Primary amoebic meningoencephalitis laboratory findings
|
Primary amoebic meningoencephalitis Microchapters |
|
Differentiating Primary Amoebic Meningoencephalitis from other Diseases |
|---|
|
Diagnosis |
|
Treatment |
|
Case Studies |
|
Primary amoebic meningoencephalitis laboratory findings On the Web |
|
American Roentgen Ray Society Images of Primary amoebic meningoencephalitis laboratory findings |
|
FDA on Primary amoebic meningoencephalitis laboratory findings |
|
CDC on Primary amoebic meningoencephalitis laboratory findings |
|
Primary amoebic meningoencephalitis laboratory findings in the news |
|
Blogs on Primary amoebic meningoencephalitis laboratory findings |
|
Directions to Hospitals Treating Primary amoebic meningoencephalitis |
|
Risk calculators and risk factors for Primary amoebic meningoencephalitis laboratory findings |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]; Associate Editor(s)-in-Chief: Hardik Patel, M.D.
Please help WikiDoc by adding more content here. It's easy! Click here to learn about editing.
Overview
Primary amoebic meningoencephalitis is diagnosed using specific laboratory tests available in only a few laboratories in the United States. Because of the rarity of the infection and difficulty in initial detection, about 75% of diagnoses are made after the death of the patient. It can be diagnosed in the laboratory by detecting Naegleria fowleri organisms, nucleic acid, or antigen in cerebrospinal fluid (CSF), biopsy, or tissue specimens.
Laboratory Findings
CSF Analysis
Cerebrospinal fluid (CSF) studies of patients infected with Naegleria fowleri typically demonstrate a pattern similar to bacterial meningitis with an elevated opening pressure, a polymorphonuclear pleocytosis, normal or low glucose, and elevated protein. However the observations of blood in the CSF and/or motile amoeba are clues to a potential diagnosis of PAM.
Direct Visualization[1][2]
The motile Naegleria fowleri can often be seen moving rapidly under a microscope when looking at a fresh sample of CSF. The amoebae can also be stained with a variety of stains, such as Giemsa-Wright or a modified trichrome stain, for identification.
- CSF: The diagnosis of Naegleria fowleri infection can be made most quickly by microscopic examination of fresh, unfrozen, unrefrigerated cerebrospinal fluid (CSF) (samples cannot be frozen or refrigerated because cold temperatures kill the amoebae). A wet mount of freshly-centrifuged CSF sediment might demonstrate actively moving trophozoites. Naegleria fowleri (15-30 µm trophozoite) moves rapidly (~1 µm/s) using eruptive pseudopods and moves sinuously in a generally linear forward direction. Additionally, Naegleria can be identified in CSF smears or cultures using hematoxylin and eosin (H&E), periodic acid-Schiff (PAS), trichrome, Giemsa, or Wright-Giemsa stains. A Gram stain should be avoided as the amoebae can be destroyed during heat fixation. A stained CSF smear will show amoeboid trophozoites with morphology typical of Naegleria (i.e., a nucleus with a large, centrally located and densely staining nucleolus). If amoebae are identified in the CSF, the diagnosis of PAM should be subsequently confirmed with culture, PCR, or immunohistochemical (IHC) tests.
- Tissue: The diagnosis can also be made from microscopic examination of hematoxylin and eosin (H&E), periodic acid-Schiff (PAS), trichrome, Giemsa, or Wright-Giemsa stained smears of brain biopsy or autopsy specimens, which might demonstrate trophozoites with morphology typical of Naegleria fowleri. The ameboid trophozoites measure 10-35 µm but when rounded are usually 10-15 µm in diameter. The cytoplasm is granular and contains many vacuoles. The single nucleus is large and has a large, dense karyosome. Naegleria fowleri does not form cysts in human tissues.
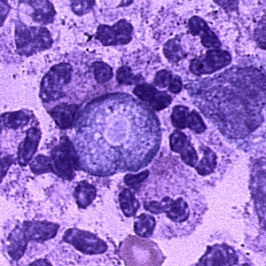 |
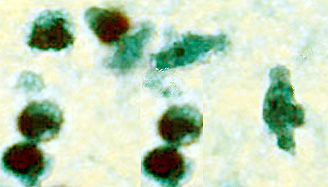 |
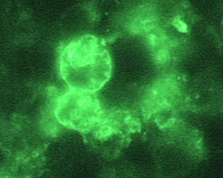 |
Copyleft images obtained courtesy of http://www.cdc.gov/parasites/naegleria/diagnosis-hcp.html
Immunohistochemistry (IHC)[1][2]
A specific antibody to Naegleria fowleri can be used in conjunction with another antibody that deposits a chemical or glows under specific types of light (indirect fluorescent antibody [IFA]) to directly stain the amebae in tissue. Serologic testing for Naegleria fowleri using indirect immunofluorescent antibody (IFA) testing is currently considered a research technique as it has not been evaluated for use as a routine diagnostic procedure. Although IFA can be performed to measure serum antibody titers in patient sera, most patients with PAM die before an immune response is mounted.
Polymerase Chain Reaction (PCR)[3][4][5]
Specific molecular tools can amplify DNA from the amoebae in CSF or tissue to specifically identify if the amoebae are present. Looking at strains or subtypes of Naegleria fowleri can be done, but little is known about the natural populations in the environment, which makes it difficult to interpret what the findings mean. An increasing number of PCR-based techniques (conventional and real-time PCR) have been described for detection and identification of free-living amebic infections in clinical specimens, but are only available in selected reference diagnostic laboratories. A real-time CLIA-certified PCR was developed at CDC for qualitative assessment of Naegleria fowleri, Acanthamoeba spp., and Balamuthia mandrillaris in clinical samples. This assay uses distinct primers and TaqMan probes for the simultaneous identification of all three free-living amoebae.
Amoeba Culture
The sample is added to a growth plate covered in bacteria that can serve as a food source for Naegleria fowleri. Incubating at higher temperatures selects for Naegleria fowleri growth, which can be seen as tracks made by the amoeba as it moves across the plate eating the bacteria. Growing Naegleria fowleri in mammalian cell culture and looking for toxic cell effects is also possible. It involves inoculating mammalian cell cultures and monitoring for cytopathogenicity, or inoculating plates of E. coli lawns and monitoring for tracks where the amoebae have moved through the lawn eating the bacteria. Further, Naegleria can be identified in cultures from clinical specimens using hematoxylin and eosin (H&E), periodic acid-Schiff (PAS), trichrome, Giemsa, or Wright-Giemsa stains. Stained cultures might demonstrate trophozoites with morphology typical of Naegleria fowleri, as previously described. In culture, trophozoites may measure more than 40 µm. A negative culture result does not rule out the presence of free-living amoebae and other tests should be performed.
Environmental Detection
Water samples can be collected, concentrated, and put into culture to grow and select for Naegleria fowleri. Samples can be tested using the serologic or molecular methods described above.
References
- ↑ 1.0 1.1 Visvesvara GS (2010). "Amebic meningoencephalitides and keratitis: challenges in diagnosis and treatment". Curr Opin Infect Dis. 23 (6): 590–4. doi:10.1097/QCO.0b013e32833ed78b. PMID 20802332.
- ↑ 2.0 2.1 da Rocha-Azevedo B, Tanowitz HB, Marciano-Cabral F (2009). "Diagnosis of infections caused by pathogenic free-living amoebae". Interdiscip Perspect Infect Dis. 2009: 251406. doi:10.1155/2009/251406. PMC 2719787. PMID 19657454.
- ↑ Qvarnstrom Y, Visvesvara GS, Sriram R, da Silva AJ (2006). "Multiplex real-time PCR assay for simultaneous detection of Acanthamoeba spp., Balamuthia mandrillaris, and Naegleria fowleri". J Clin Microbiol. 44 (10): 3589–95. doi:10.1128/JCM.00875-06. PMC 1594764. PMID 17021087.
- ↑ Robinson BS, Monis PT, Dobson PJ (2006). "Rapid, sensitive, and discriminating identification of Naegleria spp. by real-time PCR and melting-curve analysis". Appl Environ Microbiol. 72 (9): 5857–63. doi:10.1128/AEM.00113-06. PMC 1563602. PMID 16957204.
- ↑ Luan F, Xu X, Cordeiro MN, Liu H, Zhang X (2012). "QSAR modeling for the antimalarial activity of 1, 4-naphthoquinonyl derivatives as potential antimalarial agents". Curr Comput Aided Drug Des. PMID 23157413.