Pheochromocytoma pathophysiology: Difference between revisions
Ifrah Fatima (talk | contribs) |
Ifrah Fatima (talk | contribs) |
||
| Line 52: | Line 52: | ||
* '''[[Neurofibromatosis type I|Neurofibromatosis type 1]] (NF1)''' | * '''[[Neurofibromatosis type I|Neurofibromatosis type 1]] (NF1)''' | ||
|} | |} | ||
==Associated conditions== | ==Associated conditions== | ||
Revision as of 05:08, 24 July 2020
|
Pheochromocytoma Microchapters |
|
Diagnosis |
|---|
|
Treatment |
|
Case Studies |
|
Pheochromocytoma pathophysiology On the Web |
|
American Roentgen Ray Society Images of Pheochromocytoma pathophysiology |
|
Risk calculators and risk factors for Pheochromocytoma pathophysiology |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]; Associate Editor(s)-in-Chief: Ahmad Al Maradni, M.D. [2] Mohammed Abdelwahed M.D[3]
Overview
Pheochromocytoma arises from chromaffin cells of the adrenal medulla. On gross pathology, pheochromocytoma has a multinodular and a multicentric pattern of growth. On microscopic histopathological analysis, nesting (Zellballen) pattern composed of well-defined clusters of tumor cells separated by fibrovascular stroma may be seen. It may be benign, malignant, familial (multiple endocrine neoplasia 1 and type 2B) or sporadic. All of these forms have genetic origin depending on a large number of genes, for example, VHL, SDH, NF1, RET genes.
Pathophysiology
The pathophysiology associated with pheochromocytoma is as follow:[1] [2][3]
- Pheochromocytoma is a tumor which arises from the chromaffin cells of the adrenal medulla and sympathetic ganglia.
- Malignant and benign pheochromocytomas share the same biochemical and histological features.
- Pheochromocytoma leads to excessive secretion of catecholamines and subsequent stimulation of adrenergic receptors.
- Commonly secreted catecholamines include norepinephrine (predominant) and epinephrine. Some tumors may also secrete dopamine.
- Excessive secretion of catecholamines may be either continuous or intermittent.
- The exact mechanism responsible for surge in catecholamine secretion remains unclear but it has been postulated that certain medications (such as opiates, metoclopramide or beta blockers) and changes in tumor blood flow and pressure could be responsible factors.
Effects of adrenergic stimulation by pheochromocytoma
- Epinephrine acts on nearly all body tissues. Its actions vary by tissue type and tissue expression of adrenergic receptors.
- Epinephrine is a nonselective agonist of all adrenergic receptors, including the major subtypes α1, α2, β1, β2, and β3:
- Binding to α1 receptors causes vasoconstriction. α1-adrenergic receptors are present in the blood vessels of skin, the sphincters of the gastrointestinal system, kidney (renal artery) and brain. During the fight-or-flight response vasoconstriction results in decreased blood flow to these organs.
- Binding to α2 receptors inhibits insulin secretion by the pancreas, stimulates glycogenolysis in the liver and muscle, and stimulates glycolysis and inhibits insulin-mediated glycogenesis in muscle. It suppresses the release of norepinephrine by negative feedback.
- Binding to β2 receptors causes smooth muscle relaxation in the uterus, GI tract, detrusor urinae muscle of bladder wall, and bronchi. It also causes dilatation of smaller coronary arteries, hepatic artery, arteries to skeletal muscle.
- Binding to β1 receptors causes renin release from juxtaglomerular cells and lipolysis in adipose tissue. It Increases cardiac output by:
- Increase in heart rate in sinoatrial node
- Increase in atrial cardiac muscle contractility
- Increases in contractility and automaticity of ventricular cardiac muscle
- Increases in conduction and automaticity of atrioventricular node
Genetics
- Genes involved in the pathogenesis of pheochromocytoma include:
- RET gene (MEN 2A, MEN 2B syndromes)
- VHL gene (VHL disease)
- SDHD, SDHB, and SDHC genes of the mitochondrial complex [6]
- It has autosomal dominant inheritance and has two pathways of tumor pathogenesis. Cluster 1 tumors are noradrenergic. Cluster 2 tumors are adrenergic.[7]
| Familial pheocromocytoma | |
|---|---|
| Cluster 1 (Noradrenergic) | Cluster 2 (Adrenergic) |
|
|
Associated conditions
Conditions associated with pheochromocytoma include:
- Multiple endocrine neoplasia (MEN1)
- Multiple endocrine neoplasia (MEN2B)
- Von-Hippel Lindau (VHL) disease (VHL)
| MEN 1 | MEN 2 |
|---|---|
Gross Pathology
On gross pathology, the characteristic findings of pheochromocytoma are:
- Small to large tumors usually associated with hemorrhage and necrosis.[8]
- Usually lobulated
- Bilateral when familial tumors
- Associated with hyperplasia in the adjacent medulla.
- Chromaffin reaction: fresh tumor cut section turns dark brown if add potassium dichromate at pH 5-6.
-
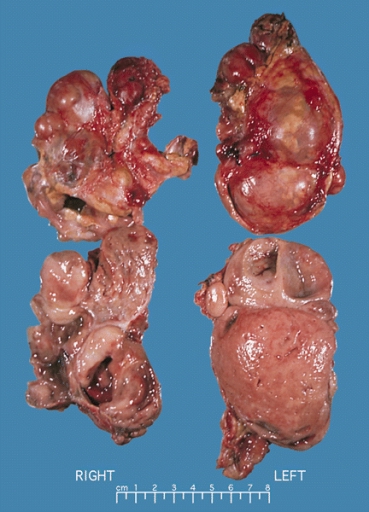
Bilateral pheochromocytoma in MEN2. Gross image. Source: https://upload.wikimedia.org/wikipedia/commons/5/5f/Bilateral_pheo_MEN2.jpg
Microscopic Pathology
On microscopic histopathological analysis, the characterisitc findings of pheochromocytoma typically include:
- A nesting (Zellballen) pattern- this pattern is composed of well-defined clusters of tumor cells containing eosinophilic cytoplasm separated by fibrovascular stroma.
-
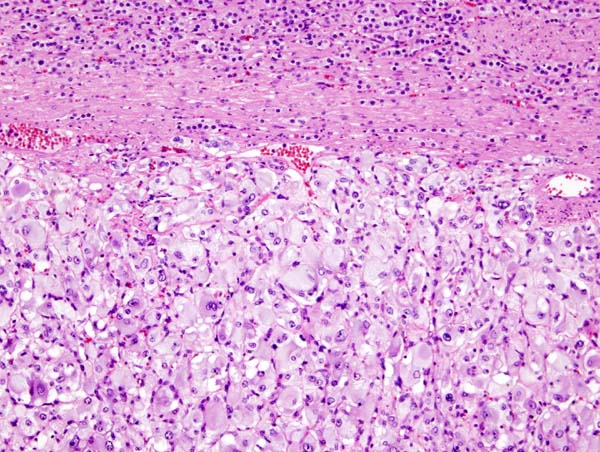
Micrograph of pheochromocytoma. Source: By Nephron - Own work, CC BY-SA 3.0, https://commons.wikimedia.org/w/index.php?curid=5938524
-
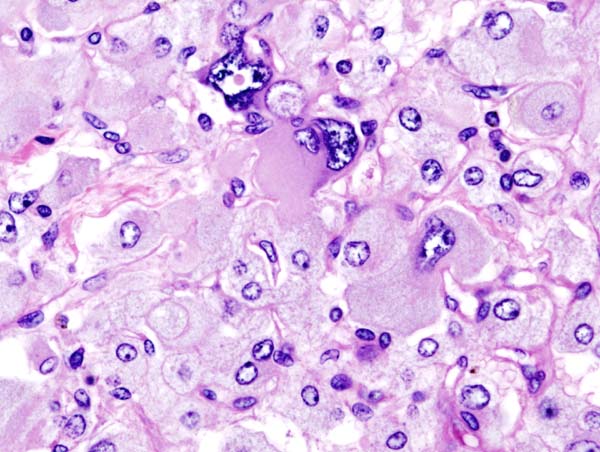
Histopathology of adrenal pheochromocytoma. Adrenectomy specimen. Source: CC BY-SA 3.0, https://commons.wikimedia.org/w/index.php?curid=535945
-
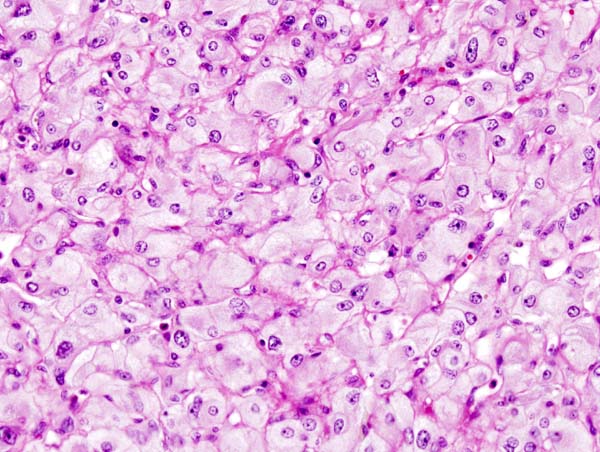
Micrograph of pheochromocytoma. Source: CC BY-SA 3.0, https://commons.wikimedia.org/w/index.php?curid=535944
Videos
{{#ev:youtube|7yjxG3KmX98}}
References
- ↑ Goldstein RE, O'Neill JA, Holcomb GW, Morgan WM, Neblett WW, Oates JA; et al. (1999). "Clinical experience over 48 years with pheochromocytoma". Ann Surg. 229 (6): 755–64, discussion 764-6. PMC 1420821. PMID 10363888.
- ↑ Raz I, Katz A, Spencer MK (1991). "Epinephrine inhibits insulin-mediated glycogenesis but enhances glycolysis in human skeletal muscle". Am J Physiol. 260 (3 Pt 1): E430–5. PMID 1900669.
- ↑ Arnall DA, Marker JC, Conlee RK, Winder WW (1986). "Effect of infusing epinephrine on liver and muscle glycogenolysis during exercise in rats". Am J Physiol. 250 (6 Pt 1): E641–9. PMID 3521311.
- ↑ Webb TA, Sheps SG, Carney JA (1980). "Differences between sporadic pheochromocytoma and pheochromocytoma in multiple endocrime neoplasia, type 2". Am. J. Surg. Pathol. 4 (2): 121–6. PMID 6103678.
- ↑ Yee JK, Moores JC, Jolly DJ, Wolff JA, Respess JG, Friedmann T (1987). "Gene expression from transcriptionally disabled retroviral vectors". Proc. Natl. Acad. Sci. U.S.A. 84 (15): 5197–201. PMC 298821. PMID 3474647.
- ↑ Gimm O (2005). "Pheochromocytoma-associated syndromes: genes, proteins and functions of RET, VHL and SDHx". Fam Cancer. 4 (1): 17–23. doi:10.1007/s10689-004-5740-1. PMID 15883706.
- ↑ King KS, Pacak K (2014). "Familial pheochromocytomas and paragangliomas". Mol Cell Endocrinol. 386 (1–2): 92–100. doi:10.1016/j.mce.2013.07.032. PMC 3917973. PMID 23933153.
- ↑ Sajjanar AB, Athanikar VS, Dinesh US, Nanjappa B, Patil PB (2015). "Non Functional Unilateral Adrenal Myelolipoma, A Case Report". J Clin Diagn Res. 9 (6): ED03–4. doi:10.7860/JCDR/2015/13209.6070. PMC 4525519. PMID 26266130.