Neuraminidase: Difference between revisions
m (Robot: Automated text replacement (-{{WikiDoc Cardiology Network Infobox}} +, -<references /> +{{reflist|2}}, -{{reflist}} +{{reflist|2}})) |
imported>Nbauman (describe what's going on in that illustration) |
||
| (One intermediate revision by one other user not shown) | |||
| Line 1: | Line 1: | ||
{{ | [[File:Neuraminidase Ribbon Diagram.jpg|thumb|235 px|Neuraminidase ribbon diagram. The neuraminic acid, which it cleaves, is the small white and red molecule in the center.]] | ||
| | [[File:Neu5Ac.png|thumb|235 px|[[N-Acetylneuraminic acid.]]]] | ||
| | '''Neuraminidase''' enzymes are [[glycoside hydrolase]] [[enzyme]]s that cleave (cut) the [[glycosidic linkage]]s of [[neuraminic acid]]s. Neuraminidase enzymes are a large family, found in a range of organisms. The best-known neuraminidase is the [[viral neuraminidase]], a drug target for the prevention of the spread of [[influenza]] infection. The viral neuraminidases are frequently used as [[antigenic determinants]] found on the surface of the influenza virus. Some variants of the influenza neuraminidase confer more virulence to the virus than others. Other homologues are found in mammalian cells, which have a range of functions. At least four mammalian [[sialidase]] homologues have been described in the [[human genome]] (see [[NEU1]], [[NEU2]], [[NEU3]], [[NEU4]]). | ||
| | |||
| | == Reaction == | ||
| | There are two major classes of Neuraminidase that cleave exo or endo poly-sialic acids: | ||
| | * Exo hydrolysis of α-(2→3)-, α-(2→6)-, α-(2→8)-glycosidic linkages of terminal sialic acid residues<ref name="pmid6762816">{{cite journal | vauthors = Schauer R | title = Chemistry, metabolism, and biological functions of sialic acids | journal = Advances in Carbohydrate Chemistry and Biochemistry | volume = 40 | issue = | pages = 131–234 | year = 1982 | pmid = 6762816 | doi = 10.1016/S0065-2318(08)60109-2 | isbn = 978-0-12-007240-8 | series = Advances in Carbohydrate Chemistry and Biochemistry }}</ref><ref name="pmid1883340">{{cite journal | vauthors = Cabezas JA | title = Some questions and suggestions on the type references of the official nomenclature (IUB) for sialidase(s) and endosialidase | journal = The Biochemical Journal | volume = 278 ( Pt 1) | issue = Pt 1 | pages = 311–2 | date = August 1991 | pmid = 1883340 | pmc = 1151486 | doi = 10.1042/bj2780311 }}</ref> | ||
| | * Endo hydrolysis of (2→8)-α-sialosyl linkages in oligo- or poly(sialic) acids<ref name="pmid1883340"/> | ||
| | {| | ||
|{{enzyme | |||
| | | Name = exo-α-sialidase | ||
| | | EC_number = 3.2.1.18 | ||
| | | CAS_number = 9001-67-6 | ||
| | | IUBMB_EC_number = 3/2/1/18 | ||
| | | GO_code = 0004308 | ||
| | | image = | ||
| | | width = | ||
| | | caption = | ||
}} | }} | ||
|{{enzyme | |||
| Name = endo-α-sialidase | |||
| EC_number = 3.2.1.129 | |||
| CAS_number = 91195-87-8 | |||
| IUBMB_EC_number = 3/2/1/129 | |||
| GO_code = 0016996 | |||
| image = | |||
| width = | |||
| caption = | |||
}} | |||
|} | |||
== Function == | |||
Neuraminidases, also called sialidases, catalyze the hydrolysis of terminal [[sialic acid]] residues from the newly formed [[Virion#Structure|virions]] and from the host cell receptors.<ref name="pmid18049471">{{cite journal | vauthors = von Itzstein M | title = The war against influenza: discovery and development of sialidase inhibitors | journal = Nature Reviews. Drug Discovery | volume = 6 | issue = 12 | pages = 967–74 | date = December 2007 | pmid = 18049471 | doi = 10.1038/nrd2400 }}</ref> Sialidase activities include assistance in the mobility of virus particles through the respiratory tract mucus and in the elution of virion progeny from the infected cell.<ref name="pmid4472498">{{cite journal | vauthors = Palese P, Tobita K, Ueda M, Compans RW | title = Characterization of temperature sensitive influenza virus mutants defective in neuraminidase | journal = Virology | volume = 61 | issue = 2 | pages = 397–410 | date = October 1974 | pmid = 4472498 | doi = 10.1016/0042-6822(74)90276-1 }}</ref><ref name="pmid7815489">{{cite journal | vauthors = Liu C, Eichelberger MC, Compans RW, Air GM | title = Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding | journal = Journal of Virology | volume = 69 | issue = 2 | pages = 1099–106 | date = February 1995 | pmid = 7815489 | pmc = 188682 | doi = | url = http://jvi.asm.org/cgi/content/abstract/69/2/1099 }}</ref> | |||
== Subtypes == | |||
[[Swiss-Prot]] lists 137 types of neuraminidase from various species as of October 18, 2006.<ref>[http://www.expasy.org/cgi-bin/sprot-search-de?neuraminidase Search in UniProt Knowledgebase (Swiss-Prot and TrEMBL) for: neuraminidase<!-- Bot generated title -->]</ref> Nine subtypes of influenza neuraminidase are known; many occur only in various species of duck and chicken. Subtypes N1 and N2 have been positively linked to epidemics in man, and strains with N3 or N7 subtypes have been identified in a number of isolated deaths.{{Citation needed|date=July 2007}} | |||
The following is a list of major classes of neuraminidase enzymes: | |||
* [[Viral neuraminidase]] | |||
* [[Bacterial neuraminidase]] | |||
* Mammalian neuraminidases: | |||
{| | |||
|-valign=top | |||
|{{infobox protein | Name = [[NEU1|sialidase 1 (lysosomal sialidase)]] | caption = | image = | width = | HGNCid = 7758 | Symbol = [[NEU1]] | AltSymbols = | EntrezGene = 4758 | OMIM = 608272 | RefSeq = NM_000434 | UniProt = Q99519 | PDB = | ECnumber = | Chromosome = 6 | Arm = p | Band = 21 | LocusSupplementaryData = }} | |||
|{{infobox protein | Name = [[NEU2|sialidase 2 (cytosolic sialidase)]] | caption = | image = | width = | HGNCid = 7759 | Symbol = [[NEU2]] | AltSymbols = | EntrezGene = 4759 | OMIM = 605528 | RefSeq = NM_005383 | UniProt = Q9Y3R4 | PDB = | ECnumber = | Chromosome = 2 | Arm = q | Band = 37 | LocusSupplementaryData = }} | |||
|{{infobox protein | Name = [[NEU3|sialidase 3 (membrane sialidase)]] | caption = | image = | width = | HGNCid = 7760 | Symbol = [[NEU3]] | AltSymbols = | EntrezGene = 10825 | OMIM = 604617 | RefSeq = NM_006656 | UniProt = Q9UQ49 | PDB = | ECnumber = | Chromosome = 11 | Arm = q | Band = 13.5 | LocusSupplementaryData = }} | |||
|{{infobox protein | Name = [[NEU4|sialidase 4]] | caption = | image = | width = | HGNCid = 21328 | Symbol = [[NEU4]] | AltSymbols = | EntrezGene = 129807 | OMIM = 608527 | RefSeq = NM_080741 | UniProt = Q8WWR8 | PDB = | ECnumber = | Chromosome = 2 | Arm = q | Band = 37.3 | LocusSupplementaryData = }} | |||
|} | |||
== Structure == | |||
{{main| Viral neuraminidase}} | |||
[[Influenza]] neuraminidase is a mushroom-shaped projection on the surface of the influenza virus. It has a head consisting of four co-planar and roughly spherical subunits, and a hydrophobic region that is embedded within the interior of the virus' membrane. It comprises a single polypeptide chain that is oriented in the opposite direction to the hemagglutinin antigen. The composition of the polypeptide is a single chain of six conserved polar amino acids, followed by hydrophilic, variable amino acids. β-Sheets predominate as the secondary level of protein conformation. | |||
Recent emergence of [[oseltamivir]] and [[zanamivir]] resistant [[human influenza]] A([[H1N1]]) H274Y has emphasized the need for suitable expression systems to obtain large quantities of highly pure and stable, [[Recombinant virus|recombinant]] neuraminidase through two separate artificial tetramerization domains that facilitate the formation of catalytically active neuraminidase homotetramers from [[yeast]] and [[Staphylothermus]] marinus, which allow for secretion of [[FLAG-tag]]ged [[proteins]] and further purification.<ref>Schmidt PM, Attwood RM, Mohr PG, Barrett SA, McKimm-Breschkin JL (2011) "A Generic System for the Expression and Purification of Soluble and Stable Influenza Neuraminidase". ''PLoS ONE'' 6(2): e16284. [http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0016284 ] {{doi|10.1371/journal.pone.0016284}}</ref> | |||
[[ | |||
== | == Mechanism == | ||
{{multiple image | |||
|direction = vertical | |||
|width = 350 | |||
|image1 = Figure1(Yao).gif|caption1 = Proposed mechanism of catalysis of influenza virus sialidase 4 (Link to glycosidase mechanism) | |||
|image2 = Figure2(Yao).gif|caption2 = Proposed transition state for the sialidase mechanism 7 | |||
|image3 = Figure3(Yao).gif|caption3 = Chemical structures of 4- substituted Neu5Ac2en derivatives 8 | |||
}} | |||
The enzymatic mechanism of influenza virus sialidase has been studied by Taylor et al., shown in Figure 1. The enzyme catalysis process has four steps. The first step involves the distortion of the α-sialoside from a <sup>2</sup>C<sub>5</sub> chair conformation (the lowest-energy form in solution) to a pseudoboat conformation when the sialoside binds to the sialidase. The second step leads to an oxocarbocation intermediate, the sialosyl cation. The third step is the formation of Neu5Ac initially as the α-anomer, and then mutarotation and release as the more thermodynamically-stable β-Neu5Ac.<ref name="pmid8126701">{{cite journal | vauthors = Taylor NR, von Itzstein M | title = Molecular modeling studies on ligand binding to sialidase from influenza virus and the mechanism of catalysis | journal = Journal of Medicinal Chemistry | volume = 37 | issue = 5 | pages = 616–24 | date = March 1994 | pmid = 8126701 | doi = 10.1021/jm00031a011 }}</ref> | |||
Neuraminidase | == Inhibitors == | ||
{{main| Neuraminidase inhibitors}} | |||
[[Neuraminidase inhibitor]]s are useful for combating [[influenza]] infection: [[zanamivir]], administered by inhalation; [[oseltamivir]], administered orally; [[peramivir]] administered parenterally, that is through intravenous or intramuscular injection; and [[laninamivir]] which is in phase III clinical trials. | |||
There are two major proteins on the surface of influenza virus particles. One is the lectin haemagglutinin protein with three relatively shallow sialic acid-binding sites and the other is enzyme sialidase with the active site in a pocket. Because of the relative deep active site in which low-molecular-weight inhibitors can make multiple favorable interactions and approachable methods of designing transition-state analogues in the hydrolysis of sialosides, the sialidase becomes more attractive anti-influenza drug target than the haemagglutinin.<ref name="isbn0-19-928278-1">{{cite book | last1 = Drickamer | first1 = Kurt | last2 = Taylor | first2 = Maureen P. | name-list-format = vanc | title = Introduction to glycobiology | edition = | publisher = Oxford University Press | location = Oxford [Oxfordshire] | year = 2006 | origyear = | pages = 177–178 | isbn = 0-19-928278-1 }}</ref> After the X-ray crystal structures of several influenza virus sialidases were available, the structure-based inhibitor design was applied to discover potent inhibitors of this enzyme.<ref>{{cite journal | title = Anti-Influenza Virus Drug Design: Sialidase Inhibitors | year = 2001 | journal = Australian Journal of Chemistry | pages = 663–670 | volume = 54 | issue = 11 | doi = 10.1071/CH01173 | last1 = Dyason | first1 = Jeffrey C. | last2 = Itzstein | first2 = Mark von | name-list-format = vanc }}</ref> | |||
The unsaturated sialic acid (N-acetylneuraminic acid [Neu5ac]) derivative 2-deoxy-2, 3-didehydro-D-N-acetylneuraminic acid (Neu5Ac2en), a sialosyl cation transition-state (Figure 2) analogue, is believed the most potent inhibitor core template. Structurally modified Neu5Ac2en derivatives may give more effective inhibitors.<ref name="isbn0-8247-5355-0">{{cite book | last = Fgedi | first = Pťer | name-list-format = vanc | title = The organic chemistry of sugars | edition = | publisher = Taylor & Francis | location = Washington, DC | year = 2006 | origyear = | pages = 822–823 | quote = | isbn = 0-8247-5355-0 }}</ref> | |||
== | Many Neu5Ac2en-based compounds have been synthesized and tested for their influenza virus sialidase inhibitory potential. For example: | ||
The 4-substituted Neu5Ac2en derivatives (Figure 3), 4-amino-Neu5Ac2en (Compound 1), which showed two orders of magnitude better inhibition of influenza virus sialidase than Neu5Ac2en5 and 4-guanidino-Neu5Ac2en (Compound 2), known as Zanamivir, which is now marketed for treatment of influenza virus as a drug, have been designed by von Itzstein and coworkers.<ref name="pmid8050102">{{cite journal | vauthors = von Itzstein M, Wu WY, Jin B | title = The synthesis of 2,3-didehydro-2,4-dideoxy-4-guanidinyl-N-acetylneuraminic acid: a potent influenza virus sialidase inhibitor | journal = Carbohydrate Research | volume = 259 | issue = 2 | pages = 301–5 | date = June 1994 | pmid = 8050102 | doi = 10.1016/0008-6215(94)84065-2 }}</ref> A series of amide-linked C9 modified Neu5Ac2en have been reported by Megesh and colleagues as NEU1 inhibitors.<ref name="pmid18068975">{{cite journal | vauthors = Magesh S, Moriya S, Suzuki T, Miyagi T, Ishida H, Kiso M | title = Design, synthesis, and biological evaluation of human sialidase inhibitors. Part 1: selective inhibitors of lysosomal sialidase (NEU1) | journal = Bioorganic & Medicinal Chemistry Letters | volume = 18 | issue = 2 | pages = 532–7 | date = January 2008 | pmid = 18068975 | doi = 10.1016/j.bmcl.2007.11.084 }}</ref> | |||
==See also== | == See also == | ||
*[[Glycoside hydrolase]] | *[[Glycoside hydrolase]] | ||
*[[ | *[[Neuraminidase inhibitors]] | ||
{{-}} | |||
== References == | |||
{{reflist|33em}} | |||
== | == External links == | ||
{{ | * {{MeshName|Neuraminidase}} | ||
*[https://web.archive.org/web/20090514142638/http://gsbs.utmb.edu/microbook/ch058.htm Orthomyxoviruses], Robert B. Couch, UTMB. Article includes a good clear line drawing of a neuraminidase on an influenza virus. | |||
{{Glycoprotein metabolism enzymes}} | |||
{{Sugar hydrolases}} | |||
{{Enzymes}} | |||
{{Portal bar|Molecular and Cellular Biology|border=no}} | |||
[[Category: | [[Category:Carbohydrate chemistry]] | ||
[[Category:EC 3.2.1]] | [[Category:EC 3.2.1]] | ||
[[Category: | [[Category:Glycobiology]] | ||
[[Category | [[Category:Neuraminidase inhibitors]] | ||
Latest revision as of 20:25, 31 October 2018
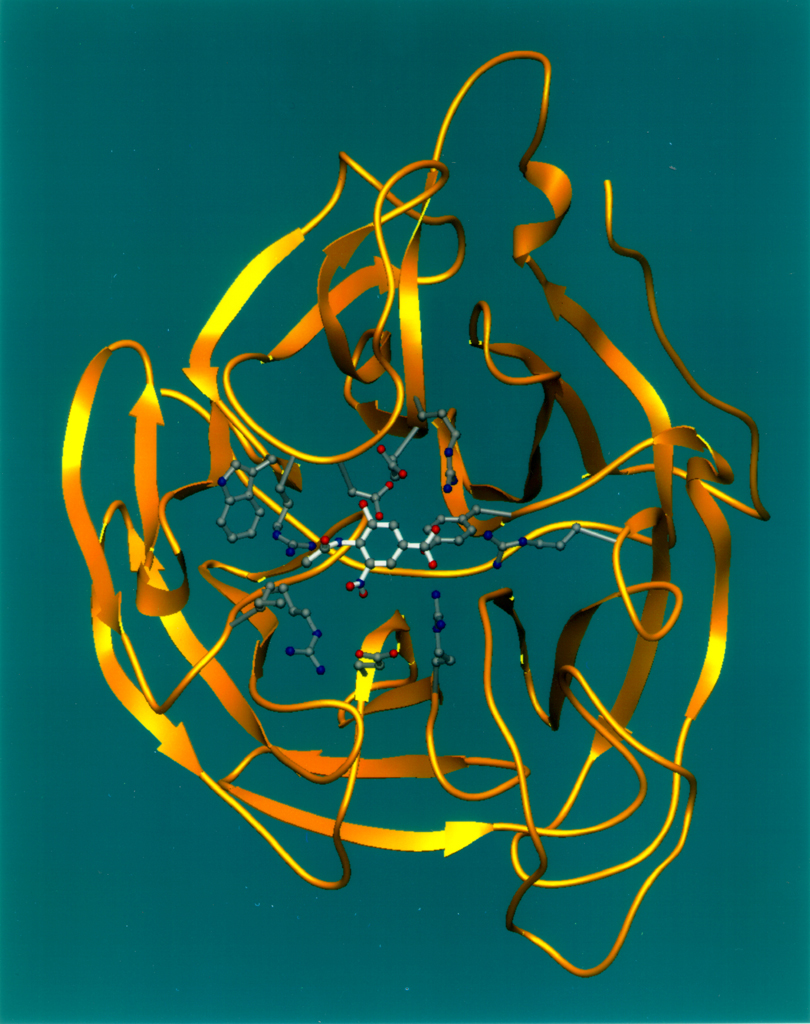
Neuraminidase enzymes are glycoside hydrolase enzymes that cleave (cut) the glycosidic linkages of neuraminic acids. Neuraminidase enzymes are a large family, found in a range of organisms. The best-known neuraminidase is the viral neuraminidase, a drug target for the prevention of the spread of influenza infection. The viral neuraminidases are frequently used as antigenic determinants found on the surface of the influenza virus. Some variants of the influenza neuraminidase confer more virulence to the virus than others. Other homologues are found in mammalian cells, which have a range of functions. At least four mammalian sialidase homologues have been described in the human genome (see NEU1, NEU2, NEU3, NEU4).
Reaction
There are two major classes of Neuraminidase that cleave exo or endo poly-sialic acids:
- Exo hydrolysis of α-(2→3)-, α-(2→6)-, α-(2→8)-glycosidic linkages of terminal sialic acid residues[1][2]
- Endo hydrolysis of (2→8)-α-sialosyl linkages in oligo- or poly(sialic) acids[2]
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Function
Neuraminidases, also called sialidases, catalyze the hydrolysis of terminal sialic acid residues from the newly formed virions and from the host cell receptors.[3] Sialidase activities include assistance in the mobility of virus particles through the respiratory tract mucus and in the elution of virion progeny from the infected cell.[4][5]
Subtypes
Swiss-Prot lists 137 types of neuraminidase from various species as of October 18, 2006.[6] Nine subtypes of influenza neuraminidase are known; many occur only in various species of duck and chicken. Subtypes N1 and N2 have been positively linked to epidemics in man, and strains with N3 or N7 subtypes have been identified in a number of isolated deaths.[citation needed]
The following is a list of major classes of neuraminidase enzymes:
- Viral neuraminidase
- Bacterial neuraminidase
- Mammalian neuraminidases:
|
|
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Structure
Influenza neuraminidase is a mushroom-shaped projection on the surface of the influenza virus. It has a head consisting of four co-planar and roughly spherical subunits, and a hydrophobic region that is embedded within the interior of the virus' membrane. It comprises a single polypeptide chain that is oriented in the opposite direction to the hemagglutinin antigen. The composition of the polypeptide is a single chain of six conserved polar amino acids, followed by hydrophilic, variable amino acids. β-Sheets predominate as the secondary level of protein conformation.
Recent emergence of oseltamivir and zanamivir resistant human influenza A(H1N1) H274Y has emphasized the need for suitable expression systems to obtain large quantities of highly pure and stable, recombinant neuraminidase through two separate artificial tetramerization domains that facilitate the formation of catalytically active neuraminidase homotetramers from yeast and Staphylothermus marinus, which allow for secretion of FLAG-tagged proteins and further purification.[7]
Mechanism
The enzymatic mechanism of influenza virus sialidase has been studied by Taylor et al., shown in Figure 1. The enzyme catalysis process has four steps. The first step involves the distortion of the α-sialoside from a 2C5 chair conformation (the lowest-energy form in solution) to a pseudoboat conformation when the sialoside binds to the sialidase. The second step leads to an oxocarbocation intermediate, the sialosyl cation. The third step is the formation of Neu5Ac initially as the α-anomer, and then mutarotation and release as the more thermodynamically-stable β-Neu5Ac.[8]
Inhibitors
Neuraminidase inhibitors are useful for combating influenza infection: zanamivir, administered by inhalation; oseltamivir, administered orally; peramivir administered parenterally, that is through intravenous or intramuscular injection; and laninamivir which is in phase III clinical trials.
There are two major proteins on the surface of influenza virus particles. One is the lectin haemagglutinin protein with three relatively shallow sialic acid-binding sites and the other is enzyme sialidase with the active site in a pocket. Because of the relative deep active site in which low-molecular-weight inhibitors can make multiple favorable interactions and approachable methods of designing transition-state analogues in the hydrolysis of sialosides, the sialidase becomes more attractive anti-influenza drug target than the haemagglutinin.[9] After the X-ray crystal structures of several influenza virus sialidases were available, the structure-based inhibitor design was applied to discover potent inhibitors of this enzyme.[10]
The unsaturated sialic acid (N-acetylneuraminic acid [Neu5ac]) derivative 2-deoxy-2, 3-didehydro-D-N-acetylneuraminic acid (Neu5Ac2en), a sialosyl cation transition-state (Figure 2) analogue, is believed the most potent inhibitor core template. Structurally modified Neu5Ac2en derivatives may give more effective inhibitors.[11]
Many Neu5Ac2en-based compounds have been synthesized and tested for their influenza virus sialidase inhibitory potential. For example: The 4-substituted Neu5Ac2en derivatives (Figure 3), 4-amino-Neu5Ac2en (Compound 1), which showed two orders of magnitude better inhibition of influenza virus sialidase than Neu5Ac2en5 and 4-guanidino-Neu5Ac2en (Compound 2), known as Zanamivir, which is now marketed for treatment of influenza virus as a drug, have been designed by von Itzstein and coworkers.[12] A series of amide-linked C9 modified Neu5Ac2en have been reported by Megesh and colleagues as NEU1 inhibitors.[13]
See also
References
- ↑ Schauer R (1982). "Chemistry, metabolism, and biological functions of sialic acids". Advances in Carbohydrate Chemistry and Biochemistry. Advances in Carbohydrate Chemistry and Biochemistry. 40: 131–234. doi:10.1016/S0065-2318(08)60109-2. ISBN 978-0-12-007240-8. PMID 6762816.
- ↑ 2.0 2.1 Cabezas JA (August 1991). "Some questions and suggestions on the type references of the official nomenclature (IUB) for sialidase(s) and endosialidase". The Biochemical Journal. 278 ( Pt 1) (Pt 1): 311–2. doi:10.1042/bj2780311. PMC 1151486. PMID 1883340.
- ↑ von Itzstein M (December 2007). "The war against influenza: discovery and development of sialidase inhibitors". Nature Reviews. Drug Discovery. 6 (12): 967–74. doi:10.1038/nrd2400. PMID 18049471.
- ↑ Palese P, Tobita K, Ueda M, Compans RW (October 1974). "Characterization of temperature sensitive influenza virus mutants defective in neuraminidase". Virology. 61 (2): 397–410. doi:10.1016/0042-6822(74)90276-1. PMID 4472498.
- ↑ Liu C, Eichelberger MC, Compans RW, Air GM (February 1995). "Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding". Journal of Virology. 69 (2): 1099–106. PMC 188682. PMID 7815489.
- ↑ Search in UniProt Knowledgebase (Swiss-Prot and TrEMBL) for: neuraminidase
- ↑ Schmidt PM, Attwood RM, Mohr PG, Barrett SA, McKimm-Breschkin JL (2011) "A Generic System for the Expression and Purification of Soluble and Stable Influenza Neuraminidase". PLoS ONE 6(2): e16284. [1] doi:10.1371/journal.pone.0016284
- ↑ Taylor NR, von Itzstein M (March 1994). "Molecular modeling studies on ligand binding to sialidase from influenza virus and the mechanism of catalysis". Journal of Medicinal Chemistry. 37 (5): 616–24. doi:10.1021/jm00031a011. PMID 8126701.
- ↑ Drickamer K, Taylor MP (2006). Introduction to glycobiology. Oxford [Oxfordshire]: Oxford University Press. pp. 177–178. ISBN 0-19-928278-1.
- ↑ Dyason JC, Itzstein Mv (2001). "Anti-Influenza Virus Drug Design: Sialidase Inhibitors". Australian Journal of Chemistry. 54 (11): 663–670. doi:10.1071/CH01173.
- ↑ Fgedi P (2006). The organic chemistry of sugars. Washington, DC: Taylor & Francis. pp. 822–823. ISBN 0-8247-5355-0.
- ↑ von Itzstein M, Wu WY, Jin B (June 1994). "The synthesis of 2,3-didehydro-2,4-dideoxy-4-guanidinyl-N-acetylneuraminic acid: a potent influenza virus sialidase inhibitor". Carbohydrate Research. 259 (2): 301–5. doi:10.1016/0008-6215(94)84065-2. PMID 8050102.
- ↑ Magesh S, Moriya S, Suzuki T, Miyagi T, Ishida H, Kiso M (January 2008). "Design, synthesis, and biological evaluation of human sialidase inhibitors. Part 1: selective inhibitors of lysosomal sialidase (NEU1)". Bioorganic & Medicinal Chemistry Letters. 18 (2): 532–7. doi:10.1016/j.bmcl.2007.11.084. PMID 18068975.
External links
- Neuraminidase at the US National Library of Medicine Medical Subject Headings (MeSH)
- Orthomyxoviruses, Robert B. Couch, UTMB. Article includes a good clear line drawing of a neuraminidase on an influenza virus.
- Pages with broken file links
- All articles with unsourced statements
- Articles with unsourced statements from July 2007
- Articles with invalid date parameter in template
- Protein pages needing a picture
- Genes on human chromosome 6
- Genes on human chromosome 2
- Genes on human chromosome 11
- Portal templates with all redlinked portals
- Carbohydrate chemistry
- EC 3.2.1
- Glycobiology
- Neuraminidase inhibitors