Coronavirus pathophysiology
|
Coronavirus Microchapters |
|
Diagnosis |
|---|
|
Treatment |
|
Case Studies |
|
Coronavirus pathophysiology On the Web |
|
American Roentgen Ray Society Images of Coronavirus pathophysiology |
|
Risk calculators and risk factors for Coronavirus pathophysiology |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]; Associate Editor(s)-in-Chief: Aditya Govindavarjhulla, M.B.B.S. [2]
Pathophysiology
Structure
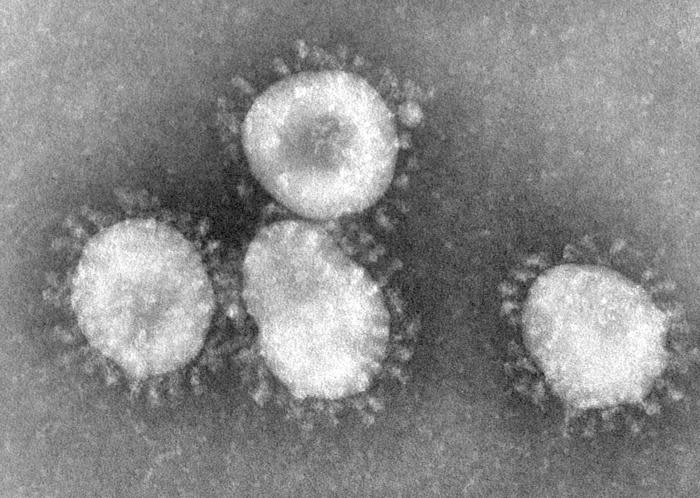
Coronaviruses are enveloped viruses with a positive-sense single-stranded RNA genome and a helical symmetry. The genomic size of coronaviruses ranges from approximately 16 to 31 kilobases, extraordinarily large for an RNA virus. The name coronavirus is derived from the greek (κορώνα, meaning crown) as the virus envelope appears under electron microscopy (E.M.) to be crowned by a characteristic ring of small bulbous structures. This morphology is actually formed by the viral spike (S) peplomers, which are proteins that populate the surface of the virus and determine host tropism. Coronaviruses are grouped in the order Nidovirales, named for the Latin (nidus, meaning nest) as all viruses in this order produce a 3' co-terminal nested set of subgenomic mRNA's during infection.
Proteins that contribute to the overall structure of all coronaviruses are the spike (S), envelope (E), membrane (M) and nucleocapsid (N). In the specific case of SARS, a defined receptor-binding domain on S mediates the attachment of the virus to its cellular receptor, angiotensin-converting enzyme 2 (ACE2).[1] Members of the group 2 coronaviruses also have a shorter spike-like protein called hemagglutinin esterase (HE) encoded in their genome, but for some reason this protein is not always brought to expression (produced) in the cell.[2]
References
- ↑ Li, Fang, et. al. (2005). "Structure of SARS Coronavirus Spike Receptor-Binding Domain Complexed with Receptor". Science. 309: 1864&ndash, 1868. doi:10.1126/science.1116480.
- ↑ de Haan CAM, Rottier PJM (2005). "Molecular Interactions in the Assembly of Coronaviruses". Advances in Virus Research. 64: 185&ndash, 186.