Serpin
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [14]
Serpins are a group of proteins with similar structures that were first identified as a set of proteins able to inhibit proteases. The name serpin is derived from this activity - serine protease inhibitors.[2]
The first members of the serpin superfamily to be extensively studied were the human plasma proteins antithrombin and antitrypsin, which play key roles in controlling blood coagulation and inflammation, respectively. Initially, research focused upon their role in human disease: antithrombin deficiency results in thrombosis and antitrypsin deficiency causes emphysema. In 1980 Hunt and Dayhoff made the surprising discovery that both these molecules share significant amino acid sequence similarity to the major protein in chicken egg white, ovalbumin, and they proposed a new protein superfamily.[3] Over 1000 serpins have now been identified, these include 36 human proteins, as well as molecules in plants, bacteria, archaea and certain viruses.[4][5] Serpins are thus the largest and most diverse family of protease inhibitors.[6]
While most serpins control proteolytic cascades, certain serpins do not inhibit enzymes, but instead perform diverse functions such as storage (ovalbumin, in egg white), hormone carriage proteins (thyroxine-binding globulin, cortisol binding globulin) and tumor suppressor genes (maspin). The term serpin is used to describe these latter members as well, despite their noninhibitory function.[7]
As serpins control processes such as coagulation and inflammation, these proteins are the target of medical research. However, serpins are also of particular interest to the structural biology and protein folding communities, because they undergo a unique and dramatic change in shape (or conformational change) when they inhibit target proteases.[8] This is unusual - most classical protease inhibitors function as simple "lock and key" molecules that bind to and block access to the protease active site (see for example, bovine pancreatic trypsin inhibitor). While the serpin mechanism of protease inhibition confers certain advantages, it also has drawbacks and serpins are vulnerable to mutations that result in protein misfolding and the formation of inactive long chain polymers (serpinopathies).[9][10] Serpin polymerisation reduces the amount of active inhibitor, as well as accumulation of serpin polymers causing cell death and organ failure. For example, the serpin antitrypsin is primarily produced in the liver, and antitrypsin polymerisation causes liver cirrhosis.[10] Understanding serpinopathies also provides insights on protein misfolding in general, a process common to many human diseases, such as Alzheimer’s and CJD.[9]
Proteases inhibited by serpins
Most inhibitory serpins target chymotrypsin-like serine proteases (see Table 1). These enzymes are defined by the presence of a nucleophilic serine residue in their catalytic site. Examples include thrombin, trypsin and human neutrophil elastase.[11]
Some serpins inhibit other classes of protease and are termed "cross class inhibitors". For example squamous cell carcinoma antigen 1 (SCCA-1) and the avian serpin myeloid and erythroid nuclear termination stage specific protein (MENT) both inhibit papain-like cysteine proteases[12][13][14]
The viral serpin crmA is a suppressor of the inflammatory response through inhibition of IL-1 and IL-18 processing by the cysteine protease caspase-1.[15] Cysteine proteases differ from serine proteases in that they are defined by the presence of a nucleophilic cysteine residue, rather than a serine residue, in their catalytic site.[16] Nonetheless, the enzymatic chemistry is similar, and serpins most likely inhibit both classes of enzyme in a similar fashion.[17]
Localisation and general biological roles
Approximately two thirds of human serpins perform extracellular roles. For example, extracellular serpins regulate the proteolytic cascades central to blood clotting (antithrombin), the inflammatory response (antitrypsin, antichymotrypsin and C1 inhibitor) and tissue remodelling (PAI-1). Non-inhibitory extracellular serpins also perform important roles. Thyroxine-binding globulin and cortisol binding globulin transport the sterol hormones thyroxine and cortisol respectively.[18][1] The protease renin cleaves off a ten amino acid N-terminal peptide from angiotensinogen to produce the peptide hormone angiotensin I.[19] Table 1 at the bottom of this article provides a brief summary of human serpin function as well as some of the diseases that result from serpin deficiency.
The first Intracellular members of the serpin superfamily were identified in the early 1990s.[20][21] As all nine serpins in Caenorhabditis elegans lack signal sequences, they are probably intracellular.[22] Based upon these data it seems likely that the ancestral serpin to human serpins was an intracellular molecule.
The protease targets of intracellular inhibitory serpins have been more difficult to identify. Characterisation is complicated by these molecules appearing to perform overlapping roles, as well as the lack of precise functional equivalents of human serpins in model organisms such as the mouse. An important function of intracellular serpins may be to protect against the inappropriate activity of proteases inside the cell.[23] For example, one of the best characterised human intracellular serpins is SERPINB9, which inhibits the cytotoxic granule protease granzyme B. In doing so, SERPINB9 may protect against inadvertent release of granzyme B and premature or unwanted activation of cell death pathways.[24]
Intracellular serpins also perform roles distinct from protease inhibition. For example, maspin, a non-inhibitory serpin, is important for preventing metastasis in breast and prostate cancers.[25][26] Another example is the avian nuclear cysteine protease inhibitor MENT, which acts as a chromatin remodelling molecule in avian red blood cells.[27][13]
Phylogenetic studies show that most intracellular serpins belong to a single clade (see table 1). Exceptions include the non-inhibitory heat shock serpin HSP47, which is a chaperone essential for proper folding of collagen and cycles between the cis-Golgi and the endoplasmic reticulum.[28]
Structure

Structural biology has played a central role in the understanding of serpin function and biology. Over eighty serpin structures, in a variety of different conformations (described below) have been determined to date. Although the function of serpins varies widely, these molecules all share a common structure (or fold).
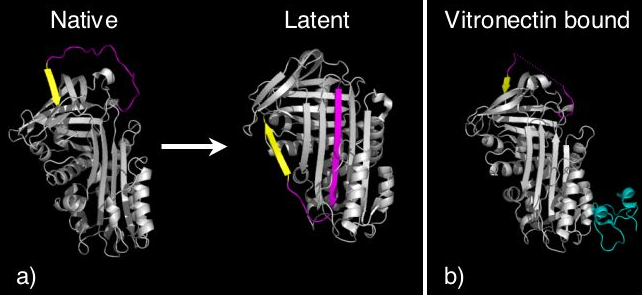
The structure of the non-inhibitory serpin ovalbumin, and the inhibitory serpin antitrypsin revealed the archetype native serpin fold.[31][32] All typically have three β-sheets (termed A, B and C) and eight or nine α-helices (hA-hI) (see figure 1). Serpins also possess an exposed region termed the reactive centre loop (RCL) that in inhibitory molecules includes the specificity determining region and forms the initial interaction with the target protease. In antitrypsin, the RCL is held at the top of the molecule and is not pre-inserted into the A β-sheet (figure 1, left panel). This conformation commonly exists in dynamic equilibrium with a partially inserted native conformation[33] seen in other inhibitory serpins (see figure 1, right panel).
Conformational change and inhibitory mechanism
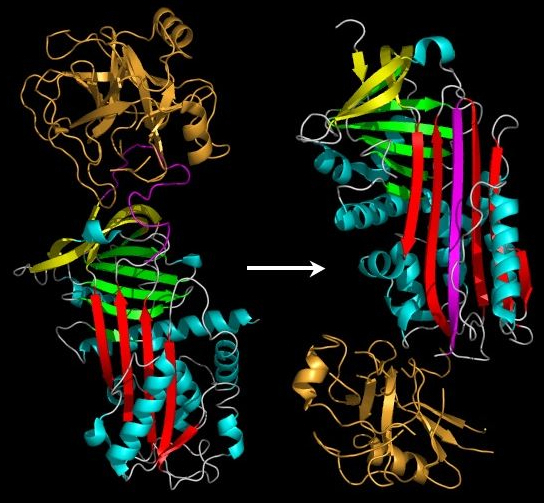
Early studies on serpins revealed that the mechanism by which these molecules inhibit target proteases appeared distinct from the lock-and-key-type mechanism utilised by small protease inhibitors such as the Kunitz-type inhibitors (eg. Basic pancreatic protease inhibitor). Indeed, serpins form covalent complexes with target proteases.[34] Structural studies on serpins also revealed that inhibitory members of the family undergo an unusual conformational change, termed the Stressed to Relaxed (S to R) transition.[31][33][35][36] During this structural transition the RCL inserts into β-sheet A (in red in figure 1 and 2) and forms an extra (fourth) β-strand. The serpin conformational change is key to the mechanism of inhibition of target proteases.
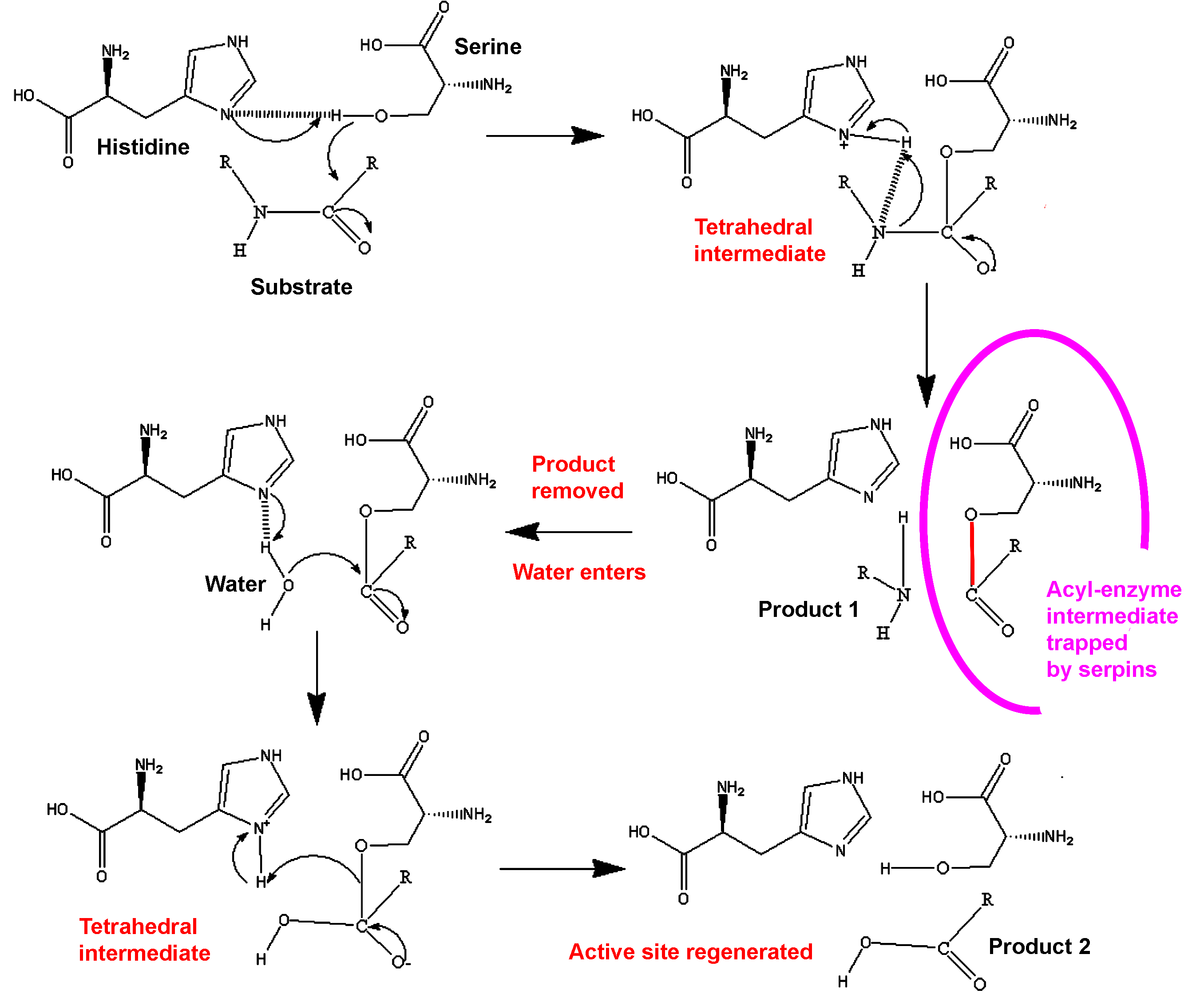
When attacking a substrate, serine proteases catalyze peptide bond cleavage in a two-step process. Initially, the catalytic serine performs a nucleophilic attack on the peptide bond of the substrate (Figure 3). This releases the new N-terminus and forms an ester-bond between the enzyme and the substrate. This covalent enzyme-substrate complex is called an acyl enzyme intermediate. Subsequently, this ester bond is hydrolysed and the new C-terminus is released. The RCL of a serpin acts as a substrate for its cognate protease. However, after the RCL is cleaved, but prior to hydrolysis of the acyl-enzyme intermediate, the serpin rapidly undergoes the S to R transition. Since the RCL is still covalently attached to the protease via the ester bond, the S to R transition causes the protease to be moved from the top to the bottom of the serpin. At the same time, the protease is distorted into a conformation where the acyl enzyme intermediate is hydrolysed extremely slowly.[8] The protease thus remains covalently attached to the target protease and is thereby inhibited. Further, since the serpin has to be cleaved to inhibit the target protases, inhibition consumes the serpin as well. Serpins are therefore irreversible enzyme inhibitors. The serpin mechanism of inhibition is illustrated in figure 2 and several movies illustrating the serpin mechanism can be seen at this link.
 |
 |
Conformational modulation of serpin activity
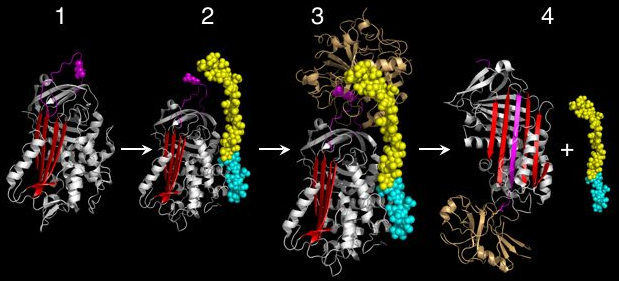
The conformational mobility of serpins provides a key advantage over static lock and key protease inhibitors. In particular, the function of inhibitory serpins can be readily controlled by specific cofactors. The X-ray crystal structures of antithrombin, heparin co-factor II, MENT and murine antichymotrypsin reveal that these serpins adopt a conformation where the first two amino acids of the RCL are inserted into the top of the A β-sheet (see figures 1 and 4). The partially inserted conformation is important because co-factors are able to conformationally switch partially inserted serpins into a fully expelled form.[38][39] This conformational rearrangement makes the serpin a more effective inhibitor.
The archetypal example of this situation is antithrombin, which circulates in plasma in a partially inserted relatively inactive state. The primary specificity determining residue (the P1 Arginine) points towards the body of the serpin and is unavailable to the protease (Figure 4). Upon binding a high affinity heparin pentasaccharide sequence within long chain heparin, antithrombin undergoes a conformational change, RCL expulsion and exposure of the P1 Arginine. The heparin pentasaccharide bound form of antithrombin is thus a more effective inhibitor of thrombin and factor Xa (figure 4).[40][41] Furthermore, both of these coagulation proteases contain binding sites (called exosites) for heparin. Heparin therefore also acts as a template for binding of both protease and serpin, further dramatically accelerating the interaction between the two parties (Figure 4). After the initial interaction, the final serpin complex is formed and the heparin moiety is released. This interaction is physiologically important. For example, after injury to the blood vessel wall heparin is exposed, and antithrombin is thus activated to control the clotting response. The understanding of the molecular basis of this interaction formed the basis of the development of Fondaparinux, a synthetic form of Heparin pentasaccharide used as an anti-clotting drug.[42]
Certain serpins spontaneously undergo the S to R transition as part of their function, to form a conformation termed the latent state (Figure 5). In latent serpins the first strand of the C-sheet has to peel off to allow full RCL insertion. Latent serpins are unable to interact with proteases and are not protease inhibitors. The transition to latency represents a control mechanism for the serpin PAI-1. PAI-1 is released in the inhibitory conformation, however, undergoes conformational change to the latent state unless it is bound to the cofactor vitronectin.[43] Thus PAI-1 contains an "auto-inactivation" mechanism. Similarly, antithrombin can also spontaneously convert to the latent state as part of its normal function. Finally, the N-terminus of tengpin[15][16], a serpin from Thermoanaerobacter tengcongensis, is required to lock the molecule in the native inhibitory state. Disruption of interactions made by the N-terminal region results in spontaneous conformational change of this serpin to the latent conformation.[44]
Serpin receptor interactions
In humans, extracellular serpin-enzyme complexes are rapidly cleared from circulation. One mechanism by which this occurs is the low density lipoprotein receptor related protein (LRP receptor), which binds to inhibitory complexes made by antithrombin, PA1-1 and neuroserpin, causing uptake and subsequent signalling events.[45] Thus, as a consequence of the conformational change during serpin-enzyme complex formation, serpins may act as signalling molecules that alert cells to the presence of protease activity.[45] The fate of intracellular serpin-enzyme complexes remains to be characterised.
Conformational change and non-inhibitory function
Certain non-inhibitory serpins also use the serpin conformational change as part of their function. For example the native (S) form of thyroxine-binding globulin has high affinity for thyroxine, whereas the cleaved (R) form has low affinity. Similarly, native (S) Cortisol Binding Globulin (CBG) has higher affinity for cortisol than its cleaved (R) counterpart. Thus, in these serpins, RCL cleavage and the S to R transition has been commandeered to allow for ligand release, rather than protease inhibition.[46][18][1]
Serpins, serpinopathies and human disease
The complexity of the serpin mechanism renders these molecules vulnerable to inactivating mutations that promote inappropriate conformational change (or misfolding) and diseases ("serpinopathies"). Well characterised serpinopathies include emphysema, cirrhosis, thrombosis and dementia. Serpins thus belong to a large group of molecules such as the prion proteins and the glutamine repeat containing proteins that are susceptible to misfolding, causing conformational disease.[9]
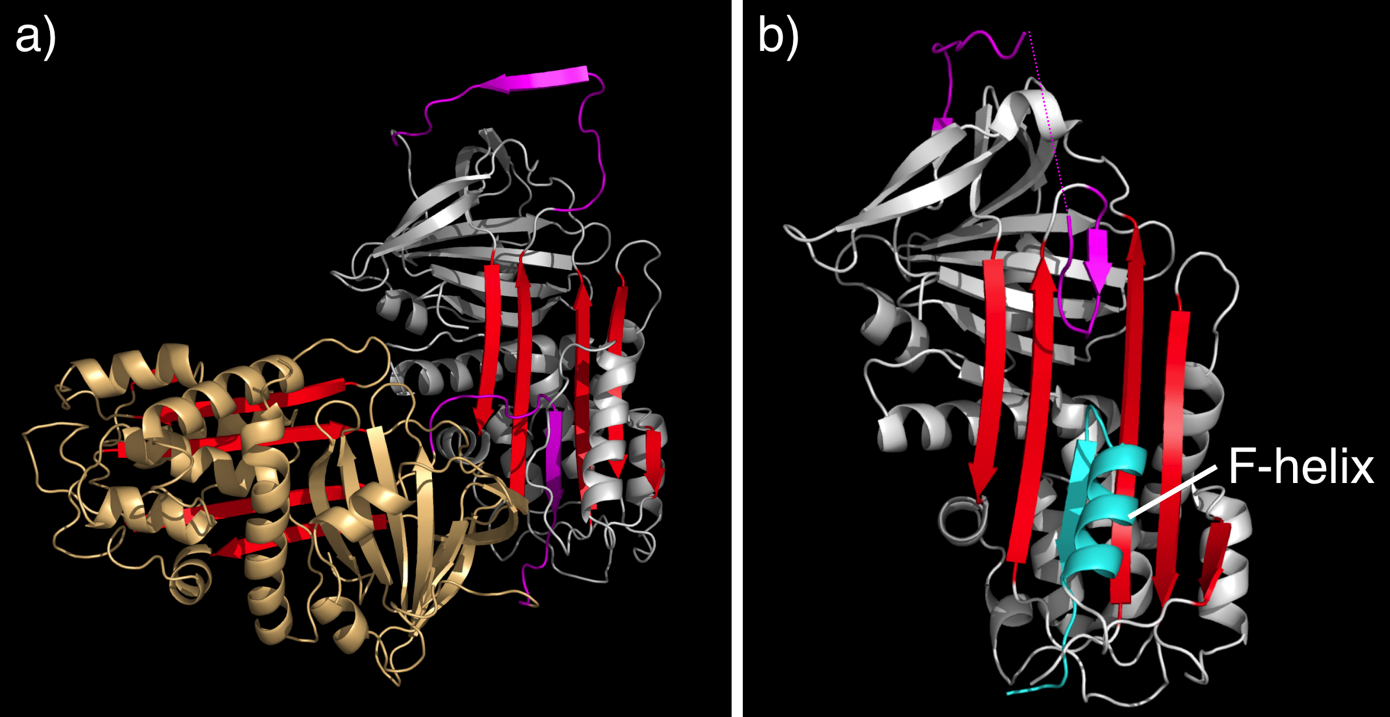
The ability to map the mutations in serpins that cause serpinopathies onto a structural framework aided understanding of the mechanism of normal serpin conformational changes, as well as serpin dysfunction.[47] In particular, many serpin mutations that cause disease localise to two distinct regions of the molecule (highlighted in figure 1a) termed the shutter and the breach. The shutter and the breach contain highly-conserved residues and underlie the path of RCL insertion.
Serpin misfolding results in two common outcomes, both of which stem from the instability of the native (S) conformation. Firstly, pathogenic mutations in serpins can promote inappropriate transition to the monmoeric latent state. This causes disease because it reduces the amount of active inhibitory serpin. For example, the disease-linked antithrombin variants wibble and wobble,[48] both promote formation of the latent state.
Secondly, and more insidiously, mutations in serpins may cause polymerisation. While the X-ray crystal structure of an intact serpin polymer remains to be determined, much biochemical, biophysical and structural data suggest that serpins "domain swap" with one another and form long-chain polymers.[10][49][50] This may occur by a RCL of one serpin inserting into the A-sheet of another serpin, to form a chain, rather than inserting into its "own" A-sheet (see figure 6a for a model). The polymeric form is inactive and causes pathology. Serpin polymerisation causes disease in two ways. Firstly, the lack of active serpin results in uncontrolled protease activity and tissue destruction, this is seen in the case of antitrypsin deficiency. Secondly, the polymers themselves clog up the endoplasmic reticulum of cells that synthesize serpins, eventually resulting in cell death and tissue damage. In the case of antitrypsin deficiency, antitrypsin polymers cause the death of liver cells, eventually resulting in liver damage and cirrhosis.
.
Finally, it is worth highlighting a structure of a disease-linked human antichymotrypsin variant that demonstrates the extraordinary flexibility of the serpin scaffold. The structure of antichymotrypsin (Leucine 55 to Proline) revealed a novel "delta" conformation that may represent an intermediate between the native and latent state (Figure 6b). In the delta conformation four residues of the RCL are inserted into the top of β-sheet A. The bottom half of the sheet is filled as a result of one of the α-helices (the F-helix) partially switching to a strand-like conformation, completing the β-sheet hydrogen bonding.[52] It is unclear whether other serpins can adopt this conformer, or whether this conformation has a functional role. However, this conformation may be important for thyroxine release by Thyroxine binding globulin.[18]
In humans, simple deficiency of many serpins (e.g. through a null mutation) may result in disease (see table 1).
Rarely, single amino acid changes in the RCL of a serpin alters the specificity of the inhibitor and allow it to target the wrong protease. For example, the Antitrypsin-Pittsburgh mutation (methionine 358 to arginine) allowed the serpin to inhibit thrombin, thus causing a bleeding disorder. [53]
Serpins are suicide inhibitors, the RCL acting as a "bait". Certain disease-linked mutations in the RCL of human serpins permit true substrate-like behaviour and cleavage without complex formation. Such variants are speculated to affect the rate or the extent of RCL insertion into the A-sheet. These mutations effectively result in serpin deficiency through a failure to properly control the target protease.[47][54]
Several non-inhibitory serpins play key roles in important human diseases. Most notably, maspin functions as a tumour suppressor in breast and prostate cancer. The mechanism of maspin function remains to be fully understood. Murine knockouts of maspin are lethal; these data suggest that maspin plays a key role in development.[55]
Evolution
Serpins were initially believed to be restricted to eukaryote organisms, but have since been found in a number of bacteria and archaea.[4][5][56] It remains unclear whether these prokaryote genes are the descendants of an ancestral prokaryotic serpin or whether they are the product of lateral gene transfer (genetic transfer between organisms not by evolutionary descent). Rawlings et al., showed that serpins are the most widely distributed and largest family of protease inhibitors.[6]
Types of serpins
Human serpins
The human genome encodes 36 serpins (see Law et al., (2006) for a recent review.[57]). Table 1 lists each human serpin, together with brief notes in regards to each molecules function and the consequence (where known) of dysfunction or deficiency.
Table 1
| Protein name | PDB | Common Name | Description | Disease | Chromosomal location |
| SERPINA1 | [17][18][19] | Alpha 1-antitrypsin | extracellular, inhibits human neutrophil elastase. | Deficiency results in emphysema, antitrypsin polymerisation results in cirrhosis. Serpinopathy.[10] The C-terminal fragment of cleaved SERPINA1 may inhibit HIV-1 infection.[58] | 14q32.1 |
| SERPINA2 | Antitrypsin-related protein | extracellular, possible pseudogene, unknown function[59] | 14q32.1 | ||
| SERPINA3 | [20][21], | Alpha 1-antichymotrypsin | extracellular, inhibits cathepsin G. | Deficiency results in emphysema. Serpinopathy[52] | 14q32.1 |
| SERPINA4 | Kallistatin | extracellular, inhibition of kallikrein, regulation of vascular function[60] | 14q32.1 | ||
| SERPINA5 | [22][61] | Protein C inhibitor | extracellular, inhibitior of active protein C | Male murine knockouts are infertile[62] | 14q32.1 |
| SERPINA6 | [23] [1] | Cortisol binding globulin | extracellular, non-inhibitory; cortisol binding | Deficiency may cause chronic fatigue[63] | 14q32.1 |
| SERPINA7 | [24][18] | Thyroxine-binding globulin | extracellular, non-inhibitory; thyroxine binding[18] | Deficiency causes hypothyroidism.[64] | Xq22.2 |
| SERPINA8 | Angiotensinogen | Extracellular; non-inhibitory, cleavage by renin results in release of angiotensin I | variants linked to hypertension[65] | 1q42-q43 | |
| SERPINA9 | Centerin | Extracellular; inhibitory, maintenance of naive B cells[66][67] | 14q32.1 | ||
| SERPINA10 | Protein Z-related protease inhibitor | extracellular, binds protein Z and inactivates factor Xa and factor XIa) | Deficiency may cause venous thromboembolic disease[68] | 14q32.1 | |
| SERPINA11 | - | probably extracellular, not characterised. | |||
| SERPINA12 | Vaspin | extracellular, insulin-sensitizing adipocytokine[69] | |||
| SERPINA13 | - | probably extracellular, not characterised | |||
| SERPINB1 | [25] | Monocyte neutrophil elastase inhibitor | Intracellular, inhibition of neutrophil elastase[70] | Murine knockout results in neutrophil survival defect and immune deficiency [71] | 6p25 |
| SERPINB2 | [26] | Plasminogen activator inhibitor-2 | Intracellular/extracellular, inhibition of extracellular uPA / unknown intracellular function | Murine knockouts viable / no obvious phenotype[72] | 18q21.3 |
| SERPINB3 | Squamous cell carcinoma antigen-1 (SCCA-1) | Intracellular, inhibitor of papain-like cysteine proteases[12] | 18q21.3 | ||
| SERPINB4 | Squamous cell carcinoma antigen-2 (SCCA-2) | Intracellular, inhibitor of cathepsin G and chymase[73] | 18q21.3 | ||
| SERPINB5 | [27] | Maspin | intracellular, non inhibitory, tumour suppressor in breast and prostate cancer | Murine knockouts lethal, important role in cancer metastasis[55] | 18q21.3 |
| SERPINB6 | PI-6 | intracellular, inhibition of cathepsin G | Murine knockout reveals mild neutropenia[74] | 6p25 | |
| SERPINB7 | Megsin | intracellular, involved in megakaryocyte maturation[75] | 18q21.3 | ||
| SERPINB8 | PI-8 | intracellular; possible furin inhibitor[76] | 18q21.3 | ||
| SERPINB9 | PI-9 | intracellular, inhibitor of the cytotoxic granule protease granzyme B | murine knockout reveals immune dysfunction[77] | 6p25 | |
| SERPINB10 | Bomapin | intracellular, unknown function[78] | A natural knockout of bomapin in mice has no obvious phenotype. | 18q21.3 | |
| SERPINB11 | intracellular, unknown function [79] | Murine Serpinb11 is an active inhibitor whereas the human orthalogue is inactive. | 18q21.3 | ||
| SERPINB12 | Yukopin | intracellular, unknown function[80] | 18q21.3 | ||
| SERPINB13 | Hurpin/Headpin | intracellular, inhibitor of papain-like cysteine proteases[81] | 18q21.3 | ||
| SERPINC1 | [28][29][30][31] [32] | Antithrombin | Extracellular, inhibitor of coagulation, specifically factor X, factor IX and thrombin | Deficiency results in thrombosis and other clotting disorders. Serpinopathy[82] | 1q23-q21 |
| SERPIND1 | [33][34][83] | Heparin cofactor II | extracellular, thrombin inhibitor[84] | Murine knockouts are lethal.[85] | 22q11 |
| SERPINE1 | [35][36] | Plasminogen activator inhibitor 1 | extracellular; inhibitor of thrombin, uPA and TPa | Cardiovascular disease, tumour progression[86] | 7q21.3-q22 |
| SERPINE2 | Protease nexin I | Extracellular, inhibition of uPA and tPA | Abnormal expression leads to human male infertility[87] | 2q33-q35 | |
| SERPINF1 | [37] | Pigment epithelium derived factor | Extracellular, non-inhibitory, potent anti-angiogenic molecule[88] | 17p13.3 | |
| SERPINF2 | Alpha 2-antiplasmin | extracellular, plasmin inhibitor, inhibitor of fibrinolysis. | Bleeding disorder[89] | 17pter-p12 | |
| SERPING1 | [38] | Complement 1-inhibitor | Extracellular, C1 esterase inhibitor.[90] | Angiodemia, serpinopathy[91] | 11q11-q13.1 |
| SERPINH1 | 47 kDa Heat shock protein (HSP47) | intracellular, non inhibitory, molecular chaperone in collagen folding. | Murine knockouts are lethal[92] | 11p15 | |
| SERPINI1 | [39] | Neuroserpin | Extracellular, inhibitor of tPA, uPA and plasmin | Mutated in dementia (FENIB). Serpinopathy[93] | 3q26 |
| SERPINI2 | Pancpin | Extracellular | possible role in inhibition of pancreatic cancer metastasis[94] | 3q26 |
Insect Serpins
Studies on Drosophila serpins reveal that Serpin-27A inhibits the Easter protease (the final protease in the Nudel, Gastrulation Defective, Snake and Easter proteolytic cascade) and thus controls dorsoventral patterning. Easter functions to cleave Spätzle (a chemokine-type ligand), which results in toll mediated signaling. In addition to its central role in embryonic patterning, toll signalling is also important for the innate immune response in insects. Accordingly, serpin-27A additionally functions to control the insect immune response.[95][96][97]
Worm Serpins
The genome of the nematode worm C. elegans contains nine serpins, however, only five of these molecules appear to function as protease inhibitors. [22] One of these serpins, SRP-6, has been shown to perform a protective function and guard against stress induced calpain-associated lysosomal disruption. Further SRP-6 functions to inhibit lysosomal cysteine proteases released after lysosomal rupture. Accordingly, worms lacking SRP-6 are sensitive to stress. Most notably, SRP-6 knockout worms die when placed in water (the hypo-osmotic stress lethal phenotype or Osl). Based on these data it is suggested that lysosomes play a general and controllable role in determining cell fate. [98]
Plant serpins
The presence of serpins in plants has long been recognised - indeed, barley Z serpin is the major protein component in beer. The genome sequence of Arabidopsis thaliana is predicted to encode 29 serpins. Plant serpins are able to inhibit serine proteases in vitro. However, the absence of close relatives of chymotrypsin-like proteases in plants suggests that these molecules may instead perform an alternative function. Indeed, Arabidopsis serpin1 inhibits metacaspase-like proteases in vivo and may control cell death pathways.[99]
Prokaryote serpins
Predicted serpin genes are sporadicly distributed in prokaryotes. In vitro studies on some of these moelcules have revealed that they are able to inhibit proteases and it is suggested that they function as inhibitors in vivo. Interestingly, several prokaryote serpins are found in extremeophiles. Accordingly, and in contrast to mammalian serpins, these molecule possess elevated resistance to heat denaturation.[100][101] The precise role of most bacterial serpins remains obscure, however, Clostridium thermocellum serpin localises to the cellulosome, a large extracellular mulitprotein complex that breaks down cellulose. It is suggested that the role of cellulosome-associated serpins may be to prevent unwanted protease activity against the cellulosome.[102]
Classification
In 2001, a serpin nomenclature was established.[7] The naming system is based upon a phylogenetic analysis of ~500 serpins.[4] This work classified the serpins into sixteen major clades, with several orphan sequences. The serpin family continues to grow - to date over 1000 serpins have been identified.
See also
References
- ↑ 1.0 1.1 1.2 1.3 Klieber MA, Underhill C, Hammond GL, Muller YA. (2007). "Corticosteroid-binding globulin: structural basis for steroid transport and proteinase-triggered release". J Biol Chem. PMID 17644521.
- ↑ R. Carrell and J. Travis. (1985). "α1-Antitrypsin and the serpins: Variation and countervariation". Trends Biochem. Sci. 10: 20-24. doi:10.1016/0968-0004(85)90011-8.
- ↑ Hunt LT, Dayhoff MO (1980). "A surprising new protein superfamily containing ovalbumin, antithrombin-III, and α1-proteinase inhibitor". Biochem Biophys Res Commun. 95 (2): 864-71. PMID 6968211.
- ↑ 4.0 4.1 4.2 Irving JA, Pike RN, Lesk AM, Whisstock. (2000). "Phylogeny of the Serpin Superfamily: Implications of Patterns of Amino Acid Conservation for Structure and Function". Genome Res. 10: 1845–64. PMID 11116082.
- ↑ 5.0 5.1 Irving J, Steenbakkers P, Lesk A, Op den Camp H, Pike R, Whisstock J (2002). "Serpins in prokaryotes". Mol Biol Evol. 19 (11): 1881–90. PMID 12411597.
- ↑ 6.0 6.1 Rawlings ND, Tolle DP, Barrett AJ. (2004). "Evolutionary families of peptidase inhibitors". Biochem J. 378: 705-16. PMID 14705960.
- ↑ 7.0 7.1 Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, Irving JA, Lomas DA, Luke CJ, Moyer RW, Pemberton PA, Remold-O'Donnell E, Salvesen GS, Travis J, Whisstock JC. (2001). "'The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature". J Biol Chem. 276: 33293-6. PMID 11435447.
- ↑ 8.0 8.1 8.2 Huntington J, Read R, Carrell R (2000). "Structure of a serpin-protease complex shows inhibition by deformation". Nature. 407 (6806): 923–6. PMID 11057674.
- ↑ 9.0 9.1 9.2 Carrell RW, Lomas DA. (1997). "Conformational disease". Lancet. 350 (9071): 134-8. PMID 9228977.
- ↑ 10.0 10.1 10.2 10.3 Lomas DA, Evans DL, Finch JT & Carrell RW (1992). "The mechanism of Z alpha 1-antitrypsin accumulation in the liver". Nature. 357: 605–607. PMID 1608473.
- ↑ Barrett AJ, Rawlings ND. (1995). "Families and clans of serine peptidases". Arch Biochem Biophys. 318 (2): 247-50. PMID 7733651.
- ↑ 12.0 12.1 Schick C, Brömme D, Bartuski A, Uemura Y, Schechter N, Silverman G (1998). "The reactive site loop of the serpin SCCA1 is essential for cysteine proteinase inhibition". Proc Natl Acad Sci U S A. 95 (23): 13465–70. PMID 9811823.
- ↑ 13.0 13.1 McGowan S, Buckle A, Irving J, Ong P, Bashtannyk-Puhalovich T, Kan W, Henderson K, Bulynko Y, Popova E, Smith A, Bottomley S, Rossjohn J, Grigoryev S, Pike R, Whisstock J (2006). "X-ray crystal structure of MENT: evidence for functional loop-sheet polymers in chromatin condensation". EMBO J. 25 (13): 3144–55. PMID 16810322.
- ↑ Ong PC, McGowan S, Pearce MC, Irving JA, Kan WT, Grigoryev SA, Turk B, Silverman GA, Brix K, Bottomley SP, Whisstock JC, Pike RN (2007). "DNA accelerates the inhibition of human cathepsin V by serpins". doi:10.1074/jbc.M706991200. PMID 17923478.
- ↑ Ray C, Black R, Kronheim S, Greenstreet T, Sleath P, Salvesen G, Pickup D (1992). "Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1 beta converting enzyme". Cell. 69 (4): 597–604. PMID 1339309.
- ↑ Barrett AJ, Rawlings ND. (2001). "Evolutionary lines of cysteine peptidases". Biol Chem. 382 (5): 727-33. PMID 11517925.
- ↑ Irving JA, Pike RN, Dai W, Bromme D, Worrall DM, Silverman GA, Coetzer TH, Dennison C, Bottomley SP, Whisstock JC. (2002). "Evidence that serpin architecture intrinsically supports papain-like cysteine protease inhibition: engineering alpha(1)-antitrypsin to inhibit cathepsin proteases". Biochemistry. 41 (15): 4998-5004. PMID 11939796.
- ↑ 18.0 18.1 18.2 18.3 18.4 Zhou A, Wei Z, Read RJ, Carrell RW. (2006). "Structural mechanism for the carriage and release of thyroxine in the blood". Proc Natl Acad Sci U S A.: 13321-6. PMID 16938877. Unknown parameter
|Volume=ignored (|volume=suggested) (help) - ↑ Campbell DJ. (2003). "The renin-angiotensin and the kallikrein-kinin systems". Int J Biochem Cell Biol. 35 (6): 784-91. PMID 12676165.
- ↑ Remold-O'Donnell E, Chin J, Alberts M. (1992). "Sequence and molecular characterization of human monocyte/neutrophil elastase inhibitor". Proc Natl Acad Sci U S A. 89 (12): 5635-9. PMID 1376927.
- ↑ Coughlin P, Sun J, Cerruti L, Salem HH, Bird P., (1993). "Cloning and molecular characterization of a human intracellular serine proteinase inhibitor". Proc Natl Acad Sci U S A. 90 (20): 9417-21. PMID 8415716.
- ↑ 22.0 22.1 Pak SC, Kumar V, Tsu C, Luke CJ, Askew YS, Askew DJ, Mills DR, Bromme D, Silverman GA. (2004). "SRP-2 is a cross-class inhibitor that participates in postembryonic development of the nematode Caenorhabditis elegans: initial characterization of the clade L serpins". J Biol Chem. 279 (15): 15448-59. PMID 14739286.
- ↑ Bird PI. (1999). "Regulation of pro-apoptotic leucocyte granule serine proteinases by intracellular serpins". Immunol Cell Biol. 77 (1): 47-57. PMID 10101686.
- ↑ Bird CH, Sutton VR, Sun J, Hirst CE, Novak A, Kumar S, Trapani JA, Bird PI (1998). "Selective regulation of apoptosis: the cytotoxic lymphocyte serpin proteinase inhibitor 9 protects against granzyme B-mediated apoptosis without perturbing the Fas cell death pathway". Mol Cell Biol. 18 (11): 6387-98. PMID 774654. Check
|pmid=value (help). - ↑ Zou Z, Anisowicz A, Hendrix MJ, Thor A, Neveu M, Sheng S, Rafidi K, Seftor E, Sager R. (1994). "Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells". Science. 263 (5146): 526-9. PMID 8290962.
- ↑ Luo JL, Tan W, Ricono JM, Korchynskyi O, Zhang M, Gonias SL, Cheresh DA, Karin M. (2007). "Nuclear cytokine-activated IKKalpha controls prostate cancer metastasis by repressing Maspin". Nature. 446 (7136): 690-4. PMID 17377533. Unknown parameter
|yesr=ignored (help) - ↑ Grigoryev SA, Bednar J, Woodcock CL. (1999). "MENT, a heterochromatin protein that mediates higher order chromatin folding, is a new serpin family member". J Biol Chem. 274 (9): 5626-36. PMID 10026180.
- ↑ Tasab M, Batten MR, Bulleid NJ (2000). "Hsp47: a molecular chaperone that interacts with and stabilizes correctly-folded procollagen". EMBO J. 19 (10): 2204-11. PMID 10811611.
- ↑ Elliott PR, Lomas DA, Carrell RW, Abrahams JP. (1996). "Inhibitory conformation of the reactive loop of alpha 1-antitrypsin". Nat Struct Biol. 3 (8): 676-81. PMID 8756325.
- ↑ Horvath A, Irving J, Rossjohn J, Law R, Bottomley S, Quinsey N, Pike R, Coughlin P, Whisstock J (2005). "The murine orthologue of human antichymotrypsin: a structural paradigm for clade A3 serpins". J Biol Chem. 280 (52): 43168–78. PMID 16141197.
- ↑ 31.0 31.1 Loebermann H, Tokuoka R, Deisenhofer J, Huber R. (1984). "Human alpha 1-proteinase inhibitor. Crystal structure analysis of two crystal modifications, molecular model and preliminary analysis of the implications for function". J Mol Biol. 177 (3): 531-57. PMID 6332197.
- ↑ Stein PE, Leslie AG, Finch JT, Turnell WG, McLaughlin PJ, Carrell RW. (1990). "Crystal structure of ovalbumin as a model for the reactive centre of serpins". Nature. 347 (6288): 99-102. PMID 2395463.
- ↑ 33.0 33.1 Whisstock J, Bottomley S (2006). "Molecular gymnastics: serpin structure, folding and misfolding". Curr Opin Struct Biol. 16 (6): 761–8. PMID 17079131.
- ↑ Egelund R, Rodenburg K, Andreasen P, Rasmussen M, Guldberg R, Petersen T (1998). "An ester bond linking a fragment of a serine proteinase to its serpin inhibitor". Biochemistry. 37 (18): 6375-9. PMID 9572853.
- ↑ Gettins P (2002). "Serpin structure, mechanism, and function". Chem Rev. 102 (12): 4751–804. PMID 12475206.
- ↑ Whisstock JC, Skinner R, Carrell RW, Lesk AM (2000). "Conformational changes in serpins: I. The native and cleaved conformations of alpha(1)-antitrypsin". J Mol Biol. 296: 685-99. PMID 10669617.
- ↑ Ye S, Cech A, Belmares R, Bergstrom R, Tong Y, Corey D, Kanost M, Goldsmith E (2001). "The structure of a Michaelis serpin-protease complex". Nat Struct Biol. 8 (11): 979–83. PMID 11685246.
- ↑ Jin L, Abrahams JP, Skinner R, Petitou M, Pike RN, Carrell RW. (1997). "The anticoagulant activation of antithrombin by heparin". Proc Natl Acad Sci U S A. 94: 14683-8. PMID 9405673.
- ↑ Whisstock JC, Pike RN, Jin L, Skinner R, Pei XY, Carrell RW, Lesk AM. (2000). "Conformational changes in serpins: II. The mechanism of activation of antithrombin by heparin". J Mol Biol. 301: 1287-305. PMID 10966821.
- ↑ Li W, Johnson DJ, Esmon CT, Huntington JA (2004). "Structure of the antithrombin-thrombin-heparin ternary complex reveals the antithrombotic mechanism of heparin". Nat. Struct. Mol. Biol. 11 (9): 857–62. doi:10.1038/nsmb811. PMID 15311269.
- ↑ Johnson DJ, Li W, Adams TE, Huntington JA (2006). "Antithrombin-S195A factor Xa-heparin structure reveals the allosteric mechanism of antithrombin activation". EMBO J. 25 (9): 2029–37. doi:10.1038/sj.emboj.7601089. PMID 16619025.
- ↑ Petitou M, van Boeckel CA (2004). "A synthetic antithrombin III binding pentasaccharide is now a drug! What comes next?". Angew. Chem. Int. Ed. Engl. 43 (24): 3118–33. doi:10.1002/anie.200300640. PMID 15199558.
- ↑ Lindahl T, Sigurdardottir O, Wiman B (1989). "Stability of plasminogen activator inhibitor 1 (PAI-1)". Thromb Haemost. 62 (2): 748–51. PMID 2479113.
- ↑ Zhang Q, Buckle AM, Law RH, Pearce MC, Cabrita LD, Lloyd GJ, Irving JA, Smith AI, Ruzyla K, Rossjohn J, Bottomley SP, Whisstock JC. (2007). "The N terminus of the serpin, tengpin, functions to trap the metastable native state". EMBO Rep. PMID 17557112.
- ↑ 45.0 45.1 Cao C, Lawrence DA, Li Y, Von Arnim CA, Herz J, Su EJ, Makarova A, Hyman BT, Strickland DK, Zhang L. (2006). "Endocytic receptor LRP together with tPA and PAI-1 coordinates Mac-1-dependent macrophage migration". EMBO J. 25 (9): 1860-70. PMID 16601674.
- ↑ Pemberton PA, Stein PE, Pepys MB, Potter JM, Carrell RW (1988). "Hormone binding globulins undergo serpin conformational change in inflammation". Nature. 336 (6196): 257-8. PMID 3143075.
- ↑ 47.0 47.1 Stein PE, Carrell RW. (1995). "What do dysfunctional serpins tell us about molecular mobility and disease?". Nat Struct Biol. 2: 96-113. PMID 7749926.
- ↑ Beauchamp NJ, Pike RN, Daly M, Butler L, Makris M, Dafforn TR, Zhou A, Fitton HL, Preston FE, Peake IR, Carrell RW (1998). "Antithrombins Wibble and Wobble (T85M/K): archetypal conformational diseases with in vivo latent-transition, thrombosis, and heparin activation". Blood. 92 (8): 2696–706. PMID 9763552.
- ↑ Huntington JA, Pannu NS, Hazes B, Read RJ, Lomas DA & Carrell RW (1999). "A 2.6 Å structure of a serpin polymer and implications for conformational disease". J Mol Biol. 293: 449–455. PMID 10543942.
- ↑ Dunstone MA, Dai W, Whisstock JC, Rossjohn J, Pike RN, Feil SC, Le Bonniec BF, Parker MW & Bottomley SP (2000). "Cleaved antitrypsin polymers at atomic resolution". Protein Sci. 9: 417–420. PMID 10716194.
- ↑ Chang WS, Whisstock J, Hopkins PC, Lesk AM, Carrell RW, Wardell MR. (1997). "Importance of the release of strand 1C to the polymerization mechanism of inhibitory serpins". Protein Sci. 6 (1): 89-98. PMID 9007980.
- ↑ 52.0 52.1 52.2 Gooptu B, Hazes B, Chang WS, Dafforn TR, Carrell RW, Read RJ, Lomas DA. (2000). "Inactive conformation of the serpin alpha(1)-antichymotrypsin indicates two-stage insertion of the reactive loop: implications for inhibitory function and conformational disease". Proc Natl Acad Sci U S A. 97 (1): 67-72. PMID 10618372.
- ↑ Owen MC, Brennan SO, Lewis JH, Carrell RW. (1983). "Mutation of antitrypsin to antithrombin. alpha 1-antitrypsin Pittsburgh (358 Met leads to Arg), a fatal bleeding disorder". N Engl J Med. 309 (12): 694-8. PMID 6604220.
- ↑ "Effects of mutations in the hinge region of serpins". Biochemistry. 32 (30): 7650-7. 1993. PMID 8347575. Text "Hopkins PC, Carrell RW, Stone SR. " ignored (help)
- ↑ 55.0 55.1 Gao F, Shi H, Daughty C, Cella N, Zhang M (2004). "Maspin plays an essential role in early embryonic development". Development. 131 (7): 1479–89. PMID 14985257.
- ↑ Cabrita LD, Irving JA, Pearce MC, Whisstock JC, Bottomley SP. (2007). "Aeropin from the extremophile Pyrobaculum aerophilum bypasses the serpin misfolding trap". J Biol Chem. PMID 17635906.
- ↑ Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, Rosado CJ, Langendorf CG, Pike RN, Bird PI, Whisstock JC (2006). "An overview of the serpin superfamily". Genome Biol. 7 (5, ): 216. PMID 16737556.
- ↑ Munch J, Standker L, Adermann K, Schulz A, Schindler M, Chinnadurai R, Pohlmann S, Chaipan C, Biet T, Peters T, Meyer B, Wilhelm D, Lu H, Jing W, Jiang S, Forssmann WG, Kirchhoff F. (2007). "Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide". Cell. 129 (2): 263-75. PMID 17448989.
- ↑ Seixas S, Suriano G, Carvalho F, Seruca R, Rocha J, Di Rienzo A. (2007). "Sequence Diversity at the Proximal 14q32.1 SERPIN Subcluster: Evidence for Natural Selection Favoring the Pseudogenization of SERPINA2". Mol Biol Evol. 24 (2): 587-98. PMID 17135331.
- ↑ Miao RQ, Agata J, Chao L, Chao J. (2002). "Kallistatin is a new inhibitor of angiogenesis and tumor growth". Blood. 100 (9): 3245-52. PMID 12384424.
- ↑ Li W, Adams TE, Kjellberg M, Stenflo J, Huntington JA. (2007). "Structure of native protein C inhibitor provides insight into its multiple functions". 282 (18): 13759-68. PMID 17337440.
- ↑ Uhrin P, Dewerchin M, Hilpert M, Chrenek P, Schofer C, Zechmeister-Machhart M, Kronke G, Vales A, Carmeliet P, Binder BR, Geiger M. (2000). "Disruption of the protein C inhibitor gene results in impaired spermatogenesis and male infertility". J Clin Invest. (12): 1531-9. PMID 11120760. Unknown parameter
|Volume=ignored (|volume=suggested) (help) - ↑ Torpy DJ, Bachmann AW, Gartside M, Grice JE, Harris JM, Clifton P, Easteal S, Jackson RV, Whitworth JA. (2004). "Association between chronic fatigue syndrome and the corticosteroid-binding globulin gene ALA SER224 polymorphism". Endocr Res. 30 (3): 417-29. PMID 15554358.
- ↑ Bartalena L, Robbins J. (1992). "Variations in thyroid hormone transport proteins and their clinical implications". Thyroid. 2 (3): 237-45. PMID 1422238.
- ↑ Jeunemaitre X, Gimenez-Roqueplo AP, Celerier J, Corvol P. (1999). "Angiotensinogen variants and human hypertension". Curr Hypertens Rep. 1 (1): 31-41. PMID 10981040.
- ↑ Frazer JK, Jackson DG, Gaillard JP, Lutter M, Liu YJ, Banchereau J, Capra JD, Pascual V. (2000). "Identification of centerin: a novel human germinal center B cell-restricted serpin". Eur J Immunol. 30 (10): 3039-48. PMID 11069088.
- ↑ Paterson MA, Horvath AJ, Pike RN, Coughlin PB. (2007). "Molecular characterization of centerin, a germinal centre cell serpin". Biochem J. 405 (3): 489-94. PMID 17447896.
- ↑ Corral J, Gonzalez-Conejero R, Soria JM, Gonzalez-Porras JR, Perez-Ceballos E, Lecumberri R, Roldan V, Souto JC, Minano A, Hernandez-Espinosa D, Alberca I, Fontcuberta J, Vicente V. (2006). "A nonsense polymorphism in the protein Z-dependent protease inhibitor increases the risk for venous thrombosis". Blood. 108 (1): 177-83. PMID 16527896.
- ↑ Hida K, Wada J, Eguchi J, Zhang H, Baba M, Seida A, Hashimoto I, Okada T, Yasuhara A, Nakatsuka A, Shikata K, Hourai S, Futami J, Watanabe E, Matsuki Y, Hiramatsu R, Akagi S, Makino H, Kanwar YS (2005 J). "Visceral adipose tissue-derived serine protease inhibitor: a unique insulin-sensitizing adipocytokine in obesity". Proc Natl Acad Sci U S A. 102 (30): 10610-5. PMID 16030142. Check date values in:
|year=(help) - ↑ Remold-O'Donnell E, Chin J, Alberts M. (1992). "Sequence and molecular characterization of human monocyte/neutrophil elastase inhibitor". Proc Natl Acad Sci U S A. 89 (12): 5635-9. PMID 1376927.
- ↑ Benarafa C, Priebe GP, Remold-O'donnell E. (2007). "The neutrophil serine protease inhibitor serpinb1 preserves lung defense functions in Pseudomonas aeruginosa infection". J Exp Med. PMID 17664292.
- ↑ Dougherty KM, Pearson JM, Yang AY, Westrick RJ, Baker MS, Ginsburg D. (1999). "The plasminogen activator inhibitor-2 gene is not required for normal murine development or survival". Proc Natl Acad Sci U S A. (2): 686-91. PMID 9892694. Unknown parameter
|volme=ignored (help) - ↑ Schick C, Kamachi Y, Bartuski AJ, Cataltepe S, Schechter NM, Pemberton PA, Silverman GA. (1997). "Squamous cell carcinoma antigen 2 is a novel serpin that inhibits the chymotrypsin-like proteinases cathepsin G and mast cell chymase". J Biol Chem. 272 (3): 1849-55. PMID 8999871.
- ↑ Scarff KL, Ung KS, Nandurkar H, Crack PJ, Bird CH, Bird PI. (2004). "Targeted disruption of SPI3/Serpinb6 does not result in developmental or growth defects, leukocyte dysfunction, or susceptibility to stroke". Mol Cell Biol. 24 (9): 4075-82. PMID 1508279.
- ↑ Miyata T, Inagi R, Nangaku M, Imasawa T, Sato M, Izuhara Y, Suzuki D, Yoshino A, Onogi H, Kimura M, Sugiyama S, Kurokawa K. (2002). "Overexpression of the serpin megsin induces progressive mesangial cell proliferation and expansion". J Clin Invest. 109 (5): 585-93. PMID 11877466.
- ↑ Dahlen JR, Jean F, Thomas G, Foster DC, Kisiel W (1998). "Inhibition of soluble recombinant furin by human proteinase inhibitor 8". J Biol Chem. 273 (4): 1851-4. PMID 9442015.
- ↑ Zhang M, Park SM, Wang Y, Shah R, Liu N, Murmann AE, Wang CR, Peter ME, Ashton-Rickardt PG. (2006). "Serine protease inhibitor 6 protects cytotoxic T cells from self-inflicted injury by ensuring the integrity of cytotoxic granules". Immunity. 24 (4): 451-61. PMID 16618603.
- ↑ Riewald M, Chuang T, Neubauer A, Riess H, Schleef RR. (1998). "Expression of bomapin, a novel human serpin, in normal/malignant hematopoiesis and in the monocytic cell lines THP-1 and AML-193". Blood. 9 (4): 1256-62. PMID 9454755.
- ↑ Askew DJ, Cataltepe D, Kumar V, Edwards C, Pace SM, Howarth RN, Pak SC, Askew Y, Bromme D, Luke CJ, Whisstock JC, Silverman GA. (2007). "Serpinb11 is a new non-inhibitory intracellular serpin: Common single nucleotide polymorphisms in the scaffold impair conformational change". J. Biol. Chem. PMID 17562709.
- ↑ Askew YS, Pak SC, Luke CJ, Askew DJ, Cataltepe S, Mills DR, Kato H, Lehoczky J, Dewar K, Birren B, Silverman GA. (2001). "SERPINB12 is a novel member of the human ov-serpin family that is widely expressed and inhibits trypsin-like serine proteinases". J Biol Chem. 276 (52): 49320-30. PMID 11604408.
- ↑ Welss T, Sun J, Irving JA, Blum R, Smith AI, Whisstock JC, Pike RN, von Mikecz A, Ruzicka T, Bird PI, Abts HF. (2003). "Hurpin is a selective inhibitor of lysosomal cathepsin L and protects keratinocytes from ultraviolet-induced apoptosis". Biochemistry. 42 (24): 7381-9. PMID 12809493.
- ↑ Bruce D, Perry DJ, Borg JY, Carrell RW, Wardell MR. (1994). "Thromboembolic disease due to thermolabile conformational changes of antithrombin Rouen-VI (187 Asn-->Asp)". J Clin Invest. 94 (6): 2265-74. PMID 7989582.
- ↑ Baglin TP, Carrell RW, Church FC, Esmon CT, Huntington JA (2002). "Crystal structures of native and thrombin-complexed heparin cofactor II reveal a multistep allosteric mechanism". Proc. Natl. Acad. Sci. U.S.A. 99 (17): 11079–84. doi:10.1073/pnas.162232399. PMID 12169660.
- ↑ Vicente CP, He L, Pavao MS, Tollefsen DM (2004). "Antithrombotic activity of dermatan sulfate in heparin cofactor II-deficient mice". Blood. 104 (13): 3965-70. PMID 15315969.
- ↑ Aihara K, Azuma H, Akaike M, Ikeda Y, Sata M, Takamori N, Yagi S, Iwase T, Sumitomo Y, Kawano H, Yamada T, Fukuda T, Matsumoto T, Sekine K, Sato T, Nakamichi Y, Yamamoto Y, Yoshimura K, Watanabe T, Nakamura T, Oomizu A, Tsukada M, Hayashi H, Sudo T, Kato S, Matsumoto T. (2007). "Strain-dependent embryonic lethality and exaggerated vascular remodeling in heparin cofactor II-deficient mice". J Clin Invest. 117 (6): 1514-26. PMID 175492.
- ↑ Gils A, Declerck PJ. (2004). "The structural basis for the pathophysiological relevance of PAI-I in cardiovascular diseases and the development of potential PAI-I inhibitors". Thromb Haemost. 91 (3): 425-37. PMID 14983217.
- ↑ Murer V, Spetz JF, Hengst U, Altrogge LM, de Agostini A, Monard D. (2001). "Male fertility defects in mice lacking the serine protease inhibitor protease nexin-1". 98 (6): 3029-33. PMID 11248026. Unknown parameter
|journa=ignored (help) - ↑ Doll JA, Stellmach VM, Bouck NP, Bergh AR, Lee C, Abramson LP, Cornwell ML, Pins MR, Borensztajn J, Crawford SE. (2003). "Pigment epithelium-derived factor regulates the vasculature and mass of the prostate and pancreas". Nat Med. 9 (6): 774-80. PMID 12740569.
- ↑ Miles LA, Plow EF, Donnelly KJ, Hougie C, Griffin JH. (1982). "A bleeding disorder due to deficiency of alpha 2-antiplasmin". Blood. 59 (6): 1246-51. PMID 7082827.
- ↑ Beinrohr L, Harmat V, Dobo J, Lorincz Z, Gal P, Zavodszky P. (2007). "C1-inhibitor serpin domain structure reveals the likely mechanism of heparin potentiation and conformational disease". J Biol Chem. PMID 17488724.
- ↑ Aulak KS, Eldering E, Hack CE, Lubbers YP, Harrison RA, Mast A, Cicardi M, Davis AE 3rd. (1993). "A hinge region mutation in C1-inhibitor (Ala436-->Thr) results in nonsubstrate-like behavior and in polymerization of the molecule". J Biol Chem. 268 (24): 18088-94. PMID 8349686.
- ↑ Nagai N, Hosokawa M, Itohara S, Adachi E, Matsushita T, Hosokawa N, Nagata K. "Embryonic lethality of molecular chaperone hsp47 knockout mice is associated with defects in collagen biosynthesis". J Cell Biol. year=2000. 150 (6): 1499-506. PMID 10995453.
- ↑ Davis RL, Shrimpton AE, Holohan PD, Bradshaw C, Feiglin D, Collins GH, Sonderegger P, Kinter J, Becker LM, Lacbawan F, Krasnewich D, Muenke M, Lawrence DA, Yerby MS, Shaw CM, Gooptu B, Elliott PR, Finch JT, Carrell RW, Lomas DA. (1999). "Familial dementia caused by polymerization of mutant neuroserpin". Nature. 401 (6751): 376-9. PMID 10517635.
- ↑ Ozaki K, Nagata M, Suzuki M, Fujiwara T, Miyoshi Y, Ishikawa O, Ohigashi H, Imaoka S, Takahashi E, Nakamura Y. (1998). "Isolation and characterization of a novel human pancreas-specific gene, pancpin, that is down-regulated in pancreatic cancer cells". 22 (3): 179-85. PMID 9624529. Unknown parameter
|jounr=ignored (help) - ↑ Rushlow C (2004). "Dorsoventral patterning: a serpin pinned down at last". Curr. Biol. 14 (1): R16–8. doi:10.1016/j.cub.2003.12.015. PMID 14711428.
- ↑ Ligoxygakis P, Roth S, Reichhart JM (2003). "A serpin regulates dorsal-ventral axis formation in the Drosophila embryo". Curr. Biol. 13 (23): 2097–102. PMID 14654000.
- ↑ Hashimoto C, Kim DR, Weiss LA, Miller JW, Morisato D (2003). "Spatial regulation of developmental signaling by a serpin". Dev. Cell. 5 (6): 945–50. PMID 14667416.
- ↑ Cliff J. Luke, Stephen C. Pak, Yuko S. Askew, Terra L. Naviglia, David J. Askew, Shila M. Nobar, Anne C. Vetica, Olivia S. Long, Simon C. Watkins, Donna B. Stolz, Robert J. Barstead, Gary L. Moulder, Dieter Brömme, and Gary A. Silverman (2007). "An Intracellular Serpin Regulates Necrosis by Inhibiting the Induction and Sequelae of Lysosomal Injury". Cell. 130: 1108-1119. doi:10.1016/j.cell.2007.07.013.
- ↑ Vercammen D, Belenghi B, van de Cotte B, Beunens T, Gavigan JA, De Rycke R, Brackenier A, Inze D, Harris JL, Van Breusegem F. (2006). "Serpin1 of Arabidopsis thaliana is a suicide inhibitor for metacaspase 9". J Mol Biol. 364 (4): 625-36. PMID 17028019.
- ↑ Irving JA, Cabrita LD, Rossjohn J, Pike RN, Bottomley SP, Whisstock JC (2003). "The 1.5 A crystal structure of a prokaryote serpin: controlling conformational change in a heated environment". Structure. 11 (4): 387-97. PMID 12679017.
- ↑ Fulton KF, Buckle AM, Cabrita LD, Irving JA, Butcher RE, Smith I, Reeve S, Lesk AM, Bottomley SP, Rossjohn J, Whisstock JC. (2005). "The high resolution crystal structure of a native thermostable serpin reveals the complex mechanism underpinning the stressed to relaxed transition". J Biol Chem. 280 (9): 8435-42. PMID 15590653.
- ↑ Kang S, Barak Y, Lamed R, Bayer EA, Morrison M. (2006). "The functional repertoire of prokaryote cellulosomes includes the serpin superfamily of serine proteinase inhibitors". Mol Microbiol. 60 (6): 1344-54. PMID 16796673.
External links
- PDB Molecule of the Month Serpin
- Serpins at the US National Library of Medicine Medical Subject Headings (MeSH)
- James Whisstock laboratory at Monash University
- Jim Huntington serpin laboratory at University of Cambridge
- Frank Church serpin group at University of North Carolina at Chapel Hill
- Paul Declerck serpin group at Katholieke Universiteit Leuven
- Merops general protease / inhibitor classification page at University of Cambridge
- The alpha one foundation provides information and support for sufferers of antitrypsin deficiency