625010 | doi=10.1111/1753-0407.12483 | pmc= | url=https://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=27625010 }}</ref><ref>Brorsson CA, Pociot F, Type 1 Diabetes Genetics Consortium. Shared genetic basis for type 1 diabetes, islet autoantibodies, and autoantibodies associated with other immune-mediated diseases in families with type 1 diabetes. Diabetes Care 2015; 38 (suppl 3): S8–13.</ref><ref>Ahlqvist E, van Zuydam NR, Groop LC, McCarthy MI. The genetics of diabetic complications. Nat Rev Nephrol 2015; 11: 277–87.</ref><ref>Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet 2013; 14: 661–73.</ref><ref name=":0">Type 1 Diabetes mellitus "Dennis Kasper, Anthony Fauci, Stephen Hauser, Dan Longo, J. Larry Jameson, Joseph Loscalzo"Harrison's Principles of Internal Medicine, 19e Accessed on December 27th,2016</ref> | *[[Genes]] associated with [[Diabetes mellitus]] include the following: <ref name="pmid27302272">{{cite journal| author=Pociot F, Lernmark Å| title=Genetic risk factors for type 1 diabetes. | journal=Lancet | year= 2016 | volume= 387 | issue= 10035 | pages= 2331-9 | pmid=27302272 | doi=10.1016/S0140-6736(16)30582-7 | pmc= | url=https://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=27302272 }}</ref><ref name="pmid27625010">{{cite journal| author=Safari-Alighiarloo N, Taghizadeh M, Tabatabaei SM, Shahsavari S, Namaki S, Khodakarim S et al.| title=Identification of new key genes for type 1 diabetes through construction and analysis of protein-protein interaction networks based on blood and pancreatic islet transcriptomes. | journal=J Diabetes | year= 2016 | volume= | issue= | pages= | pmid=27625010 | doi=10.1111/1753-0407.12483 | pmc= | url=https://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=27625010 }}</ref><ref>Brorsson CA, Pociot F, Type 1 Diabetes Genetics Consortium. Shared genetic basis for type 1 diabetes, islet autoantibodies, and autoantibodies associated with other immune-mediated diseases in families with type 1 diabetes. Diabetes Care 2015; 38 (suppl 3): S8–13.</ref><ref>Ahlqvist E, van Zuydam NR, Groop LC, McCarthy MI. The genetics of diabetic complications. Nat Rev Nephrol 2015; 11: 277–87.</ref><ref>Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet 2013; 14: 661–73.</ref><ref name=":0">Type 1 Diabetes mellitus "Dennis Kasper, Anthony Fauci, Stephen Hauser, Dan Longo, J. Larry Jameson, Joseph Loscalzo"Harrison's Principles of Internal Medicine, 19e Accessed on December 27th,2016</ref> | ||
**Currently, 58 [[Genomics|genomic regions]] are known to be associated with [[Type 1 diabetes|Type 1 DM]]. | **Currently, 58 [[Genomics|genomic regions]] are known to be associated with [[Type 1 diabetes|Type 1 DM]]. | ||
**Major susceptibility [[gene]] for [[Type 1 diabetes| | **Major susceptibility [[gene]] for [[Type 1 diabetes|type 1 diabetes]] is located on [[HLA]] region of [[Chromosome 6 (human)|chromosome 6]]. It accounts for 40-50% of the genetic risk for [[Type 1 diabetes|type 1 diabetes]]. This region encodes for class II [[major histocompatibility complex]] ([[major histocompatibility complex|MHC]]) [[molecule|molecules]]. [[major histocompatibility complex]] ([[major histocompatibility complex|MHC]]) [[molecule|molecules]] play an important role in presenting [[antigen]] to [[T helper cell]] and initiating immune response. | ||
**Other major susceptibility [[genes]] which were associated with [[Type 1 diabetes|Type 1 DM]] include [[polymorphisms]] in the [[promoter region]] of the [[insulin]] [[gene]], the [[CTLA-4|CTLA-4 gene]], [[IL-2|interleukin 2 receptor]], [[Insulin]]-[[Variable number tandem repeat|VNTR]], AIRE, [[FOXP3|FoxP3]], [[STAT3]], HIP14 and [[PTPN22]] etc.<ref name="PaschouPapadopoulou-Marketou2018">{{cite journal|last1=Paschou|first1=Stavroula A|last2=Papadopoulou-Marketou|first2=Nektaria|last3=Chrousos|first3=George P|last4=Kanaka-Gantenbein|first4=Christina|title=On type 1 diabetes mellitus pathogenesis|journal=Endocrine Connections|volume=7|issue=1|year=2018|pages=R38–R46|issn=2049-3614|doi=10.1530/EC-17-0347}}</ref><ref name="Tuomi2005">{{cite journal|last1=Tuomi|first1=T.|title=Type 1 and Type 2 Diabetes: What Do They Have in Common?|journal=Diabetes|volume=54|issue=Supplement 2|year=2005|pages=S40–S45|issn=0012-1797|doi=10.2337/diabetes.54.suppl_2.S40}}</ref> | **Other major susceptibility [[genes]] which were associated with [[Type 1 diabetes|Type 1 DM]] include [[polymorphisms]] in the [[promoter region]] of the [[insulin]] [[gene]], the [[CTLA-4|CTLA-4 gene]], [[IL-2|interleukin 2 receptor]], [[Insulin]]-[[Variable number tandem repeat|VNTR]], AIRE, [[FOXP3|FoxP3]], [[STAT3]], HIP14 and [[PTPN22]] etc.<ref name="PaschouPapadopoulou-Marketou2018">{{cite journal|last1=Paschou|first1=Stavroula A|last2=Papadopoulou-Marketou|first2=Nektaria|last3=Chrousos|first3=George P|last4=Kanaka-Gantenbein|first4=Christina|title=On type 1 diabetes mellitus pathogenesis|journal=Endocrine Connections|volume=7|issue=1|year=2018|pages=R38–R46|issn=2049-3614|doi=10.1530/EC-17-0347}}</ref><ref name="Tuomi2005">{{cite journal|last1=Tuomi|first1=T.|title=Type 1 and Type 2 Diabetes: What Do They Have in Common?|journal=Diabetes|volume=54|issue=Supplement 2|year=2005|pages=S40–S45|issn=0012-1797|doi=10.2337/diabetes.54.suppl_2.S40}}</ref> | ||
**Presence of certain [[genes]] confer protection against the development of the disease. [[Haplotype]] DQA1*0102, DQB1*0602 is extremely rare in individuals with [[Type 1 diabetes|type 1 DM]] (<1%) and appears to provide protection from [[Type 1 diabetes|type 1 | **Presence of certain [[genes]] confer protection against the development of the disease. [[Haplotype]] DQA1*0102, DQB1*0602 is extremely rare in individuals with [[Type 1 diabetes|type 1 DM]] (<1%) and appears to provide protection from [[Type 1 diabetes|type 1 diabetes]]. | ||
<SMALL><SMALL> | <SMALL><SMALL> | ||
{| class="wikitable" | {| class="wikitable" | ||
!Genes important to type 1 diabetes pathogenesis | ![[Genes]] important to [[Type 1 diabetes|type 1 diabetes]] [[pathogenesis]] | ||
!Region | !Region | ||
!Odds ratio | !Odds ratio | ||
| Line 31: | Line 31: | ||
| colspan="1" rowspan="1" |1p13.2 | | colspan="1" rowspan="1" |1p13.2 | ||
| colspan="1" rowspan="1" |1·89 | | colspan="1" rowspan="1" |1·89 | ||
| colspan="1" rowspan="1" |Regulation of innate immune response, T | | colspan="1" rowspan="1" |Regulation of innate immune response, [[T cell]] activation, and [[natural killer cell]] [[Cell growth|proliferation]] | ||
|- | |- | ||
| colspan="1" rowspan="1" |IL10 | | colspan="1" rowspan="1" |IL10 | ||
| colspan="1" rowspan="1" |1q32.1 | | colspan="1" rowspan="1" |1q32.1 | ||
| colspan="1" rowspan="1" |0·86 | | colspan="1" rowspan="1" |0·86 | ||
| colspan="1" rowspan="1" |Cytokines and inflammatory response | | colspan="1" rowspan="1" |[[Cytokine|Cytokines]] and inflammatory response | ||
|- | |- | ||
| colspan="1" rowspan="1" |AFF3 | | colspan="1" rowspan="1" |AFF3 | ||
| colspan="1" rowspan="1" |2q11.2 | | colspan="1" rowspan="1" |2q11.2 | ||
| colspan="1" rowspan="1" |1·11 | | colspan="1" rowspan="1" |1·11 | ||
| colspan="1" rowspan="1" |Regulation of transcription | | colspan="1" rowspan="1" |Regulation of [[Transcription (genetics)|transcription]] | ||
|- | |- | ||
| colspan="1" rowspan="1" |IFIH1 | | colspan="1" rowspan="1" |IFIH1 | ||
| Line 48: | Line 48: | ||
0·85 | 0·85 | ||
0·59 | 0·59 | ||
| colspan="1" rowspan="1" |Innate immune system NF-κB activation | | colspan="1" rowspan="1" |[[Innate immune system]] [[NF-κB]] activation | ||
|- | |- | ||
| colspan="1" rowspan="1" |STAT4 | | colspan="1" rowspan="1" |STAT4 | ||
| colspan="1" rowspan="1" |2q32.3 | | colspan="1" rowspan="1" |2q32.3 | ||
| colspan="1" rowspan="1" |1·10§ | | colspan="1" rowspan="1" |1·10§ | ||
| colspan="1" rowspan="1" |Cytokine-mediated signalling pathway | | colspan="1" rowspan="1" |[[Cytokine]]-mediated signalling pathway | ||
|- | |- | ||
| colspan="1" rowspan="1" |CTLA4 | | colspan="1" rowspan="1" |CTLA4 | ||
| Line 59: | Line 59: | ||
| colspan="1" rowspan="1" |0·82 | | colspan="1" rowspan="1" |0·82 | ||
0·84 | 0·84 | ||
| colspan="1" rowspan="1" |T | | colspan="1" rowspan="1" |[[T cell]] activation | ||
|- | |- | ||
| colspan="1" rowspan="1" |CCR5 | | colspan="1" rowspan="1" |CCR5 | ||
| colspan="1" rowspan="1" |3p21.31 | | colspan="1" rowspan="1" |3p21.31 | ||
| colspan="1" rowspan="1" |0·85 | | colspan="1" rowspan="1" |0·85 | ||
| colspan="1" rowspan="1" |Th1 cell development and chemokine-mediated signalling pathway | | colspan="1" rowspan="1" |[[T helper cell|Th1 cell]] development and [[chemokine]]-mediated signalling pathway | ||
|- | |- | ||
| colspan="1" rowspan="1" |IL21, IL2 | | colspan="1" rowspan="1" |IL21, IL2 | ||
| Line 72: | Line 72: | ||
1·14 | 1·14 | ||
1·15 | 1·15 | ||
| colspan="1" rowspan="1" |Cytokines and inflammatory response and Th1 or Th2 cell differentiation | | colspan="1" rowspan="1" |[[cytokine|Cytokines]] and inflammatory response and [[T helper cell|Th1 cell]] or [[T helper cell|Th2 cell]] [[differentiation]] | ||
|- | |- | ||
| colspan="1" rowspan="1" |IL7R | | colspan="1" rowspan="1" |IL7R | ||
| colspan="1" rowspan="1" |5p13.2 | | colspan="1" rowspan="1" |5p13.2 | ||
| colspan="1" rowspan="1" |1·11 | | colspan="1" rowspan="1" |1·11 | ||
| colspan="1" rowspan="1" |T | | colspan="1" rowspan="1" |[[T cell]]-mediated [[cytotoxicity]], [[immunoglobulin]] production, and [[antigen]] binding | ||
|- | |- | ||
| colspan="1" rowspan="1" |BACH2 | | colspan="1" rowspan="1" |BACH2 | ||
| Line 84: | Line 84: | ||
0·88 | 0·88 | ||
1·20 | 1·20 | ||
| colspan="1" rowspan="1" |Transcription | | colspan="1" rowspan="1" |[[Transcription (genetics)|transcription]] | ||
|- | |- | ||
| colspan="1" rowspan="1" |TNFAIP3 | | colspan="1" rowspan="1" |TNFAIP3 | ||
| Line 94: | Line 94: | ||
| colspan="1" rowspan="1" |6q25.3 | | colspan="1" rowspan="1" |6q25.3 | ||
| colspan="1" rowspan="1" |0·92 | | colspan="1" rowspan="1" |0·92 | ||
| colspan="1" rowspan="1" |Signal transduction | | colspan="1" rowspan="1" |[[Signal transduction]] | ||
|- | |- | ||
| colspan="1" rowspan="1" |IKZF1 | | colspan="1" rowspan="1" |IKZF1 | ||
| Line 106: | Line 106: | ||
1·12 | 1·12 | ||
0·90 | 0·90 | ||
| colspan="1" rowspan="1" |Regulation of transcription | | colspan="1" rowspan="1" |Regulation of [[Transcription (genetics)|transcription]] | ||
|- | |- | ||
| colspan="1" rowspan="1" |IL2RA | | colspan="1" rowspan="1" |IL2RA | ||
| Line 115: | Line 115: | ||
0·62 | 0·62 | ||
0·82 | 0·82 | ||
| colspan="1" rowspan="1" |Alternative mRNA splicing Th1 or Th2 cell differentiation | | colspan="1" rowspan="1" |Alternative [[Messenger RNA|mRNA]] splicing [[T helper cell|Th1]] or [[T helper cell|Th2 cell]] [[differentiation]] | ||
|- | |- | ||
| colspan="1" rowspan="1" |PRKCQ | | colspan="1" rowspan="1" |PRKCQ | ||
| colspan="1" rowspan="1" |10p15.1 | | colspan="1" rowspan="1" |10p15.1 | ||
| colspan="1" rowspan="1" |0·69 | | colspan="1" rowspan="1" |0·69 | ||
| colspan="1" rowspan="1" |Apoptotic process, inflammatory response, innate immune response, and T | | colspan="1" rowspan="1" |Apoptotic process, inflammatory response, [[innate immune response]], and [[T cell]]-receptor signalling pathway | ||
|- | |- | ||
| colspan="1" rowspan="1" |NRP1 | | colspan="1" rowspan="1" |NRP1 | ||
| colspan="1" rowspan="1" |10p11.22 | | colspan="1" rowspan="1" |10p11.22 | ||
| colspan="1" rowspan="1" |1·11 | | colspan="1" rowspan="1" |1·11 | ||
| colspan="1" rowspan="1" |Signal transduction | | colspan="1" rowspan="1" |[[Signal transduction]] | ||
|- | |- | ||
| colspan="1" rowspan="1" |INS | | colspan="1" rowspan="1" |INS | ||
| Line 132: | Line 132: | ||
0·63 | 0·63 | ||
0·63 | 0·63 | ||
| colspan="1" rowspan="1" |Insulin signalling pathway | | colspan="1" rowspan="1" |[[Insulin]] signalling pathway | ||
|- | |- | ||
| colspan="1" rowspan="1" |BAD | | colspan="1" rowspan="1" |BAD | ||
| colspan="1" rowspan="1" |11q13.1 | | colspan="1" rowspan="1" |11q13.1 | ||
| colspan="1" rowspan="1" |0·92 | | colspan="1" rowspan="1" |0·92 | ||
| colspan="1" rowspan="1" |Apoptosis | | colspan="1" rowspan="1" |[[Apoptosis]] | ||
|- | |- | ||
| colspan="1" rowspan="1" |CD69 | | colspan="1" rowspan="1" |CD69 | ||
| Line 143: | Line 143: | ||
| colspan="1" rowspan="1" |0·87 | | colspan="1" rowspan="1" |0·87 | ||
1·10 | 1·10 | ||
| colspan="1" rowspan="1" |Signal transduction | | colspan="1" rowspan="1" |[[Signal transduction]] | ||
|- | |- | ||
| colspan="1" rowspan="1" |ITGB7 | | colspan="1" rowspan="1" |ITGB7 | ||
| colspan="1" rowspan="1" |12q13.13 | | colspan="1" rowspan="1" |12q13.13 | ||
| colspan="1" rowspan="1" |1·19 | | colspan="1" rowspan="1" |1·19 | ||
| colspan="1" rowspan="1" |Response to virus and regulation of immune response | | colspan="1" rowspan="1" |Response to [[virus]] and regulation of immune response | ||
|- | |- | ||
| colspan="1" rowspan="1" |ERBB3 | | colspan="1" rowspan="1" |ERBB3 | ||
| colspan="1" rowspan="1" |12q13.2 | | colspan="1" rowspan="1" |12q13.2 | ||
| colspan="1" rowspan="1" |1·25 | | colspan="1" rowspan="1" |1·25 | ||
| colspan="1" rowspan="1" |Regulation of transcription, innate immune response, and lipid metabolism | | colspan="1" rowspan="1" |Regulation of [[Transcription (genetics)|transcription]], [[Innate immune system|innate immune response]], and [[lipid metabolism]] | ||
|- | |- | ||
| colspan="1" rowspan="1" |CYP27B1 | | colspan="1" rowspan="1" |CYP27B1 | ||
| colspan="1" rowspan="1" |12q14.1 | | colspan="1" rowspan="1" |12q14.1 | ||
| colspan="1" rowspan="1" |0·82 | | colspan="1" rowspan="1" |0·82 | ||
| colspan="1" rowspan="1" |Metabolism of lipids, lipoproteins, steroid hormones, and vitamin D | | colspan="1" rowspan="1" |[[lipid metabolism|Metabolism of lipids]], [[lipoprotein|lipoproteins]], steroid [[hormone|hormones]], and [[vitamin D]] | ||
|- | |- | ||
| colspan="1" rowspan="1" |SH2B3 | | colspan="1" rowspan="1" |SH2B3 | ||
| Line 165: | Line 165: | ||
0·76 | 0·76 | ||
0·76 | 0·76 | ||
| colspan="1" rowspan="1" |Signal transduction | | colspan="1" rowspan="1" |[[Signal transduction]] | ||
|- | |- | ||
| colspan="1" rowspan="1" |GPR183 | | colspan="1" rowspan="1" |GPR183 | ||
| colspan="1" rowspan="1" |13q32.3 | | colspan="1" rowspan="1" |13q32.3 | ||
| colspan="1" rowspan="1" |1·12 | | colspan="1" rowspan="1" |1·12 | ||
| colspan="1" rowspan="1" |Humoral immune response | | colspan="1" rowspan="1" |[[Humoral immunity|Humoral immune response]] | ||
|- | |- | ||
| colspan="1" rowspan="1" |DLK1 | | colspan="1" rowspan="1" |DLK1 | ||
| Line 176: | Line 176: | ||
| colspan="1" rowspan="1" |0·88 | | colspan="1" rowspan="1" |0·88 | ||
0·90 | 0·90 | ||
| colspan="1" rowspan="1" |Regulation of gene expression | | colspan="1" rowspan="1" |Regulation of [[gene expression]] | ||
|- | |- | ||
| colspan="1" rowspan="1" |RASGRP1 | | colspan="1" rowspan="1" |RASGRP1 | ||
| Line 182: | Line 182: | ||
| colspan="1" rowspan="1" |0·85 | | colspan="1" rowspan="1" |0·85 | ||
1·15 | 1·15 | ||
| colspan="1" rowspan="1" |Inflammatory response to antigenic stimulus and cytokine production | | colspan="1" rowspan="1" |Inflammatory response to [[antigen|antigenic]] stimulus and [[cytokine]] production | ||
|- | |- | ||
| colspan="1" rowspan="1" |CTSH | | colspan="1" rowspan="1" |CTSH | ||
| Line 189: | Line 189: | ||
0·78 | 0·78 | ||
0·90 | 0·90 | ||
| colspan="1" rowspan="1" |Immune response-regulating signalling pathway T | | colspan="1" rowspan="1" |Immune response-regulating signalling pathway [[T cell]]-mediated [[cytotoxicity]] adaptive immune response | ||
|- | |- | ||
| colspan="1" rowspan="1" |CLEC16A | | colspan="1" rowspan="1" |CLEC16A | ||
| Line 203: | Line 203: | ||
0·90 | 0·90 | ||
1·24 | 1·24 | ||
| colspan="1" rowspan="1" |Inflammatory response and regulation of defence response to virus | | colspan="1" rowspan="1" |Inflammatory response and regulation of defence response to [[virus]] | ||
|- | |- | ||
| colspan="1" rowspan="1" |ORMDL3 | | colspan="1" rowspan="1" |ORMDL3 | ||
| colspan="1" rowspan="1" |17q12 | | colspan="1" rowspan="1" |17q12 | ||
| colspan="1" rowspan="1" |0·90 | | colspan="1" rowspan="1" |0·90 | ||
| colspan="1" rowspan="1" |Protein binding | | colspan="1" rowspan="1" |[[Protein]] binding | ||
|- | |- | ||
| colspan="1" rowspan="1" |PTPN2 | | colspan="1" rowspan="1" |PTPN2 | ||
| colspan="1" rowspan="1" |18p11.21 | | colspan="1" rowspan="1" |18p11.21 | ||
| colspan="1" rowspan="1" |1·20 | | colspan="1" rowspan="1" |1·20 | ||
| colspan="1" rowspan="1" |Cytokine signalling and B | | colspan="1" rowspan="1" |[[Cytokine]] signalling and [[B cell]] and [[T cell]] [[differentiation]] | ||
|- | |- | ||
| colspan="1" rowspan="1" |CD226 | | colspan="1" rowspan="1" |CD226 | ||
| colspan="1" rowspan="1" |18q22.2 | | colspan="1" rowspan="1" |18q22.2 | ||
| colspan="1" rowspan="1" |1·13 | | colspan="1" rowspan="1" |1·13 | ||
| colspan="1" rowspan="1" |Immunoregulation and adaptive immune system | | colspan="1" rowspan="1" |Immunoregulation and [[adaptive immune system]] | ||
|- | |- | ||
| colspan="1" rowspan="1" |TYK2 | | colspan="1" rowspan="1" |TYK2 | ||
| Line 225: | Line 225: | ||
0·87 | 0·87 | ||
0·67 | 0·67 | ||
| colspan="1" rowspan="1" |Cytokine-mediated signalling pathway, intracellular signal transduction, and type I interferon signalling pathway | | colspan="1" rowspan="1" |[[Cytokine]]-mediated signalling pathway, intracellular [[signal transduction]], and type I [[interferon]] signalling pathway | ||
|- | |- | ||
| colspan="1" rowspan="1" |FUT2 | | colspan="1" rowspan="1" |FUT2 | ||
| Line 237: | Line 237: | ||
| colspan="1" rowspan="1" |21q22.3 | | colspan="1" rowspan="1" |21q22.3 | ||
| colspan="1" rowspan="1" |1·16 | | colspan="1" rowspan="1" |1·16 | ||
| colspan="1" rowspan="1" |Regulation of cytokine production | | colspan="1" rowspan="1" |Regulation of [[cytokine]] production | ||
Regulation of T | Regulation of [[T cel]]l receptor signalling pathway | ||
|- | |- | ||
| colspan="1" rowspan="1" |C1QTNF6 | | colspan="1" rowspan="1" |C1QTNF6 | ||
| colspan="1" rowspan="1" |22q12.3 | | colspan="1" rowspan="1" |22q12.3 | ||
| colspan="1" rowspan="1" |1·11 | | colspan="1" rowspan="1" |1·11 | ||
| colspan="1" rowspan="1" |B | | colspan="1" rowspan="1" |[[B cell]] receptor signalling pathway, [[chemokine]] signalling pathway, and [[natural killer cell]]-mediated [[cytotoxicity]] | ||
|} | |} | ||
</SMALL></SMALL> | </SMALL></SMALL> | ||
| Line 315: | Line 315: | ||
* [[Vitiligo]] | * [[Vitiligo]] | ||
* [[Addison's disease]] | * [[Addison's disease]] | ||
==Gross Pathology== | |||
On gross pathology, [feature1], [feature2], and [feature3] are characteristic findings of [disease name]. | |||
==Microscopic Pathology== | |||
On microscopic histopathological analysis, [feature1], [feature2], and [feature3] are characteristic findings of [disease name]. | |||
==References== | ==References== | ||
Revision as of 21:04, 22 August 2020
|
Diabetes mellitus type 1 Microchapters |
|
Differentiating Diabetes mellitus type 1 from other Diseases |
|
Diagnosis |
|
Treatment |
|
Cardiovascular Disease and Risk Management |
|
Case Studies |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]; Associate Editor(s)-in-Chief: Priyamvada Singh, M.B.B.S. [2]; Cafer Zorkun, M.D., Ph.D. [3]Vishal Devarkonda, M.B.B.S[4]
Overview
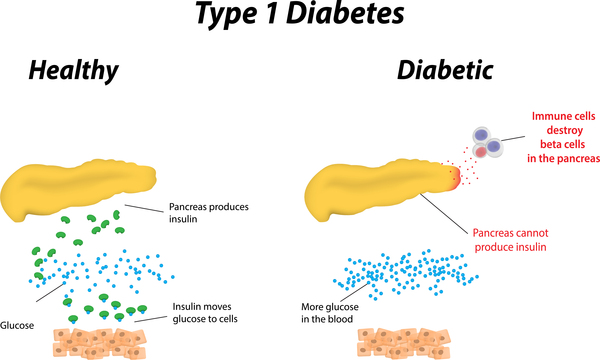
Type 1 diabetes is a disorder characterized by abnormally high blood sugar levels. Type 1 DM is the result of interactions of genetic, environmental, and immunologic factors that ultimately lead to the destruction of the pancreatic beta cells and insulin deficiency.
Pathophysiology
- Type 1 diabetes is a disorder characterized by abnormally high blood sugar levels.
- In this form of diabetes, specialized cells in the pancreas called beta cells stop producing insulin. Insulin controls how much glucose (a type of sugar) is passed from the blood into cells for conversion to energy. Lack of insulin results in the inability to use glucose for energy or to control the amount of sugar in the blood.
Pathogenesis
- Type 1 DM is the result of interactions of genetic, environmental, and immunologic factors that ultimately lead to the destruction of the pancreatic beta cells and insulin deficiency.
- Concordance of type 1 DM in identical twins ranges between 40 and 60%, indicating the presence of additional modifying factors.
Genetics
- Genes associated with Diabetes mellitus include the following: [1][2][3][4][5][6]
- Currently, 58 genomic regions are known to be associated with Type 1 DM.
- Major susceptibility gene for type 1 diabetes is located on HLA region of chromosome 6. It accounts for 40-50% of the genetic risk for type 1 diabetes. This region encodes for class II major histocompatibility complex (MHC) molecules. major histocompatibility complex (MHC) molecules play an important role in presenting antigen to T helper cell and initiating immune response.
- Other major susceptibility genes which were associated with Type 1 DM include polymorphisms in the promoter region of the insulin gene, the CTLA-4 gene, interleukin 2 receptor, Insulin-VNTR, AIRE, FoxP3, STAT3, HIP14 and PTPN22 etc.[7][8]
- Presence of certain genes confer protection against the development of the disease. Haplotype DQA1*0102, DQB1*0602 is extremely rare in individuals with type 1 DM (<1%) and appears to provide protection from type 1 diabetes.
| Genes important to type 1 diabetes pathogenesis | Region | Odds ratio | Gene funtion |
|---|---|---|---|
| PTPN22 | 1p13.2 | 1·89 | Regulation of innate immune response, T cell activation, and natural killer cell proliferation |
| IL10 | 1q32.1 | 0·86 | Cytokines and inflammatory response |
| AFF3 | 2q11.2 | 1·11 | Regulation of transcription |
| IFIH1 | 2q24.2 | 0·85
0·85 0·59 |
Innate immune system NF-κB activation |
| STAT4 | 2q32.3 | 1·10§ | Cytokine-mediated signalling pathway |
| CTLA4 | 2q33.2 | 0·82
0·84 |
T cell activation |
| CCR5 | 3p21.31 | 0·85 | Th1 cell development and chemokine-mediated signalling pathway |
| IL21, IL2 | 4q27 | 1·13
1·12 1·14 1·15 |
Cytokines and inflammatory response and Th1 cell or Th2 cell differentiation |
| IL7R | 5p13.2 | 1·11 | T cell-mediated cytotoxicity, immunoglobulin production, and antigen binding |
| BACH2 | 6q15 | 1·10
0·88 1·20 |
transcription |
| TNFAIP3 | 6q23.3 | 1·12 | Inflammatory response |
| TAGAP | 6q25.3 | 0·92 | Signal transduction |
| IKZF1 | 7p12.2 | 0·89 | Immune-cell regulation |
| GLIS3 | 9p24.2 | 1·12
1·12 0·90 |
Regulation of transcription |
| IL2RA | 10p15.1 | 1·20
0·73 0·52 0·62 0·82 |
Alternative mRNA splicing Th1 or Th2 cell differentiation |
| PRKCQ | 10p15.1 | 0·69 | Apoptotic process, inflammatory response, innate immune response, and T cell-receptor signalling pathway |
| NRP1 | 10p11.22 | 1·11 | Signal transduction |
| INS | 11p15.5 | 0·42
0·63 0·63 |
Insulin signalling pathway |
| BAD | 11q13.1 | 0·92 | Apoptosis |
| CD69 | 12p13.31 | 0·87
1·10 |
Signal transduction |
| ITGB7 | 12q13.13 | 1·19 | Response to virus and regulation of immune response |
| ERBB3 | 12q13.2 | 1·25 | Regulation of transcription, innate immune response, and lipid metabolism |
| CYP27B1 | 12q14.1 | 0·82 | Metabolism of lipids, lipoproteins, steroid hormones, and vitamin D |
| SH2B3 | 12q24.12 | 1·24
0·76 0·76 |
Signal transduction |
| GPR183 | 13q32.3 | 1·12 | Humoral immune response |
| DLK1 | 14q32.2 | 0·88
0·90 |
Regulation of gene expression |
| RASGRP1 | 15q14 | 0·85
1·15 |
Inflammatory response to antigenic stimulus and cytokine production |
| CTSH | 15q25.1 | 0·81
0·78 0·90 |
Immune response-regulating signalling pathway T cell-mediated cytotoxicity adaptive immune response |
| CLEC16A | 16p13.13 | 0·83
0·82 1·14 |
Unknown |
| IL27 | 16p11.2 | 1·19
0·90 1·24 |
Inflammatory response and regulation of defence response to virus |
| ORMDL3 | 17q12 | 0·90 | Protein binding |
| PTPN2 | 18p11.21 | 1·20 | Cytokine signalling and B cell and T cell differentiation |
| CD226 | 18q22.2 | 1·13 | Immunoregulation and adaptive immune system |
| TYK2 | 19p13.2 | 0·82
0·87 0·67 |
Cytokine-mediated signalling pathway, intracellular signal transduction, and type I interferon signalling pathway |
| FUT2 | 19q13.33 | 0·87
0·75 0·87 |
Metabolic pathways |
| UBASH3A | 21q22.3 | 1·16 | Regulation of cytokine production
Regulation of T cell receptor signalling pathway |
| C1QTNF6 | 22q12.3 | 1·11 | B cell receptor signalling pathway, chemokine signalling pathway, and natural killer cell-mediated cytotoxicity |
Environment
Environmental factors were found to influence type 1 DM through various pathways. Some were found to confer protection against type 1 DM, while others were associated with the progression and promotion of Type 1 DM, including:[9][10]
| Triggers | Protective factors | |
|---|---|---|
| Prenatal triggers |
|
|
| Postnatal triggers |
|
|
| Promoters of progression |
|
Immunological
Several studies have found that abnormalities in the humoral and cellular arm of the immune system, were identified to be associated with Type 1 DM, these include:[11][12][13][14][15]
- Islet cell autoantibodies
- Activated lymphocytes in the islets, peripancreatic lymph nodes, and systemic circulation
- T lymphocytes that proliferate when stimulated with islet proteins
- Release of cytokines within the insulitis
- An enzyme named glutamic acid decarboxylase (GAD65) found in β cells has similar amino acid sequence with the Coxsackie B4 P2-C protein, which augments the response of humoral immunity.
- Autoantibodies against IA-2 and zinc transporter (ZnT8) have been positive in 60% and 60-80% of type 1 DM at the time of diagnose, respectively.
Associated conditions
Conditions associated with type 1 DM include:[6][16]
- Autoimmune thyroid disease (ATD)
- Celiac disease (CD)
- Autoimmune gastritis (AIG)
- Pernicious anemia (PA)
- Vitiligo
- Addison's disease
Gross Pathology
On gross pathology, [feature1], [feature2], and [feature3] are characteristic findings of [disease name].
Microscopic Pathology
On microscopic histopathological analysis, [feature1], [feature2], and [feature3] are characteristic findings of [disease name].
References
- ↑ Pociot F, Lernmark Å (2016). "Genetic risk factors for type 1 diabetes". Lancet. 387 (10035): 2331–9. doi:10.1016/S0140-6736(16)30582-7. PMID 27302272.
- ↑ Safari-Alighiarloo N, Taghizadeh M, Tabatabaei SM, Shahsavari S, Namaki S, Khodakarim S; et al. (2016). "Identification of new key genes for type 1 diabetes through construction and analysis of protein-protein interaction networks based on blood and pancreatic islet transcriptomes". J Diabetes. doi:10.1111/1753-0407.12483. PMID 27625010.
- ↑ Brorsson CA, Pociot F, Type 1 Diabetes Genetics Consortium. Shared genetic basis for type 1 diabetes, islet autoantibodies, and autoantibodies associated with other immune-mediated diseases in families with type 1 diabetes. Diabetes Care 2015; 38 (suppl 3): S8–13.
- ↑ Ahlqvist E, van Zuydam NR, Groop LC, McCarthy MI. The genetics of diabetic complications. Nat Rev Nephrol 2015; 11: 277–87.
- ↑ Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet 2013; 14: 661–73.
- ↑ 6.0 6.1 Type 1 Diabetes mellitus "Dennis Kasper, Anthony Fauci, Stephen Hauser, Dan Longo, J. Larry Jameson, Joseph Loscalzo"Harrison's Principles of Internal Medicine, 19e Accessed on December 27th,2016
- ↑ Paschou, Stavroula A; Papadopoulou-Marketou, Nektaria; Chrousos, George P; Kanaka-Gantenbein, Christina (2018). "On type 1 diabetes mellitus pathogenesis". Endocrine Connections. 7 (1): R38–R46. doi:10.1530/EC-17-0347. ISSN 2049-3614.
- ↑ Tuomi, T. (2005). "Type 1 and Type 2 Diabetes: What Do They Have in Common?". Diabetes. 54 (Supplement 2): S40–S45. doi:10.2337/diabetes.54.suppl_2.S40. ISSN 0012-1797.
- ↑ Volume 387, Issue 10035, 4–10 June 2016, Pages 2340–2348 Series Environmental risk factors for type 1 diabetes Prof Marian Rewers, MDa, Prof Johnny Ludvigsson, MD
- ↑ Butalia S, Kaplan GG, Khokhar B, Rabi DM (2016). "Environmental Risk Factors and Type 1 Diabetes: Past, Present, and Future". Can J Diabetes. 40 (6): 586–593. doi:10.1016/j.jcjd.2016.05.002. PMID 27545597.
- ↑ Jaberi-Douraki M, Pietropaolo M, Khadra A (2015). "Continuum model of T-cell avidity: Understanding autoreactive and regulatory T-cell responses in type 1 diabetes". J Theor Biol. 383: 93–105. doi:10.1016/j.jtbi.2015.07.032. PMC 4567915. PMID 26271890.
- ↑ Rydén A, Ludvigsson J, Fredrikson M, Faresjö M (2014). "General immune dampening is associated with disturbed metabolism at diagnosis of type 1 diabetes". Pediatr Res. 75 (1–1): 45–50. doi:10.1038/pr.2013.167. PMID 24105410.
- ↑ Type 1 Diabetes mellitus "Dennis Kasper, Anthony Fauci, Stephen Hauser, Dan Longo, J. Larry Jameson, Joseph Loscalzo"Harrison's Principles of Internal Medicine, 19e Accessed on December 27th,2016
- ↑ Paschou, Stavroula A; Papadopoulou-Marketou, Nektaria; Chrousos, George P; Kanaka-Gantenbein, Christina (2018). "On type 1 diabetes mellitus pathogenesis". Endocrine Connections. 7 (1): R38–R46. doi:10.1530/EC-17-0347. ISSN 2049-3614.
- ↑ Ellis TM, Schatz DA, Ottendorfer EW, Lan MS, Wasserfall C, Salisbury PJ; et al. (1998). "The relationship between humoral and cellular immunity to IA-2 in IDDM". Diabetes. 47 (4): 566–9. doi:10.2337/diabetes.47.4.566. PMID 9568688.
- ↑ Witek PR, Witek J, Pańkowska E (2012). "[Type 1 diabetes-associated autoimmune diseases: screening, diagnostic principles and management]". Med Wieku Rozwoj. 16 (1): 23–34. PMID 22516771.
Template:WH Template:WS Template:WH Template:WS