Human Immunodeficiency Virus (HIV): Difference between revisions
Ahmed Younes (talk | contribs) |
|||
| (97 intermediate revisions by 9 users not shown) | |||
| Line 1: | Line 1: | ||
<div style="-webkit-user-select: none;"> | |||
{|class="infobox" style="position: fixed; top: 65%; right: 10px; margin: 0 0 0 0; border: 0; float: right; | |||
{| class=" | |||
|- | |- | ||
| | | {{#ev:youtube|https://https://www.youtube.com/watch?v=5g1ijpBI6Dk|350}} | ||
}} | |||
|- | |- | ||
|} | |} | ||
__NOTOC__ | |||
{{AIDS}} | |||
{{CMG}}; {{AE}} {{Ammu}}, {{AL}} | |||
==Overview== | |||
AIDS is caused by the human immunodeficiency virus (HIV). HIV is a [[retrovirus]] classified into the family of ''[[Retroviridae]]'' and the sub family ''[[orthoretroviridae]]''.<ref name=HIV>{{cite web | title =HIV monograph| url = http://monographs.iarc.fr/ENG/Monographs/vol100B/mono100B-10.pdf }}</ref>. Two main subspecies of HIV exist: HIV-1, and HIV-2. HIV-1 is composed of two copies of single-stranded [[RNA]] enclosed by a conical [[capsid]] comprising the viral protein p24. The [[genome]] consists of several major [[genes]] that code for structural and functional [[protein]]s. These include the ''gag'', ''pol'', ''env'', ''tat'', and ''nef'' [[gene]]s. The [[genome]] and [[proteins]] of [[HIV]] have been the subject of extensive research since the discovery of the [[virus]] in 1983. It is a well known fact that no two HIV [[genome]]s are the same, not even from the same person, causing some to speculate that HIV is a "[[Quasispecies model|quasispecies]]" of a [[virus]].<ref>WainHobson, S., 1989. HIV genome variability in vivo. AIDS 3: supp 1; 139.</ref> A major requirement for all [[retroviruses]] is [[reverse transcriptase]] that transcribes the [[viral RNA]] into [[double-stranded DNA]] and [[integrase]] that integrates this newly formed [[DNA]] into the host [[genome]]. | |||
HIV | ==Taxonomy== | ||
* HIV 1 and HIV 2 are classified into the family of ''[[Retroviridae]]'' and sub family ''orthoretroviridae''.<ref name=HIV>{{cite web | title =HIV monograph| url = http://monographs.iarc.fr/ENG/Monographs/vol100B/mono100B-10.pdf }}</ref> | |||
* Retrovirus are enveloped [[RNA]] viruses which requires a [[DNA]] intermediate to replicate. | |||
* HIV 1 and HIV 2 belongs to the genus [[Lentivirus]] (Lentus which in latin means slow) | |||
* The [[retrovirus]] rely on [[enzyme]] [[reverse transcriptase]] to transcribe their genome from [[RNA]] to [[DNA]]. | |||
* [[Integrase]] incorporates the [[DNA]] into the host [[DNA]] and becomes a part of cellular [[DNA]] replicating with it. | |||
HIV | ==Origin== | ||
* Both HIV-1 and HIV-2 are of primate origin. The origin of HIV-1 is the Central Common Chimpanzee (''Pan troglodytes troglodytes'') found in southern Cameroon.<ref name=Keele>{{ | |||
cite journal | cite journal | ||
| author= | | author=Keele BF, van Heuverswyn F, Li YY, et al | title=Chimpanzee Reservoirs of Pandemic and Nonpandemic HIV-1 | ||
| journal=Science | year=2006 |volume=313 |issue=5786 |pages=523–6 | |||
| journal= | | url= http://www.sciencemag.org/cgi/content/abstract/1126531 | ||
| pmid= | | pmid=16728595 | doi=10.1126/science.1126531 | ||
}}</ref> | }}</ref> | ||
* It is believed that HIV-2 originated from the Sooty Mangabey (''Cercocebus atys''), an Old World monkey of Guinea Bissau, Gabon, and Cameroon. | |||
* Most experts believe that HIV probably transferred to humans as a result of direct contact with primates, for instance during hunting or butchery.<ref name=CohenOrigns>{{ | |||
{{ | |||
cite journal | |||
| author=Cohen J | |||
| title=Vaccine Theory of AIDS Origins Disputed at Royal Society | |||
| journal=Science | year=2000 | pages=1850–1851 | volume= 289| issue=5486 | | |||
pmid=11012346 | doi=10.1126/science.289.5486.1850 | |||
}}</ref> | }}</ref> | ||
==Structure== | |||
[[Image:HIV Viron.png|thumb|left|200px|Figure 1. Diagram of HIV - By US National Institute of Health]] | |||
* HIV is different in structure from other [[retrovirus]]es. It is around 120 [[nanometer|nm]] in diameter (120 billionths of a meter; around 60 times smaller than a red blood cell) and roughly spherical. | |||
* HIV-1 is composed of two copies of single-stranded [[RNA]] enclosed by a conical [[capsid]] comprising the viral protein [[#p24, p6, p7, p21|p24]], typical of [[lentivirus]]es (Figure 1). The RNA component is 9749 nucleotides long. This is in turn surrounded by a [[cell membrane|plasma membrane]] of host-cell origin. The single-strand [[RNA]] is tightly bound to the [[nucleocapsid proteins]], [[#p24, p6, p7, p21|p7]] and [[enzymes]] that are indispensable for the development of the [[virion]], such as [[reverse transcriptase]] and [[integrase]]. The nucleocapsid (p7 and p6) associates with the genomic [[RNA]] (one molecule per hexamer) and protects the [[RNA]] from digestion by [[nuclease]]s. A matrix composed of an association of the viral protein p17 surrounds the [[capsid]], ensuring the integrity of the [[virion particle]]. Also enclosed within the virion particle are [[#Vif|Vif]], [[#Vpr|Vpr]], [[#Nef|Nef]], p7 and viral [[#protease|protease]] (Figure 1). The envelope is formed when the capsid buds from the host cell, taking some of the host-cell membrane with it. The envelope includes the glycoproteins [[#gp120|gp120]] and [[#gp41|gp41]]. | |||
* Recently, an Anglo-German team compiled a 3D structure of HIV by combining multiple images. It is hoped that this new information would contribute to scientific understanding of the virus, and help in the creation of a cure. Oxford University's Professor Stephen D. Fuller said the 3D map would assist in understanding how the virus grows. <ref>BBC News: [http://news.bbc.co.uk/1/hi/health/4642940.stm 3D Structure of HIV Revealed]</ref> The validity of this work remains a matter of debate <ref>The SIV Surface Spike Imaged by Electron Tomography: One Leg or Three? Subramaniam S PLoS Pathogens Vol. 2, No. 8, e91 doi:10.1371/journal.ppat.0020091</ref>, with a conflicting model produced by another team led by Florida State University Professor [[Kenneth Roux]] in the US <ref>Distribution and three-dimensional structure of AIDS virus envelope spikes Ping Zhu, Jun Liu, Julian Bess, Jr, Elena Chertova, Jeffrey D. Lifson, Henry Grisé, Gilad A. Ofek, Kenneth A. Taylor and Kenneth H. Roux, Nature 441, 847-852 (15 June 2006) | doi:10.1038/nature04817; Received 8 March 2006; Accepted 24 April 2006; Published online 24 May 2006</ref>. | |||
== | ==Genome organization== | ||
* HIV has several major [[gene]]s coding for structural proteins that are found in all [[retroviruses]], and several nonstructural ("accessory") genes that are unique to HIV. The ''gag'' gene provides the basic physical infrastructure of the [[virus]], and ''pol'' provides the basic mechanism by which retroviruses reproduce, while the others help HIV to enter the host [[cell]] and enhance its reproduction. Though they may be altered by mutation, all of these genes except ''tev'' exist in all known variants of HIV; see [[HIV#Genetic variability|Genetic variability of HIV]]. | |||
:* ''gag'' (Group-specific Antigen): codes for [[#p24|p24]], the viral capsid; [[#p6 and p7|p6 and p7]], the nucleocapsid proteins; and [[#p17|p17]], a matrix protein. | |||
:* ''pol'': Codes for viral [[enzyme]]s, the most important of which are [[#reverse transcriptase|reverse transcriptase]], [[#integrase|integrase]], and [[#protease|protease]] which cleaves the proteins derived from ''gag'' and ''pol'' into functional proteins. | |||
:* ''env'' (for "envelope"): Codes for the precursor to [[#gp120|gp120]] and [[#gp41|gp41]], proteins embedded in the viral envelope which enable the virus to attach to and fuse with target cells. | |||
:* ''tat'', ''rev'', ''nef'', ''vif'', ''vpr'', ''vpu'': Each of these genes codes for a single protein with the same names; see [[#Tat|Tat]], [[#Rev|Rev]], [[#Nef|Nef]], [[#Vif|Vif]], [[#Vpr|Vpr]], [[#Vpu|Vpu]]. | |||
:* ''tev'': This gene is only present in a few HIV-1 isolates. It is a fusion of parts of the ''tat'', ''env'', and ''rev'' genes, and codes for a protein with some of the properties of [[#Tat|Tat]], but little or none of the properties of [[#Rev|Rev]]. | |||
==Protein function== | |||
===Gag=== | |||
* These proteins are encoded by the ''gag'' [[gene]], and provide structural elements of the [[virus]]. | |||
====p24==== | |||
* p24 makes up the viral [[capsid]]. | |||
* When a [[Western blot]] test is used to detect HIV [[infection]], p24 is one of the three major proteins tested for, along with [[#gp120|gp120]]/gp160 and [[#gp41|gp41]]. | |||
====p6, p7, and p17==== | |||
* p6 and p7 provide the [[nucleocapsid]]. | |||
* p17 provides a protective [[matrix]]. | |||
===Pol=== | |||
====Reverse transcriptase==== | |||
{{main|Reverse transcriptase}} | |||
* Common to all [[retroviruses]], this enzyme transcribes the viral [[RNA]] into double-stranded [[DNA]]. | |||
====Integrase==== | |||
{{main|Integrase}} | |||
* This enzyme integrates the [[DNA]] produced by reverse transcriptase into the host's [[genome]]. | |||
====Protease==== | |||
* A [[protease]] is any enzyme that cuts proteins into segments. HIV's ''gag'' and ''pol'' genes do not produce their proteins in their final form, but as larger combination proteins; the specific protease used by HIV cleaves these into separate functional units. [[Antiretroviral drug#Protease inhibitors (PIs)|Protease inhibitor]] drugs block this step. | |||
===Env=== | |||
* The ''env'' gene does not actually code for gp120 and gp41, but for a precursor to both, gp160. During HIV reproduction, the host cell's own enzymes cleave gp160 into gp120 and gp41. See [[HIV#Replication cycle of HIV|Replication cycle of HIV]]. | |||
====gp120==== | |||
{{main|gp120}} | |||
* Exposed on the surface of the [[viral envelope]], the [[glycoprotein]] gp120 binds to the [[CD4]] [[receptor (biochemistry)|receptor]] on any target cell that has such a receptor, particularly the [[helper T-cell]]. See [[HIV#HIV tropism|HIV tropism]] and [[HIV#Replication cycle of HIV|Replication cycle of HIV]]. | |||
* Since [[CD4]] receptor binding is the most obvious step in HIV infection, gp120 was among the first targets of [[HIV vaccine]] research. These efforts have been hampered by its chemical properties, which make it difficult for antibodies to bind to gp120; also, it can easily be shed from the [[virus]] due to its loose binding with gp41. | |||
====gp41==== | |||
* The [[glycoprotein]] gp41 is non-[[covalent bond|covalently]] bound to gp120, and provides the second step by which [[viral entry|HIV enters]] the [[cell]]. It is originally buried within the viral envelope, but when gp120 binds to a [[CD4]] receptor, gp120 changes its [[chemical conformation|conformation]] causing gp41 to become exposed, where it can assist in fusion with the host [[cell]]. | |||
* [[Fusion inhibitor]] drugs such as [[enfuvirtide]] block the fusion process by binding to gp41. | |||
===Transactivators=== | |||
====Tat==== | |||
* Stands for "Trans-Activator of Transcription". Tat consists of between 86 and 101 amino acids depending on the subtype.<ref>Jeang, K. T. (1996) In: Human Retroviruses and AIDS: [http://hiv.lanl.gov/content/hiv-db/COMPENDIUM/1996/PART-III/1.pdf A Compilation and Analysis of Nucleic Acid and Amino Acid Sequences.] Los Alamos National Laboratory (Ed.) pp. III-3–III-18</ref> Tat helps HIV reproduce by compensating for a defect in its genome: the HIV [[RNA]] initially has a hairpin-structured portion which prevents full transcription occurring. However, a small number of [[RNA]] transcripts will be made, which allow the Tat protein to be produced. Tat then binds to and phosphorylates cellular factors, eliminating the effect of the hairpin [[RNA]] structure and allowing transcription of the HIV [[DNA]].<ref>Kim JB, Sharp PA. (2001) [http://www.jbc.org/cgi/content/full/276/15/12317 Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase]. ''J. Biol. Chem.'' '''276''', 12317-12323 PMID 11145967</ref> This itself increases the rate of transcription, providing a [[positive feedback]] cycle. This in turn allows HIV to have an explosive response once a threshold amount of Tat is produced, a useful tool for defeating the body's [[response]]. Tat also appears to play a more direct role in the HIV disease process. The protein is released by infected cells in culture, and is found in the blood of HIV-1 infected patients.<ref name="Xiao">Xiao, H., Neuveut, C., Tiffany, H. L., Benkirane, M., Rich, E. A., Murphy, P. M. and Jeang, K. T. (2000) [http://www.ncbi.nlm.nih.gov/entrez/utils/lofref.fcgi?PrId=3494&uid=11027346&db=pubmed&url=http://www.pubmedcentral.gov/articlerender.fcgi?tool=pubmed&pubmedid=11027346 Selective CXCR4 antagonism by Tat: implications for in vivo expansion of coreceptor use by HIV-1]. ''Proc. Natl. Acad. Sci. U.S.A.'' '''97''', 11466-11471 PMID 11027346</ref> It can be absorbed by [[cells]] that are not infected with HIV, and can act directly as a [[toxin]] producing cell death via [[apoptosis]] in uninfected "bystander" [[T cell]]s, assisting in progression toward [[AIDS]].<ref>Campbell GR, Pasquier E, Watkins J, Bourgarel-Rey V, Peyrot V, Esquieu D, Barbier P, de Mareuil J, Braguer D, Kaleebu P, Yirrell DL, Loret EP. (2004) [http://www.ncbi.nlm.nih.gov/entrez/utils/lofref.fcgi?PrId=3051&uid=15331610&db=pubmed&url=http://www.jbc.org/cgi/pmidlookup?view=long&pmid=15331610 The glutamine-rich region of the HIV-1 Tat protein is involved in T-cell apoptosis]. ''J. Biol. Chem.'' '''279''', 48197-48204 PMID 15331610</ref> By interacting with the [[CXCR4]] receptor, Tat also appears to encourage the reproduction of less virulent M-tropic strains of HIV early in the course of infection, allowing the more rapidly pathogenic T-tropic strains to emerge later.<ref name="Xiao"> </ref> | |||
====Rev==== | |||
{| style="float:right" | |||
|[[Image:Rev-mediated HIV mRNA transport.png|thumb|left|300px|'''Rev-mediated HIV mRNA transport.''' Rev (red) binds the Rev response element (RRE, blue) to mediate export of unspliced and singly spliced mRNA from the nucleus to the cytoplasm. - De I, Vossman, CC BY 2.5, https://commons.wikimedia.org/w/index.php?curid=2401724]] | |||
|} | |||
* Stands for "Regulator of Virion". This protein allows fragments of HIV [[mRNA]] that contain a Rev Response Unit (RRE) to be exported from the nucleus to the [[cytoplasm]]. In the absence of the ''rev'' gene, [[RNA]] splicing machinery in the nucleus quickly splices the [[RNA]] so that only the smaller, regulatory proteins can be produced; in the presence of ''rev'', [[RNA]] is exported from the nucleus before it can be spliced, so that the structural proteins and [[RNA]] [[genome]] can be produced. Again, this mechanism allows a positive feedback loop to allow [[HIV]] to overwhelm the host's defenses, and provides time-dependent regulation of replication (a common process in viral infections)<ref name="Strebel">Strebel, K (2003) Virus-host interactions: role of HIV proteins Vif, Tat, and Rev. ''AIDS'' '''17 Suppl 4''', S25-S34 PMID 15080177</ref> | |||
====Vpr==== | |||
* Stands for "Viral Protein R". Vpr, a 96 amino acid 14-kDa protein, plays an important role in regulating nuclear import of the HIV-1 pre-integration complex, and is required for virus replication in non-dividing [[cell]]s such as [[macrophages]]. Vpr also induces [[cell cycle]] arrest and [[apoptosis]] in proliferating cells, which can result in [[immune dysfunction]].<ref>Bukrinsky M, Adzhubei A. (1999) Viral protein R of HIV-1. ''Rev Med Virol'' '''9''', 39-49 PMID 10371671</ref><ref>Muthumani K., et al (2006) The HIV-1 Vpr and glucocorticoid receptor complex: A gain of function interaction that prevents the nuclear localization of PARP-1. Nat Cell Biol. Feb;8(2):170-9.</ref> | |||
* Vpr is also immunosuppressive due to its ability to sequester a proinflammatory transcriptional activator in the [[cytoplasm]]. HIV-2 contains both a Vpr [[protein]] and a related (by sequence homology) Vpx protein (Viral Protein X). Two functions of Vpr in HIV-1 are split between Vpr and Vpx in HIV-2, with the HIV-2 Vpr protein inducing [[cell cycle]] arrest and the Vpx protein required for nuclear import. | |||
===Other regulatory proteins=== | |||
====Nef==== | |||
* Stands for "Negative Regulatory Factor". The expression of Nef early in the viral [[life cycle]] ensures T cell activation and the establishment of a persistent state of infection, two basic attributes of HIV [[infection]]. Nef also promotes the survival of infected cells by downmodulating the expression of several surface molecules important in [[host]] [[immune]] function. These include [[major histocompatibility complex]]-I (MHC I) and MHC II present on [[antigen presenting cell]]s (APCs) and target cells, [[CD4]] and [[CD28]] present on CD4+ T cells. One group of patients in Sydney were infected with a nef-deleted virus and took much longer than expected to progress to AIDS.<ref>Learmont JC, Geczy AF, Mills J, Ashton LJ, Raynes-Greenow CH, Garsia RJ, Dyer WB, McIntyre L, Oelrichs RB, Rhodes DI, Deacon NJ, Sullivan JS. (1999) [http://content.nejm.org/cgi/content/abstract/340/22/1715 Immunologic and virologic status after 14 to 18 years of infection with an attenuated strain of HIV-1]. A report from the Sydney Blood Bank Cohort. ''N Engl J Med'' '''340''', 1715-1722 PMID 10352163</ref> | |||
* A ''nef''-deleted virus vaccine has not been trialed in humans and has failed in nonhuman animals.HIV-1 Nef-induced FasL induction and bystander killing requires p38 MAPK activation. | |||
====Vif==== | |||
* Stands for "[[Viral infectivity factor]]". Vif is a 23-[[kilodalton]] protein that is essential for viral replication.<ref name="Strebel"> </ref> Vif inhibits the cellular protein, [[APOBEC3G]], from entering the virion during budding from a host cell by targeting it for proteasomal degredation. Vif hijacks the cellular Cullin5 E3 ubiquitin ligase in order to target APOBEC3G for degradation. In the absence of Vif, APOBEC3G causes hypermutation of the viral genome, rendering it dead-on-arrival at the next host cell. APOBEC3G is thus a host defence to retroviral infection which HIV-1 has overcome by the acquisition of Vif. | |||
====Vpu==== | |||
* Stands for "Viral Protein U". Vpu is involved in viral budding, enhancing [[virion]] release from the [[cell]]. | |||
==Tropism== | ==Tropism== | ||
* '''HIV tropism''' refers to the [[cell type]] that the [[human immunodeficiency virus]] (HIV) infects and replicates in. HIV [[tropism]] of a patient's [[virus]] is measured by the [[Trofile assay]]. | |||
* HIV can infect a variety of [[cell (biology)|cell]]s such as [[Helper T cell|CD4+ helper T-cells]] and [[macrophage]]s that express the [[CD4]] molecule on their surface. HIV-1 entry to [[macrophages]] and T helper [[cells]] is mediated not only through interaction of the [[virion]] envelope glycoproteins ([[gp120]]) with the CD4 molecule on the target cells but also with its [[chemokine]] coreceptors. | |||
Macrophage (M-tropic) strains of HIV-1, or non- | * [[Macrophage]] (M-tropic) strains of HIV-1, or non-syncitia-inducing strains (NSI) use the beta-chemokine [[receptor]] [[CCR5]] for entry and are thus able to replicate in [[macrophages]] and [[CD4+]] [[T-cell]]s <ref name=Coakley> | ||
{{cite journal | |||
cite journal | | author=Coakley, E., Petropoulos, C. J. and Whitcomb, J. M. | title=Assessing chemokine co-receptor usage in HIV | journal=Curr. Opin. Infect. Dis. | year=2005 | pages=9-15 | volume=18 | issue=1 | id={{PMID|15647694}} | ||
| author=Coakley, E., Petropoulos, C. J. and Whitcomb, J. M. | title=Assessing | }}</ref>. The normal [[ligand]]s for this receptor, [[RANTES]], macrophage inflammatory protein (MIP)-1-beta and MIP-1-alpha, are able to suppress HIV-1 infection ''in vitro''. This CCR5 coreceptor is used by almost all primary HIV-1 isolates regardless of viral genetic subtype. | ||
}}</ref> This CCR5 coreceptor is used by almost all primary HIV-1 isolates regardless of viral genetic subtype | * T-tropic isolates, or [[syncitia]]-inducing (SI) strains replicate in primary CD4+ T-cells as well as in [[macrophages]] and use the alpha-chemokine receptor, [[CXCR4]], for entry <ref name=Coakley> | ||
{{cite journal | |||
T-tropic isolates, or [[syncitia]]-inducing (SI) strains replicate in primary CD4 | | author=Coakley, E., Petropoulos, C. J. and Whitcomb, J. M. | title=Assessing chemokine co-receptor usage in HIV | journal=Curr. Opin. Infect. Dis. | year=2005 | pages=9-15 | volume=18 | issue=1 | id={{PMID|15647694}} | ||
}}</ref>. The alpha-chemokine, [[Stromal cell-derived factor-1|SDF-1]], a ligand for [[CXCR4]], suppresses replication of T-tropic HIV-1 isolates. It does this by down regulating the expression of [[CXCR4]] on the surface of these [[cell]]s. | |||
{{cite journal | * Viruses that use only the [[CCR5]] receptor are termed R5, those that only use [[CXCR4]] are termed X4, and those that use both, X4R5. However, the use of coreceptor alone does not explain viral [[tropism]], as not all '''R5 viruses''' are able to use CCR5 on [[macrophages]] for a productive [[infection]] <ref name=Coakley> | ||
| author= | {{cite journal | ||
| author=Coakley, E., Petropoulos, C. J. and Whitcomb, J. M. | title=Assessing chemokine co-receptor usage in HIV | journal=Curr. Opin. Infect. Dis. | year=2005 | pages=9-15 | volume=18 | issue=1 | id={{PMID|15647694}} | |||
}}</ref>. | |||
* HIV can also infect a subtype of [[dendritic cells]] <ref name=Knight> | |||
{{cite journal | |||
| author=Knight, S. C., Macatonia, S. E. and Patterson, S. | title=HIV I infection of dendritic cells | journal=Int. Rev. Immunol. | year=1990 | pages=163-175 | volume=6 | issue=2-3 | id={{PMID|2152500}} | |||
}}</ref>, MDC-1, which probably constitute a major reservoir that maintains infection when T helper cell numbers have declined to extremely low levels. | |||
}}</ref> | |||
The | |||
{{cite journal | |||
| author= | |||
}}</ref> | |||
{{cite journal | |||
| author= | |||
{{ | |||
| | |||
}}</ref> | |||
==Replication cycle== | ==Replication cycle== | ||
[[Image:Hiventrytocell.jpg|left|thumb|Schematic representation of the key structural features of HIV-1 entry into T cells. The two bottom images show alternate models for entry into cells.]] | [[File:HIV Life Cycle.jpg|thumb|600px|none|Image obtained from AIDSinfo.org http://aidsinfo.nih.gov/education-materials/fact-sheets/19/73/the-hiv-life-cycle]] | ||
[[Image:Hiv gross.png| | <br> | ||
'''Steps in the HIV Replication Cycle''' | |||
# Fusion of the HIV cell to the host cell surface. | |||
# HIV RNA, reverse transcriptase, integrase, and other viral proteins enter the host cell. | |||
# Viral DNA is formed by reverse transcription. | |||
# Viral DNA is transported across the nucleus and integrates into the host DNA. | |||
# New viral RNA is used as genomic RNA and to make viral proteins. | |||
# New viral RNA and proteins move to cell surface and a new, immature, HIV virus forms. | |||
# The virus matures by protease releasing individual HIV proteins. | |||
{| | |||
|[[Image:Hiventrytocell.jpg|left|thumb|350px|Schematic representation of the key structural features of HIV-1 entry into T cells. The two bottom images show alternate models for entry into cells. - By Rachid Sougrat, Alberto Bartesaghi, Jeffrey D. Lifson, Adam E. Bennett, Julian W. Bess, Daniel J. Zabransky, Sriram Subramaniam - Sougrat R, Bartesaghi A, Lifson JD, et al (May 2007). "Electron tomography of the contact between T cells and SIV/HIV-1: implications for viral entry". PLoS Pathog. 3 (5): e63. PMID 17480119. doi:10.1371/journal.ppat.0030063Direct link to image: http://www.plospathogens.org/article/showImageLarge.action?uri=info%3Adoi%2F10.1371%2Fjournal.ppat.0030063.g008, CC BY 2.5, https://commons.wikimedia.org/w/index.php?curid=4198856]] | |||
|[[Image:Hiv gross.png|left|thumbnail|250px|The HIV replication cycle - By Translated by Raul654 - Originally from GFDL image Image:Hiv gross german.png, CC BY-SA 3.0, https://commons.wikimedia.org/w/index.php?curid=32862]] | |||
|} | |||
===Entry to the cell=== | ===Entry to the cell=== | ||
* HIV enters [[macrophages]] and CD4<SUP>+</SUP> T cells by the [[adsorption]] of [[glycoproteins]] on its surface to receptors on the target [[cell]] followed by fusion of the viral [[envelope]] with the [[cell membrane]] and the release of the HIV [[capsid]] into the [[cell]].<ref name=Chan2>{{cite journal |author=Chan D, Kim P |title=HIV entry and its inhibition |journal=Cell |volume=93 |issue=5 |pages=681-4 |year=1998 |pmid=9630213}}</ref><ref name=Wyatt>{{cite journal |author=Wyatt R, Sodroski J |title=The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens |journal=Science |volume=280 |issue=5371 |pages=1884-8 |year=1998 | doi=10.1126/science.280.5371.1884 |pmid=9632381}}</ref> | |||
HIV enters macrophages and CD4<SUP>+</SUP> T cells by the [[adsorption]] of [[glycoproteins]] on its surface to receptors on the target cell followed by fusion of the viral envelope with the cell membrane and the release of the HIV capsid into the cell.<ref name=Chan2>{{cite journal |author=Chan D, Kim P |title=HIV entry and its inhibition |journal=Cell |volume=93 |issue=5 |pages=681-4 |year=1998 |pmid=9630213}}</ref><ref name=Wyatt>{{cite journal |author=Wyatt R, Sodroski J |title=The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens |journal=Science |volume=280 |issue=5371 |pages=1884-8 |year=1998 | doi=10.1126/science.280.5371.1884 |pmid=9632381}}</ref> | * Entry to the [[cell]] begins through interaction of the trimeric [[envelope]] complex (gp160 spike, discussed above) and both [[CD4]] and a chemokine receptor (generally either [[CCR5]] or [[CXCR4]], but others are known to interact) on the [[cell]] surface.<ref name=Chan2/><ref name=Wyatt/> The [[gp160]] spike contains binding domains for both [[CD4]] and [[chemokine]] receptors.<ref name=Chan2/><ref name=Wyatt/> The first step in fusion involves the high-affinity attachment of the [[CD4]] binding domains of [[gp120]] to [[CD4]]. Once [[gp120]] is bound with the [[CD4]] protein, the envelope complex undergoes a structural change, exposing the [[chemokine]] binding domains of [[gp120]] and allowing them to interact with the target chemokine [[receptor]].<ref name=Chan2/><ref name=Wyatt/> This allows for a more stable two-pronged attachment, which allows the N-terminal fusion peptide gp41 to penetrate the cell membrane.<ref name=Chan2/><ref name=Wyatt/> Repeat sequences in [[gp41]], HR1 and HR2 then interact, causing the collapse of the extracellular portion of [[gp41]] into a hairpin. This loop structure brings the virus and [[cell membranes]] close together, allowing fusion of the membranes and subsequent entry of the [[viral capsid]].<ref name=Chan2/><ref name=Wyatt/> | ||
* Once HIV has bound to the target cell, the HIV [[RNA]] and various [[enzymes]], including [[reverse transcriptase]], [[integrase]], [[ribonuclease]] and [[protease]], are injected into the cell.<ref name=Chan2/> | |||
Entry to the cell begins through interaction of the trimeric envelope complex (gp160 spike, discussed above) and both [[CD4]] and a chemokine receptor (generally either CCR5 or CXCR4, but others are known to interact) on the cell surface.<ref name=Chan2/><ref name=Wyatt/> The gp160 spike contains binding domains for both CD4 and chemokine receptors.<ref name=Chan2/><ref name=Wyatt/> The first step in fusion involves the high-affinity attachment of the CD4 binding domains of [[gp120]] to CD4. Once gp120 is bound with the CD4 protein, the envelope complex undergoes a structural change, exposing the chemokine binding domains of gp120 and allowing them to interact with the target chemokine receptor.<ref name=Chan2/><ref name=Wyatt/> This allows for a more stable two-pronged attachment, which allows the N-terminal fusion peptide gp41 to penetrate the cell membrane.<ref name=Chan2/><ref name=Wyatt/> Repeat sequences in gp41, HR1 and HR2 then interact, causing the collapse of the extracellular portion of gp41 into a hairpin. This loop structure brings the virus and cell membranes close together, allowing fusion of the membranes and subsequent entry of the viral capsid.<ref name=Chan2/><ref name=Wyatt/> | * HIV can infect [[dendritic cells]] (DCs) by this CD4-CCR5 route, but another route using mannose-specific C-type lectin receptors such as [[DC-SIGN]] can also be used.<ref name=Pope_2003>{{cite journal |author=Pope M, Haase A |title=Transmission, acute HIV-1 infection and the quest for strategies to prevent infection |journal=Nat Med |volume=9 |issue=7 |pages=847-52 |year=2003 |pmid=12835704}}</ref> DCs are one of the first cells encountered by the virus during sexual transmission. They are currently thought to play an important role by transmitting HIV to T cells once the virus has been captured in the [[mucosa]] by DCs.<ref name=Pope_2003 /> | ||
Once HIV has bound to the target cell, the HIV [[RNA]] and various [[enzymes]], including reverse transcriptase, integrase, ribonuclease and protease, are injected into the cell.<ref name=Chan2/> | |||
HIV can infect [[dendritic cells]] (DCs) by this CD4-CCR5 route, but another route using mannose-specific C-type lectin receptors such as [[DC-SIGN]] can also be used.<ref name=Pope_2003>{{cite journal |author=Pope M, Haase A |title=Transmission, acute HIV-1 infection and the quest for strategies to prevent infection |journal=Nat Med |volume=9 |issue=7 |pages=847-52 |year=2003 |pmid=12835704}}</ref> DCs are one of the first cells encountered by the virus during sexual transmission. They are currently thought to play an important role by transmitting HIV to T cells once the virus has been captured in the [[mucosa]] by DCs.<ref name=Pope_2003 /> | |||
===Replication and transcription=== | ===Replication and transcription=== | ||
Once the viral capsid enters the cell, an [[enzyme]] called ''[[reverse transcriptase]]'' liberates the single-stranded (+)[[RNA]] from the attached viral proteins and copies it into a complementary [[DNA]].<ref name=Zheng> | * Once the viral capsid enters the cell, an [[enzyme]] called ''[[reverse transcriptase]]'' liberates the single-stranded (+)[[RNA]] from the attached viral proteins and copies it into a complementary [[DNA]].<ref name=Zheng> | ||
{{cite journal | {{cite journal | ||
| author=Zheng, Y. H., Lovsin, N. and Peterlin, B. M. | title=Newly identified host factors modulate HIV replication | journal=Immunol. Lett. | year=2005 | pages=225-234 | volume=97 | issue=2 | pmid=15752562 | | author=Zheng, Y. H., Lovsin, N. and Peterlin, B. M. | title=Newly identified host factors modulate HIV replication | journal=Immunol. Lett. | year=2005 | pages=225-234 | volume=97 | issue=2 | pmid=15752562 | ||
}}</ref> This process of reverse transcription is extremely error-prone and it is during this step that mutations may occur. Such mutations may cause [[Resistance to antiviral drugs|drug resistance]]. The [[reverse transcriptase]] then makes a complementary [[DNA]] strand to form a double-stranded viral DNA intermediate (vDNA). This vDNA is then transported into the [[cell nucleus]]. The integration of the viral DNA into the host cell's [[genome]] is carried out by another viral enzyme called ''[[integrase]]''.<ref name=Zheng/> | |||
}}</ref> This process of reverse transcription is extremely error-prone and it is during this step that mutations may occur. Such mutations may cause [[Resistance to antiviral drugs|drug resistance]]. The reverse transcriptase then makes a complementary DNA strand to form a double-stranded viral DNA intermediate (vDNA). This vDNA is then transported into the [[cell nucleus]]. The integration of the viral DNA into the host cell's [[genome]] is carried out by another viral enzyme called ''[[integrase]]''.<ref name=Zheng/> | * This integrated viral DNA may then lie dormant, in the latent stage of HIV infection.<ref name=Zheng/> To actively produce the virus, certain cellular [[transcription factors]] need to be present, the most important of which is [[NF-kB|NF-''κ''B]] (NF kappa B), which is upregulated when T cells become activated.<ref name=Hiscott> | ||
This integrated viral DNA may then lie dormant, in the latent stage of HIV infection.<ref name=Zheng/> To actively produce the virus, certain cellular [[transcription factors]] need to be present, the most important of which is [[NF-kB|NF-''κ''B]] (NF kappa B), which is upregulated when T cells become activated.<ref name=Hiscott> | |||
{{cite journal | {{cite journal | ||
| author=Hiscott J, Kwon H, Genin P. | title=Hostile takeovers: viral appropriation of the NF-kappaB pathway | journal=J Clin Invest. | year=2001 | pages=143-151 | volume=107 | issue=2 | pmid=11160127 | | author=Hiscott J, Kwon H, Genin P. | title=Hostile takeovers: viral appropriation of the NF-kappaB pathway | journal=J Clin Invest. | year=2001 | pages=143-151 | volume=107 | issue=2 | pmid=11160127 | ||
}}</ref> This means that those cells most likely to be killed by HIV are those currently fighting infection. | }}</ref> This means that those cells most likely to be killed by HIV are those currently fighting infection. | ||
* In this replication process, the integrated [[provirus]] is copied to [[mRNA]] which is then [[Splicing (genetics)|spliced]] into smaller pieces. These small pieces produce the regulatory proteins [[HIV structure and genome#Tat|Tat]] (which encourages new virus production) and [[HIV structure and genome#Rev|Rev]]. As Rev accumulates it gradually starts to inhibit [[mRNA]] splicing.<ref name=Pollard> | |||
In this replication process, the integrated [[provirus]] is copied to [[mRNA]] which is then [[Splicing (genetics)|spliced]] into smaller pieces. These small pieces produce the regulatory proteins [[HIV structure and genome#Tat|Tat]] (which encourages new virus production) and [[HIV structure and genome#Rev|Rev]]. As Rev accumulates it gradually starts to inhibit [[mRNA]] splicing.<ref name=Pollard> | |||
{{cite journal | {{cite journal | ||
| author=Pollard, V. W. and Malim, M. H. | title=The HIV-1 Rev protein | journal=Annu. Rev. Microbiol. | year=1998 | pages=491-532 | volume=52 | issue= | pmid=9891806 | | author=Pollard, V. W. and Malim, M. H. | title=The HIV-1 Rev protein | journal=Annu. Rev. Microbiol. | year=1998 | pages=491-532 | volume=52 | issue= | pmid=9891806 | ||
}}</ref> At this stage, the structural proteins Gag and Env are produced from the full-length mRNA. The full-length RNA is actually the virus genome; it binds to the Gag protein and is packaged into new virus particles. | }}</ref> At this stage, the structural proteins Gag and Env are produced from the full-length mRNA. The full-length RNA is actually the virus genome; it binds to the Gag protein and is packaged into new virus particles. | ||
* HIV-1 and HIV-2 appear to package their [[RNA]] differently; HIV-1 will bind to any appropriate RNA whereas HIV-2 will preferentially bind to the mRNA which was used to create the [[Gag]] protein itself. This may mean that HIV-1 is better able to mutate (HIV-1 infection progresses to AIDS faster than HIV-2 infection and is responsible for the majority of global infections). | |||
HIV-1 and HIV-2 appear to package their RNA differently; HIV-1 will bind to any appropriate RNA whereas HIV-2 will preferentially bind to the mRNA which was used to create the Gag protein itself. This may mean that HIV-1 is better able to mutate (HIV-1 infection progresses to AIDS faster than HIV-2 infection and is responsible for the majority of global infections). | |||
===Assembly and release=== | ===Assembly and release=== | ||
The final step of the viral cycle, assembly of new HIV-1 virons, begins at the plasma membrane of the host cell. The Env polyprotein (gp160) goes through the [[endoplasmic reticulum]] and is transported to the [[Golgi apparatus|Golgi]] complex where it is cleaved by [[HIV-1 protease|protease]] and processed into the two HIV envelope glycoproteins gp41 and gp120. These are transported to the [[plasma membrane]] of the host cell where gp41 anchors the gp120 to the membrane of the infected cell. | * The final step of the viral cycle, assembly of new HIV-1 virons, begins at the plasma membrane of the host cell. The Env polyprotein (gp160) goes through the [[endoplasmic reticulum]] and is transported to the [[Golgi apparatus|Golgi]] complex where it is cleaved by [[HIV-1 protease|protease]] and processed into the two HIV envelope [[glycoproteins]] gp41 and gp120. These are transported to the [[plasma membrane]] of the [[host]] cell where gp41 anchors the [[gp120]] to the membrane of the infected [[cell]]. | ||
* The Gag (p55) and Gag-Pol (p160) polyproteins also associate with the inner surface of the plasma membrane along with the HIV genomic [[RNA]] as the forming virion begins to bud from the host [[cell]]. | |||
The Gag (p55) and Gag-Pol (p160) polyproteins also associate with the inner surface of the plasma membrane along with the HIV genomic RNA as the forming virion begins to bud from the host cell. | * Maturation either occurs in the forming bud or in the immature [[virion]] after it buds from the host cell. During maturation, HIV [[proteases]] cleave the [[polyproteins]] into individual functional HIV [[protein]]s and [[enzyme]]s. The various structural components then assemble to produce a mature HIV [[virion]].<ref name=Gelderblom>{{cite book | ||
| last = Gelderblom | first = H. R | year = 1997 | title = '''HIV Sequence Compendium''' | chapter = Fine structure of HIV and SIV | chapterurl = http://hiv.lanl.gov/content/hiv-db/COMPENDIUM/1997/partIII/Gelderblom.pdf | editor = Los Alamos National Laboratory (ed.) | edition = | pages = 31-44 | publisher = [[Los Alamos National Laboratory]] | location = [[Los Alamos, New Mexico]] | format = [[PDF|PDF format]]}}</ref> This cleavage step can be inhibited by protease inhibitors. The mature [[virus]] is then able to infect another [[cell]]. | |||
Maturation either occurs in the forming bud or in the immature virion after it buds from the host cell. During maturation, HIV proteases cleave the polyproteins into individual functional HIV | |||
| last = Gelderblom | first = H. R | year = 1997 | title = '''HIV Sequence Compendium''' | chapter = Fine structure of HIV and SIV | chapterurl = http://hiv.lanl.gov/content/hiv-db/COMPENDIUM/1997/partIII/Gelderblom.pdf | editor = Los Alamos National Laboratory (ed.) | edition = | pages = 31-44 | publisher = [[Los Alamos National Laboratory]] | location = [[Los Alamos, New Mexico]] | format = [[PDF|PDF format]]}}</ref> This cleavage step can be inhibited by protease inhibitors. The mature virus is then able to infect another cell. | |||
==Genetic variability== | ==Genetic variability== | ||
* HIV differs from many viruses in that it has very high genetic variability. This diversity is a result of its fast [[HIV#Life cycle of HIV|replication cycle]], with the generation of 10<sup>9</sup> to 10<sup>10</sup> virions every day, coupled with a high mutation rate of approximately 3 x 10<sup>-5</sup> per nucleotide base per cycle of replication and [[Genetic recombination|recombinogenic]] properties of reverse transcriptase.<ref name=RobertsonDL> | |||
HIV differs from many viruses in that it has very high genetic variability. This diversity is a result of its fast [[HIV#Life cycle of HIV|replication cycle]], with the generation of 10<sup>9</sup> to 10<sup>10</sup> virions every day, coupled with a high mutation rate of approximately 3 x 10<sup>-5</sup> per nucleotide base per cycle of replication and [[Genetic recombination|recombinogenic]] properties of reverse transcriptase.<ref name=RobertsonDL> | |||
{{cite journal | {{cite journal | ||
| author=Robertson DL, Hahn BH, Sharp PM. | title=Recombination in AIDS viruses | journal=J Mol Evol. | year=1995 | pages=249-259 | volume=40 | issue=3 | pmid=7723052 | | author=Robertson DL, Hahn BH, Sharp PM. | title=Recombination in AIDS viruses | journal=J Mol Evol. | year=1995 | pages=249-259 | volume=40 | issue=3 | pmid=7723052}}</ref> | ||
* This complex scenario leads to the generation of many variants of HIV in a single infected patient in the course of one day.<ref name=RobertsonDL/> This variability is compounded when a single [[cell]] is simultaneously infected by two or more different strains of HIV. When simultaneous infection occurs, the genome of progeny [[virions]] may be composed of [[RNA]] strands from two different strains. This hybrid [[virion]] then infects a new cell where it undergoes replication. As this happens, the [[reverse transcriptase]], by jumping back and forth between the two different [[RNA]] templates, will generate a newly synthesized retroviral [[DNA]] sequence that is a recombinant between the two parental genomes.<ref name=RobertsonDL/> This recombination is most obvious when it occurs between subtypes.<ref name=RobertsonDL/> | |||
}}</ref> This complex scenario leads to the generation of many variants of HIV in a single infected patient in the course of one day.<ref name=RobertsonDL/> This variability is compounded when a single cell is simultaneously infected by two or more different strains of HIV. When simultaneous infection occurs, the genome of progeny virions may be composed of RNA strands from two different strains. This hybrid virion then infects a new cell where it undergoes replication. As this happens, the reverse transcriptase, by jumping back and forth between the two different RNA templates, will generate a newly synthesized retroviral DNA sequence that is a recombinant between the two parental genomes.<ref name=RobertsonDL/> This recombination is most obvious when it occurs between subtypes.<ref name=RobertsonDL/> | * The closely related [[simian immunodeficiency virus]] (SIV) exhibits a somewhat different behavior: in its natural hosts, African green monkeys and sooty mangabeys, the retrovirus is present in high levels in the [[blood]], but evokes only a mild immune response,<ref>{{cite journal |author=Holzammer S, Holznagel E, Kaul A, Kurth R, Norley S |title=High virus loads in naturally and experimentally SIVagm-infected African green monkeys |journal=Virology |volume=283 |issue=2 |pages=324–31 |year=2001 |pmid=11336557 |doi=10.1006/viro.2001.0870}}</ref> does not cause the development of simian AIDS,<ref>Kurth, R. and Norley, S. (1996) Why don't the natural hosts of SIV develop simian AIDS?, ''J. NIH Res.'' '''8''', 33-37.</ref> and does not undergo the extensive mutation and recombination typical of HIV.<ref>{{cite journal |author=Baier M, Dittmar MT, Cichutek K, Kurth R |title=Development of vivo of genetic variability of simian immunodeficiency virus |journal=Proc. Natl. Acad. Sci. U.S.A. |volume=88 |issue=18 |pages=8126–30 |year=1991 |pmid=1896460 |doi=}}</ref> By contrast, infection of heterologous hosts (rhesus or cynomologus macaques) with SIV results in the generation of genetic diversity that is on the same order as HIV in infected humans; these heterologous hosts also develop simian AIDS.<ref>{{cite journal |author=Daniel MD, King NW, Letvin NL, Hunt RD, Sehgal PK, Desrosiers RC |title=A new type D retrovirus isolated from macaques with an immunodeficiency syndrome |journal=Science |volume=223 |issue=4636 |pages=602–5 |year=1984 |pmid=6695172 |doi=10.1126/science.6695172}}</ref> The relationship, if any, between genetic diversification, [[immune]] response, and disease progression is unknown. | ||
* Three groups of HIV-1 have been identified on the basis of differences in ''env'': M, N, and O.<ref name=Thomson> | |||
The closely related [[simian immunodeficiency virus]] (SIV) exhibits a somewhat different behavior: in its natural hosts, African green monkeys and sooty mangabeys, the retrovirus is present in high levels in the blood, but evokes only a mild immune response,<ref>{{cite journal |author=Holzammer S, Holznagel E, Kaul A, Kurth R, Norley S |title=High virus loads in naturally and experimentally SIVagm-infected African green monkeys |journal=Virology |volume=283 |issue=2 |pages=324–31 |year=2001 |pmid=11336557 |doi=10.1006/viro.2001.0870}}</ref> does not cause the development of simian AIDS,<ref>Kurth, R. and Norley, S. (1996) Why don't the natural hosts of SIV develop simian AIDS?, ''J. NIH Res.'' '''8''', 33-37.</ref> and does not undergo the extensive mutation and recombination typical of HIV.<ref>{{cite journal |author=Baier M, Dittmar MT, Cichutek K, Kurth R |title=Development of vivo of genetic variability of simian immunodeficiency virus |journal=Proc. Natl. Acad. Sci. U.S.A. |volume=88 |issue=18 |pages=8126–30 |year=1991 |pmid=1896460 |doi=}}</ref> By contrast, infection of heterologous hosts (rhesus or cynomologus macaques) with SIV results in the generation of genetic diversity that is on the same order as HIV in infected humans; these heterologous hosts also develop simian AIDS.<ref>{{cite journal |author=Daniel MD, King NW, Letvin NL, Hunt RD, Sehgal PK, Desrosiers RC |title=A new type D retrovirus isolated from macaques with an immunodeficiency syndrome |journal=Science |volume=223 |issue=4636 |pages=602–5 |year=1984 |pmid=6695172 |doi=10.1126/science.6695172}}</ref> The relationship, if any, between genetic diversification, immune response, and disease progression is unknown. | |||
Three groups of HIV-1 have been identified on the basis of differences in ''env'': M, N, and O.<ref name=Thomson> | |||
{{cite journal | {{cite journal | ||
| author=Thomson, M. M., Perez-Alvarez, L. and Najera, R. | title=Molecular epidemiology of HIV-1 genetic forms and its significance for vaccine development and therapy | journal=Lancet Infect. Dis. | year=2002 | pages=461-471 | volume=2 | issue=8 | pmid=12150845 | | author=Thomson, M. M., Perez-Alvarez, L. and Najera, R. | title=Molecular epidemiology of HIV-1 genetic forms and its significance for vaccine development and therapy | journal=Lancet Infect. Dis. | year=2002 | pages=461-471 | volume=2 | issue=8 | pmid=12150845 | ||
}}</ref> Group M is the most prevalent and is subdivided into eight subtypes (or [[clade]]s), based on the whole genome, which are geographically distinct.<ref name=Carr>{{cite book | }}</ref> Group M is the most prevalent and is subdivided into eight subtypes (or [[clade]]s), based on the whole genome, which are geographically distinct.<ref name=Carr>{{cite book | ||
| last = Carr | first = J. K. | coauthors = Foley, B. T., Leitner, T., Salminen, M., Korber, B. and McCutchan, F. | year = 1998 | title = '''HIV Sequence Compendium''' | chapter = Reference Sequences Representing the Principal Genetic Diversity of HIV-1 in the Pandemic | chapterurl = http://hiv.lanl.gov/content/hiv-db/COMPENDIUM/1998/III/Carr.pdf | editor = Los Alamos National Laboratory (ed.) | edition = | pages = 10-19 | publisher = [[Los Alamos National Laboratory]] | location = [[Los Alamos, New Mexico]] | format=[[PDF|PDF format]]}}</ref> The most prevalent are subtypes B (found mainly in North America and Europe), A and D (found mainly in Africa), and C (found mainly in Africa and Asia); these subtypes form branches in the phylogenetic tree representing the lineage of the M group of HIV-1. Coinfection with distinct subtypes gives rise to circulating recombinant forms (CRFs). In 2000, the last year in which an analysis of global subtype prevalence was made, 47.2% of infections worldwide were of subtype C, 26.7% were of subtype A/CRF02_AG, 12.3% were of subtype B, 5.3% were of subtype D, 3.2% were of CRF_AE, and the remaining 5.3% were composed of other subtypes and CRFs.<ref name=Osmanov> | | last = Carr | first = J. K. | coauthors = Foley, B. T., Leitner, T., Salminen, M., Korber, B. and McCutchan, F. | year = 1998 | title = '''HIV Sequence Compendium''' | chapter = Reference Sequences Representing the Principal Genetic Diversity of HIV-1 in the Pandemic | chapterurl = http://hiv.lanl.gov/content/hiv-db/COMPENDIUM/1998/III/Carr.pdf | editor = Los Alamos National Laboratory (ed.) | edition = | pages = 10-19 | publisher = [[Los Alamos National Laboratory]] | location = [[Los Alamos, New Mexico]] | format=[[PDF|PDF format]]}}</ref> The most prevalent are subtypes B (found mainly in North America and Europe), A and D (found mainly in Africa), and C (found mainly in Africa and Asia); these subtypes form branches in the phylogenetic tree representing the lineage of the M group of HIV-1. Coinfection with distinct subtypes gives rise to circulating recombinant forms (CRFs). In 2000, the last year in which an analysis of global subtype prevalence was made, 47.2% of infections worldwide were of subtype C, 26.7% were of subtype A/CRF02_AG, 12.3% were of subtype B, 5.3% were of subtype D, 3.2% were of CRF_AE, and the remaining 5.3% were composed of other subtypes and CRFs.<ref name=Osmanov> | ||
{{cite journal | {{cite journal | ||
| author=Osmanov S, Pattou C, Walker N, Schwardlander B, Esparza J; WHO-UNAIDS Network for HIV Isolation and Characterization. | title=Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000 | journal=Acquir. Immune. Defic. Syndr. | year=2002 | pages=184-190 | volume=29 | issue=2 | pmid=11832690 | | author=Osmanov S, Pattou C, Walker N, Schwardlander B, Esparza J; WHO-UNAIDS Network for HIV Isolation and Characterization. | title=Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000 | journal=Acquir. Immune. Defic. Syndr. | year=2002 | pages=184-190 | volume=29 | issue=2 | pmid=11832690 | ||
}}</ref> Most HIV-1 research is focused on subtype B; few laboratories focus on the other subtypes.<ref name=Perrin> | }}</ref> Most HIV-1 research is focused on subtype B; few laboratories focus on the other subtypes.<ref name=Perrin> | ||
{{cite journal | {{cite journal | ||
| author=Perrin L, Kaiser L, Yerly S. | title=Travel and the spread of HIV-1 genetic variants | journal=Lancet Infect Dis. | year=2003 | pages=22-27 | volume=3 | issue=1 | pmid=12505029 | | author=Perrin L, Kaiser L, Yerly S. | title=Travel and the spread of HIV-1 genetic variants | journal=Lancet Infect Dis. | year=2003 | pages=22-27 | volume=3 | issue=1 | pmid=12505029 | ||
}}</ref> | }}</ref> | ||
* The genetic sequence of HIV-2 is only partially homologous to HIV-1 and more closely resembles that of SIV than HIV-1. | |||
The genetic sequence of HIV-2 is only partially homologous to HIV-1 and more closely resembles that of SIV than HIV-1. | {| style="float:center" | ||
| [[Image:HIV-SIV-phylogenetic-tree.png|thumb|left|300px|The [[phylogenetic tree]] of the SIV and HIV (click on image for a detailed description). - By Kuiken, C., Foley, B., Hahn, B., Marx, P., McCutchan, F., Mellors, J. W., Mullins, J., Wolinsky, S. & Korber, B. (1999). A compilation and analysis of nucleic acid and amino acid sequences. In Human Retroviruses and AIDS. Los Alamos, New Mexico: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory. - Theoretical Biology and Biophysics Group, Los Alamos National Laboratory.http://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/99compendium.htmlhttp://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/1999/1/intro.pdf, Public Domain, https://commons.wikimedia.org/w/index.php?curid=3915158]] | |||
| [[Image:subtype.png|thumb|left|500px|Map showing HIV-1 subtype prevalence. The bigger the pie chart, the more infections are present. - By The original uploader was Grcampbell at English WikipediaLater version(s) were uploaded by Renegadeviking, Emmojo666 at en.wikipedia.(Original text: en.User:Grcampbell) - Based on Osmanov S, Pattou C, Walker N, Schwardlander B, Esparza J; WHO-UNAIDS Network for HIV Isolation and Characterization. (2002) Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J Acquir Immune Defic Syndr. 29(2):184-90., CC BY-SA 3.0, https://commons.wikimedia.org/w/index.php?curid=3745709]] | |||
[[Image: | |||
| | |||
|} | |} | ||
==References== | ==References== | ||
| Line 874: | Line 190: | ||
==External links== | ==External links== | ||
*[http:// | *[http://pathmicro.med.sc.edu/lecture/hiv9.htm Genome at sc.edu] | ||
[[Category:HIV/AIDS]] | [[Category:HIV/AIDS]] | ||
| Line 927: | Line 196: | ||
[[Category:Sexually transmitted diseases]] | [[Category:Sexually transmitted diseases]] | ||
[[Category:Immunodeficiency]] | [[Category:Immunodeficiency]] | ||
Latest revision as of 21:45, 22 September 2017
| https://https://www.youtube.com/watch?v=5g1ijpBI6Dk%7C350}} |
|
AIDS Microchapters |
|
Diagnosis |
|
Treatment |
|
Case Studies |
|
Human Immunodeficiency Virus (HIV) On the Web |
|
American Roentgen Ray Society Images of Human Immunodeficiency Virus (HIV) |
|
Risk calculators and risk factors for Human Immunodeficiency Virus (HIV) |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]; Associate Editor(s)-in-Chief: Ammu Susheela, M.D. [2], Alejandro Lemor, M.D. [3]
Overview
AIDS is caused by the human immunodeficiency virus (HIV). HIV is a retrovirus classified into the family of Retroviridae and the sub family orthoretroviridae.[1]. Two main subspecies of HIV exist: HIV-1, and HIV-2. HIV-1 is composed of two copies of single-stranded RNA enclosed by a conical capsid comprising the viral protein p24. The genome consists of several major genes that code for structural and functional proteins. These include the gag, pol, env, tat, and nef genes. The genome and proteins of HIV have been the subject of extensive research since the discovery of the virus in 1983. It is a well known fact that no two HIV genomes are the same, not even from the same person, causing some to speculate that HIV is a "quasispecies" of a virus.[2] A major requirement for all retroviruses is reverse transcriptase that transcribes the viral RNA into double-stranded DNA and integrase that integrates this newly formed DNA into the host genome.
Taxonomy
- HIV 1 and HIV 2 are classified into the family of Retroviridae and sub family orthoretroviridae.[1]
- Retrovirus are enveloped RNA viruses which requires a DNA intermediate to replicate.
- HIV 1 and HIV 2 belongs to the genus Lentivirus (Lentus which in latin means slow)
- The retrovirus rely on enzyme reverse transcriptase to transcribe their genome from RNA to DNA.
- Integrase incorporates the DNA into the host DNA and becomes a part of cellular DNA replicating with it.
Origin
- Both HIV-1 and HIV-2 are of primate origin. The origin of HIV-1 is the Central Common Chimpanzee (Pan troglodytes troglodytes) found in southern Cameroon.[3]
- It is believed that HIV-2 originated from the Sooty Mangabey (Cercocebus atys), an Old World monkey of Guinea Bissau, Gabon, and Cameroon.
- Most experts believe that HIV probably transferred to humans as a result of direct contact with primates, for instance during hunting or butchery.[4]
Structure

- HIV is different in structure from other retroviruses. It is around 120 nm in diameter (120 billionths of a meter; around 60 times smaller than a red blood cell) and roughly spherical.
- HIV-1 is composed of two copies of single-stranded RNA enclosed by a conical capsid comprising the viral protein p24, typical of lentiviruses (Figure 1). The RNA component is 9749 nucleotides long. This is in turn surrounded by a plasma membrane of host-cell origin. The single-strand RNA is tightly bound to the nucleocapsid proteins, p7 and enzymes that are indispensable for the development of the virion, such as reverse transcriptase and integrase. The nucleocapsid (p7 and p6) associates with the genomic RNA (one molecule per hexamer) and protects the RNA from digestion by nucleases. A matrix composed of an association of the viral protein p17 surrounds the capsid, ensuring the integrity of the virion particle. Also enclosed within the virion particle are Vif, Vpr, Nef, p7 and viral protease (Figure 1). The envelope is formed when the capsid buds from the host cell, taking some of the host-cell membrane with it. The envelope includes the glycoproteins gp120 and gp41.
- Recently, an Anglo-German team compiled a 3D structure of HIV by combining multiple images. It is hoped that this new information would contribute to scientific understanding of the virus, and help in the creation of a cure. Oxford University's Professor Stephen D. Fuller said the 3D map would assist in understanding how the virus grows. [5] The validity of this work remains a matter of debate [6], with a conflicting model produced by another team led by Florida State University Professor Kenneth Roux in the US [7].
Genome organization
- HIV has several major genes coding for structural proteins that are found in all retroviruses, and several nonstructural ("accessory") genes that are unique to HIV. The gag gene provides the basic physical infrastructure of the virus, and pol provides the basic mechanism by which retroviruses reproduce, while the others help HIV to enter the host cell and enhance its reproduction. Though they may be altered by mutation, all of these genes except tev exist in all known variants of HIV; see Genetic variability of HIV.
- pol: Codes for viral enzymes, the most important of which are reverse transcriptase, integrase, and protease which cleaves the proteins derived from gag and pol into functional proteins.
Protein function
Gag
p24
- p24 makes up the viral capsid.
- When a Western blot test is used to detect HIV infection, p24 is one of the three major proteins tested for, along with gp120/gp160 and gp41.
p6, p7, and p17
- p6 and p7 provide the nucleocapsid.
- p17 provides a protective matrix.
Pol
Reverse transcriptase
- Common to all retroviruses, this enzyme transcribes the viral RNA into double-stranded DNA.
Integrase
Protease
- A protease is any enzyme that cuts proteins into segments. HIV's gag and pol genes do not produce their proteins in their final form, but as larger combination proteins; the specific protease used by HIV cleaves these into separate functional units. Protease inhibitor drugs block this step.
Env
- The env gene does not actually code for gp120 and gp41, but for a precursor to both, gp160. During HIV reproduction, the host cell's own enzymes cleave gp160 into gp120 and gp41. See Replication cycle of HIV.
gp120
- Exposed on the surface of the viral envelope, the glycoprotein gp120 binds to the CD4 receptor on any target cell that has such a receptor, particularly the helper T-cell. See HIV tropism and Replication cycle of HIV.
- Since CD4 receptor binding is the most obvious step in HIV infection, gp120 was among the first targets of HIV vaccine research. These efforts have been hampered by its chemical properties, which make it difficult for antibodies to bind to gp120; also, it can easily be shed from the virus due to its loose binding with gp41.
gp41
- The glycoprotein gp41 is non-covalently bound to gp120, and provides the second step by which HIV enters the cell. It is originally buried within the viral envelope, but when gp120 binds to a CD4 receptor, gp120 changes its conformation causing gp41 to become exposed, where it can assist in fusion with the host cell.
- Fusion inhibitor drugs such as enfuvirtide block the fusion process by binding to gp41.
Transactivators
Tat
- Stands for "Trans-Activator of Transcription". Tat consists of between 86 and 101 amino acids depending on the subtype.[8] Tat helps HIV reproduce by compensating for a defect in its genome: the HIV RNA initially has a hairpin-structured portion which prevents full transcription occurring. However, a small number of RNA transcripts will be made, which allow the Tat protein to be produced. Tat then binds to and phosphorylates cellular factors, eliminating the effect of the hairpin RNA structure and allowing transcription of the HIV DNA.[9] This itself increases the rate of transcription, providing a positive feedback cycle. This in turn allows HIV to have an explosive response once a threshold amount of Tat is produced, a useful tool for defeating the body's response. Tat also appears to play a more direct role in the HIV disease process. The protein is released by infected cells in culture, and is found in the blood of HIV-1 infected patients.[10] It can be absorbed by cells that are not infected with HIV, and can act directly as a toxin producing cell death via apoptosis in uninfected "bystander" T cells, assisting in progression toward AIDS.[11] By interacting with the CXCR4 receptor, Tat also appears to encourage the reproduction of less virulent M-tropic strains of HIV early in the course of infection, allowing the more rapidly pathogenic T-tropic strains to emerge later.[10]
Rev
 |
- Stands for "Regulator of Virion". This protein allows fragments of HIV mRNA that contain a Rev Response Unit (RRE) to be exported from the nucleus to the cytoplasm. In the absence of the rev gene, RNA splicing machinery in the nucleus quickly splices the RNA so that only the smaller, regulatory proteins can be produced; in the presence of rev, RNA is exported from the nucleus before it can be spliced, so that the structural proteins and RNA genome can be produced. Again, this mechanism allows a positive feedback loop to allow HIV to overwhelm the host's defenses, and provides time-dependent regulation of replication (a common process in viral infections)[12]
Vpr
- Stands for "Viral Protein R". Vpr, a 96 amino acid 14-kDa protein, plays an important role in regulating nuclear import of the HIV-1 pre-integration complex, and is required for virus replication in non-dividing cells such as macrophages. Vpr also induces cell cycle arrest and apoptosis in proliferating cells, which can result in immune dysfunction.[13][14]
- Vpr is also immunosuppressive due to its ability to sequester a proinflammatory transcriptional activator in the cytoplasm. HIV-2 contains both a Vpr protein and a related (by sequence homology) Vpx protein (Viral Protein X). Two functions of Vpr in HIV-1 are split between Vpr and Vpx in HIV-2, with the HIV-2 Vpr protein inducing cell cycle arrest and the Vpx protein required for nuclear import.
Other regulatory proteins
Nef
- Stands for "Negative Regulatory Factor". The expression of Nef early in the viral life cycle ensures T cell activation and the establishment of a persistent state of infection, two basic attributes of HIV infection. Nef also promotes the survival of infected cells by downmodulating the expression of several surface molecules important in host immune function. These include major histocompatibility complex-I (MHC I) and MHC II present on antigen presenting cells (APCs) and target cells, CD4 and CD28 present on CD4+ T cells. One group of patients in Sydney were infected with a nef-deleted virus and took much longer than expected to progress to AIDS.[15]
- A nef-deleted virus vaccine has not been trialed in humans and has failed in nonhuman animals.HIV-1 Nef-induced FasL induction and bystander killing requires p38 MAPK activation.
Vif
- Stands for "Viral infectivity factor". Vif is a 23-kilodalton protein that is essential for viral replication.[12] Vif inhibits the cellular protein, APOBEC3G, from entering the virion during budding from a host cell by targeting it for proteasomal degredation. Vif hijacks the cellular Cullin5 E3 ubiquitin ligase in order to target APOBEC3G for degradation. In the absence of Vif, APOBEC3G causes hypermutation of the viral genome, rendering it dead-on-arrival at the next host cell. APOBEC3G is thus a host defence to retroviral infection which HIV-1 has overcome by the acquisition of Vif.
Vpu
- Stands for "Viral Protein U". Vpu is involved in viral budding, enhancing virion release from the cell.
Tropism
- HIV tropism refers to the cell type that the human immunodeficiency virus (HIV) infects and replicates in. HIV tropism of a patient's virus is measured by the Trofile assay.
- HIV can infect a variety of cells such as CD4+ helper T-cells and macrophages that express the CD4 molecule on their surface. HIV-1 entry to macrophages and T helper cells is mediated not only through interaction of the virion envelope glycoproteins (gp120) with the CD4 molecule on the target cells but also with its chemokine coreceptors.
- Macrophage (M-tropic) strains of HIV-1, or non-syncitia-inducing strains (NSI) use the beta-chemokine receptor CCR5 for entry and are thus able to replicate in macrophages and CD4+ T-cells [16]. The normal ligands for this receptor, RANTES, macrophage inflammatory protein (MIP)-1-beta and MIP-1-alpha, are able to suppress HIV-1 infection in vitro. This CCR5 coreceptor is used by almost all primary HIV-1 isolates regardless of viral genetic subtype.
- T-tropic isolates, or syncitia-inducing (SI) strains replicate in primary CD4+ T-cells as well as in macrophages and use the alpha-chemokine receptor, CXCR4, for entry [16]. The alpha-chemokine, SDF-1, a ligand for CXCR4, suppresses replication of T-tropic HIV-1 isolates. It does this by down regulating the expression of CXCR4 on the surface of these cells.
- Viruses that use only the CCR5 receptor are termed R5, those that only use CXCR4 are termed X4, and those that use both, X4R5. However, the use of coreceptor alone does not explain viral tropism, as not all R5 viruses are able to use CCR5 on macrophages for a productive infection [16].
- HIV can also infect a subtype of dendritic cells [17], MDC-1, which probably constitute a major reservoir that maintains infection when T helper cell numbers have declined to extremely low levels.
Replication cycle
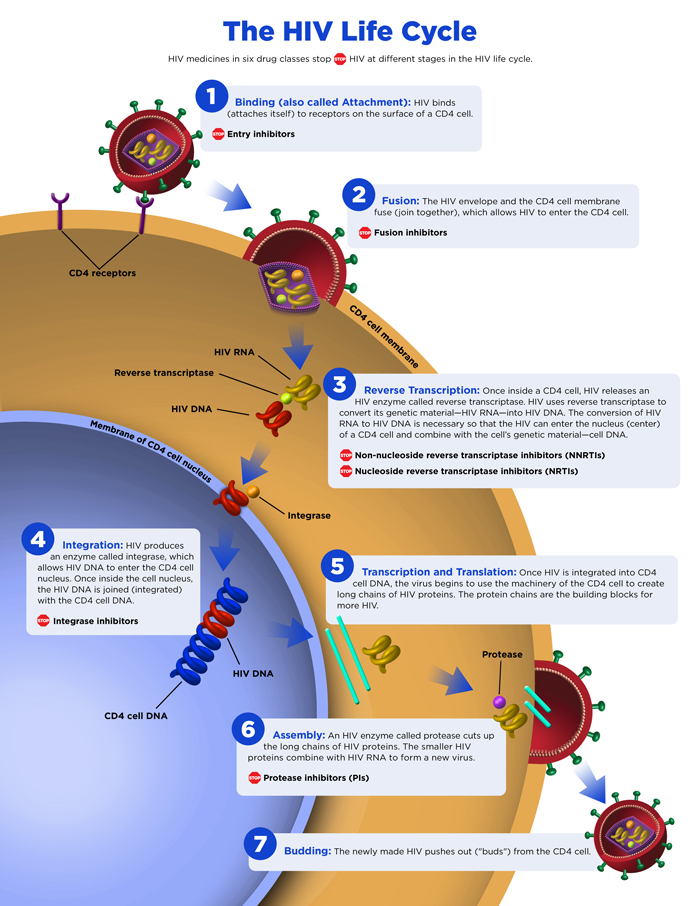
Steps in the HIV Replication Cycle
- Fusion of the HIV cell to the host cell surface.
- HIV RNA, reverse transcriptase, integrase, and other viral proteins enter the host cell.
- Viral DNA is formed by reverse transcription.
- Viral DNA is transported across the nucleus and integrates into the host DNA.
- New viral RNA is used as genomic RNA and to make viral proteins.
- New viral RNA and proteins move to cell surface and a new, immature, HIV virus forms.
- The virus matures by protease releasing individual HIV proteins.
 |
 |
Entry to the cell
- HIV enters macrophages and CD4+ T cells by the adsorption of glycoproteins on its surface to receptors on the target cell followed by fusion of the viral envelope with the cell membrane and the release of the HIV capsid into the cell.[18][19]
- Entry to the cell begins through interaction of the trimeric envelope complex (gp160 spike, discussed above) and both CD4 and a chemokine receptor (generally either CCR5 or CXCR4, but others are known to interact) on the cell surface.[18][19] The gp160 spike contains binding domains for both CD4 and chemokine receptors.[18][19] The first step in fusion involves the high-affinity attachment of the CD4 binding domains of gp120 to CD4. Once gp120 is bound with the CD4 protein, the envelope complex undergoes a structural change, exposing the chemokine binding domains of gp120 and allowing them to interact with the target chemokine receptor.[18][19] This allows for a more stable two-pronged attachment, which allows the N-terminal fusion peptide gp41 to penetrate the cell membrane.[18][19] Repeat sequences in gp41, HR1 and HR2 then interact, causing the collapse of the extracellular portion of gp41 into a hairpin. This loop structure brings the virus and cell membranes close together, allowing fusion of the membranes and subsequent entry of the viral capsid.[18][19]
- Once HIV has bound to the target cell, the HIV RNA and various enzymes, including reverse transcriptase, integrase, ribonuclease and protease, are injected into the cell.[18]
- HIV can infect dendritic cells (DCs) by this CD4-CCR5 route, but another route using mannose-specific C-type lectin receptors such as DC-SIGN can also be used.[20] DCs are one of the first cells encountered by the virus during sexual transmission. They are currently thought to play an important role by transmitting HIV to T cells once the virus has been captured in the mucosa by DCs.[20]
Replication and transcription
- Once the viral capsid enters the cell, an enzyme called reverse transcriptase liberates the single-stranded (+)RNA from the attached viral proteins and copies it into a complementary DNA.[21] This process of reverse transcription is extremely error-prone and it is during this step that mutations may occur. Such mutations may cause drug resistance. The reverse transcriptase then makes a complementary DNA strand to form a double-stranded viral DNA intermediate (vDNA). This vDNA is then transported into the cell nucleus. The integration of the viral DNA into the host cell's genome is carried out by another viral enzyme called integrase.[21]
- This integrated viral DNA may then lie dormant, in the latent stage of HIV infection.[21] To actively produce the virus, certain cellular transcription factors need to be present, the most important of which is NF-κB (NF kappa B), which is upregulated when T cells become activated.[22] This means that those cells most likely to be killed by HIV are those currently fighting infection.
- In this replication process, the integrated provirus is copied to mRNA which is then spliced into smaller pieces. These small pieces produce the regulatory proteins Tat (which encourages new virus production) and Rev. As Rev accumulates it gradually starts to inhibit mRNA splicing.[23] At this stage, the structural proteins Gag and Env are produced from the full-length mRNA. The full-length RNA is actually the virus genome; it binds to the Gag protein and is packaged into new virus particles.
- HIV-1 and HIV-2 appear to package their RNA differently; HIV-1 will bind to any appropriate RNA whereas HIV-2 will preferentially bind to the mRNA which was used to create the Gag protein itself. This may mean that HIV-1 is better able to mutate (HIV-1 infection progresses to AIDS faster than HIV-2 infection and is responsible for the majority of global infections).
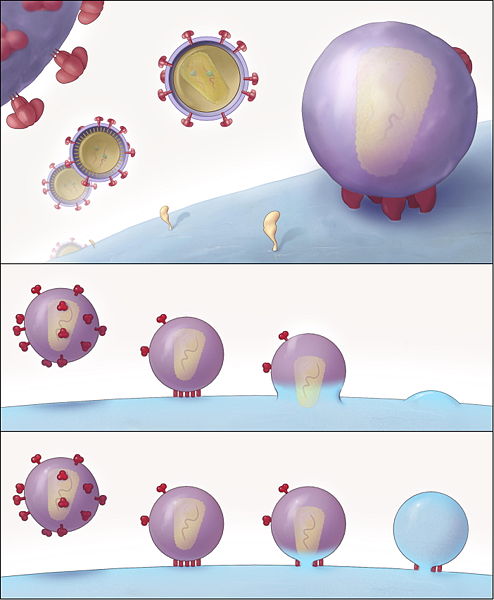
Assembly and release
- The final step of the viral cycle, assembly of new HIV-1 virons, begins at the plasma membrane of the host cell. The Env polyprotein (gp160) goes through the endoplasmic reticulum and is transported to the Golgi complex where it is cleaved by protease and processed into the two HIV envelope glycoproteins gp41 and gp120. These are transported to the plasma membrane of the host cell where gp41 anchors the gp120 to the membrane of the infected cell.
- The Gag (p55) and Gag-Pol (p160) polyproteins also associate with the inner surface of the plasma membrane along with the HIV genomic RNA as the forming virion begins to bud from the host cell.
- Maturation either occurs in the forming bud or in the immature virion after it buds from the host cell. During maturation, HIV proteases cleave the polyproteins into individual functional HIV proteins and enzymes. The various structural components then assemble to produce a mature HIV virion.[24] This cleavage step can be inhibited by protease inhibitors. The mature virus is then able to infect another cell.
Genetic variability
- HIV differs from many viruses in that it has very high genetic variability. This diversity is a result of its fast replication cycle, with the generation of 109 to 1010 virions every day, coupled with a high mutation rate of approximately 3 x 10-5 per nucleotide base per cycle of replication and recombinogenic properties of reverse transcriptase.[25]
- This complex scenario leads to the generation of many variants of HIV in a single infected patient in the course of one day.[25] This variability is compounded when a single cell is simultaneously infected by two or more different strains of HIV. When simultaneous infection occurs, the genome of progeny virions may be composed of RNA strands from two different strains. This hybrid virion then infects a new cell where it undergoes replication. As this happens, the reverse transcriptase, by jumping back and forth between the two different RNA templates, will generate a newly synthesized retroviral DNA sequence that is a recombinant between the two parental genomes.[25] This recombination is most obvious when it occurs between subtypes.[25]
- The closely related simian immunodeficiency virus (SIV) exhibits a somewhat different behavior: in its natural hosts, African green monkeys and sooty mangabeys, the retrovirus is present in high levels in the blood, but evokes only a mild immune response,[26] does not cause the development of simian AIDS,[27] and does not undergo the extensive mutation and recombination typical of HIV.[28] By contrast, infection of heterologous hosts (rhesus or cynomologus macaques) with SIV results in the generation of genetic diversity that is on the same order as HIV in infected humans; these heterologous hosts also develop simian AIDS.[29] The relationship, if any, between genetic diversification, immune response, and disease progression is unknown.
- Three groups of HIV-1 have been identified on the basis of differences in env: M, N, and O.[30] Group M is the most prevalent and is subdivided into eight subtypes (or clades), based on the whole genome, which are geographically distinct.[31] The most prevalent are subtypes B (found mainly in North America and Europe), A and D (found mainly in Africa), and C (found mainly in Africa and Asia); these subtypes form branches in the phylogenetic tree representing the lineage of the M group of HIV-1. Coinfection with distinct subtypes gives rise to circulating recombinant forms (CRFs). In 2000, the last year in which an analysis of global subtype prevalence was made, 47.2% of infections worldwide were of subtype C, 26.7% were of subtype A/CRF02_AG, 12.3% were of subtype B, 5.3% were of subtype D, 3.2% were of CRF_AE, and the remaining 5.3% were composed of other subtypes and CRFs.[32] Most HIV-1 research is focused on subtype B; few laboratories focus on the other subtypes.[33]
- The genetic sequence of HIV-2 is only partially homologous to HIV-1 and more closely resembles that of SIV than HIV-1.
 |
 |
References
- ↑ 1.0 1.1 "HIV monograph" (PDF).
- ↑ WainHobson, S., 1989. HIV genome variability in vivo. AIDS 3: supp 1; 139.
- ↑ Keele BF, van Heuverswyn F, Li YY; et al. (2006). "Chimpanzee Reservoirs of Pandemic and Nonpandemic HIV-1". Science. 313 (5786): 523–6. doi:10.1126/science.1126531. PMID 16728595.
- ↑ Cohen J (2000). "Vaccine Theory of AIDS Origins Disputed at Royal Society". Science. 289 (5486): 1850&ndash, 1851. doi:10.1126/science.289.5486.1850. PMID 11012346.
- ↑ BBC News: 3D Structure of HIV Revealed
- ↑ The SIV Surface Spike Imaged by Electron Tomography: One Leg or Three? Subramaniam S PLoS Pathogens Vol. 2, No. 8, e91 doi:10.1371/journal.ppat.0020091
- ↑ Distribution and three-dimensional structure of AIDS virus envelope spikes Ping Zhu, Jun Liu, Julian Bess, Jr, Elena Chertova, Jeffrey D. Lifson, Henry Grisé, Gilad A. Ofek, Kenneth A. Taylor and Kenneth H. Roux, Nature 441, 847-852 (15 June 2006) | doi:10.1038/nature04817; Received 8 March 2006; Accepted 24 April 2006; Published online 24 May 2006
- ↑ Jeang, K. T. (1996) In: Human Retroviruses and AIDS: A Compilation and Analysis of Nucleic Acid and Amino Acid Sequences. Los Alamos National Laboratory (Ed.) pp. III-3–III-18
- ↑ Kim JB, Sharp PA. (2001) Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J. Biol. Chem. 276, 12317-12323 PMID 11145967
- ↑ 10.0 10.1 Xiao, H., Neuveut, C., Tiffany, H. L., Benkirane, M., Rich, E. A., Murphy, P. M. and Jeang, K. T. (2000) Selective CXCR4 antagonism by Tat: implications for in vivo expansion of coreceptor use by HIV-1. Proc. Natl. Acad. Sci. U.S.A. 97, 11466-11471 PMID 11027346
- ↑ Campbell GR, Pasquier E, Watkins J, Bourgarel-Rey V, Peyrot V, Esquieu D, Barbier P, de Mareuil J, Braguer D, Kaleebu P, Yirrell DL, Loret EP. (2004) The glutamine-rich region of the HIV-1 Tat protein is involved in T-cell apoptosis. J. Biol. Chem. 279, 48197-48204 PMID 15331610
- ↑ 12.0 12.1 Strebel, K (2003) Virus-host interactions: role of HIV proteins Vif, Tat, and Rev. AIDS 17 Suppl 4, S25-S34 PMID 15080177
- ↑ Bukrinsky M, Adzhubei A. (1999) Viral protein R of HIV-1. Rev Med Virol 9, 39-49 PMID 10371671
- ↑ Muthumani K., et al (2006) The HIV-1 Vpr and glucocorticoid receptor complex: A gain of function interaction that prevents the nuclear localization of PARP-1. Nat Cell Biol. Feb;8(2):170-9.
- ↑ Learmont JC, Geczy AF, Mills J, Ashton LJ, Raynes-Greenow CH, Garsia RJ, Dyer WB, McIntyre L, Oelrichs RB, Rhodes DI, Deacon NJ, Sullivan JS. (1999) Immunologic and virologic status after 14 to 18 years of infection with an attenuated strain of HIV-1. A report from the Sydney Blood Bank Cohort. N Engl J Med 340, 1715-1722 PMID 10352163
- ↑ 16.0 16.1 16.2 Coakley, E., Petropoulos, C. J. and Whitcomb, J. M. (2005). "Assessing chemokine co-receptor usage in HIV". Curr. Opin. Infect. Dis. 18 (1): 9–15. PMID 15647694.
- ↑ Knight, S. C., Macatonia, S. E. and Patterson, S. (1990). "HIV I infection of dendritic cells". Int. Rev. Immunol. 6 (2–3): 163–175. PMID 2152500.
- ↑ 18.0 18.1 18.2 18.3 18.4 18.5 18.6 Chan D, Kim P (1998). "HIV entry and its inhibition". Cell. 93 (5): 681–4. PMID 9630213.
- ↑ 19.0 19.1 19.2 19.3 19.4 19.5 Wyatt R, Sodroski J (1998). "The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens". Science. 280 (5371): 1884–8. doi:10.1126/science.280.5371.1884. PMID 9632381.
- ↑ 20.0 20.1 Pope M, Haase A (2003). "Transmission, acute HIV-1 infection and the quest for strategies to prevent infection". Nat Med. 9 (7): 847–52. PMID 12835704.
- ↑ 21.0 21.1 21.2 Zheng, Y. H., Lovsin, N. and Peterlin, B. M. (2005). "Newly identified host factors modulate HIV replication". Immunol. Lett. 97 (2): 225–234. PMID 15752562.
- ↑ Hiscott J, Kwon H, Genin P. (2001). "Hostile takeovers: viral appropriation of the NF-kappaB pathway". J Clin Invest. 107 (2): 143–151. PMID 11160127.
- ↑ Pollard, V. W. and Malim, M. H. (1998). "The HIV-1 Rev protein". Annu. Rev. Microbiol. 52: 491–532. PMID 9891806.
- ↑ Gelderblom, H. R (1997). "Fine structure of HIV and SIV" (PDF). In Los Alamos National Laboratory (ed.). HIV Sequence Compendium (PDF format)
|format=requires|url=(help). Los Alamos, New Mexico: Los Alamos National Laboratory. pp. 31–44. - ↑ 25.0 25.1 25.2 25.3 Robertson DL, Hahn BH, Sharp PM. (1995). "Recombination in AIDS viruses". J Mol Evol. 40 (3): 249–259. PMID 7723052.
- ↑ Holzammer S, Holznagel E, Kaul A, Kurth R, Norley S (2001). "High virus loads in naturally and experimentally SIVagm-infected African green monkeys". Virology. 283 (2): 324–31. doi:10.1006/viro.2001.0870. PMID 11336557.
- ↑ Kurth, R. and Norley, S. (1996) Why don't the natural hosts of SIV develop simian AIDS?, J. NIH Res. 8, 33-37.
- ↑ Baier M, Dittmar MT, Cichutek K, Kurth R (1991). "Development of vivo of genetic variability of simian immunodeficiency virus". Proc. Natl. Acad. Sci. U.S.A. 88 (18): 8126–30. PMID 1896460.
- ↑ Daniel MD, King NW, Letvin NL, Hunt RD, Sehgal PK, Desrosiers RC (1984). "A new type D retrovirus isolated from macaques with an immunodeficiency syndrome". Science. 223 (4636): 602–5. doi:10.1126/science.6695172. PMID 6695172.
- ↑ Thomson, M. M., Perez-Alvarez, L. and Najera, R. (2002). "Molecular epidemiology of HIV-1 genetic forms and its significance for vaccine development and therapy". Lancet Infect. Dis. 2 (8): 461–471. PMID 12150845.
- ↑ Carr, J. K. (1998). "Reference Sequences Representing the Principal Genetic Diversity of HIV-1 in the Pandemic" (PDF). In Los Alamos National Laboratory (ed.). HIV Sequence Compendium (PDF format)
|format=requires|url=(help). Los Alamos, New Mexico: Los Alamos National Laboratory. pp. 10–19. Unknown parameter|coauthors=ignored (help) - ↑ Osmanov S, Pattou C, Walker N, Schwardlander B, Esparza J; WHO-UNAIDS Network for HIV Isolation and Characterization. (2002). "Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000". Acquir. Immune. Defic. Syndr. 29 (2): 184–190. PMID 11832690.
- ↑ Perrin L, Kaiser L, Yerly S. (2003). "Travel and the spread of HIV-1 genetic variants". Lancet Infect Dis. 3 (1): 22–27. PMID 12505029.