Polymerase chain reaction
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]

Overview
The polymerase chain reaction (PCR) is a biochemistry and molecular biology method of nucleic acid amplification technique[1] for exponentially amplifying a fragment of DNA, via enzymatic replication, without using a living organism (such as E. coli or yeast). PCR can be used for amplification of a single or few copies of a piece of DNA across several orders of magnitude, generating millions or more copies of the DNA piece. As PCR is an in vitro technique, it can be performed without restrictions on the form of DNA, and it can be extensively modified to perform a wide array of genetic manipulations.
Developed in 1983 by Kary Mullis, PCR is now a common technique used in medical and biological research labs for a variety of tasks, such as the sequencing of genes and the diagnosis of hereditary diseases, the identification of genetic fingerprints (used in forensics and paternity testing), the detection and diagnosis of infectious diseases, and the creation of transgenic organisms. Mullis won the Nobel Prize for his work on PCR.
PCR principle and procedure

PCR is used to amplify specific regions of a DNA strand. This can be a single gene, just a part of a gene, or a non-coding sequence. Most PCR methods typically amplify DNA fragments of up to 10 kilo base pairs (kb), although some techniques allow for amplification of fragments up to 40 kb in size.[2]
PCR, as currently practiced, requires several basic components [3]. These components are:
- DNA template that contains the region of the DNA fragment to be amplified
- One or more primers, which are complementary to the DNA regions at the 5' (five prime) and 3' (three prime) ends of the DNA region that is to be amplified.
- a DNA polymerase (e.g. Taq polymerase or another DNA polymerase with a temperature optimum at around 70°C), used to synthesize a DNA copy of the region to be amplified
- Deoxynucleotide triphosphates, (dNTPs) from which the DNA polymerase builds the new DNA
- Buffer solution, which provides a suitable chemical environment for optimum activity and stability of the DNA polymerase
- Divalent cations, magnesium or manganese ions; generally Mg2+ is used, but Mn2+ can be utilized for PCR-mediated DNA mutagenesis, as higher Mn2+ concentration increases the error rate during DNA synthesis [4]
- Monovalent cation potassium ions
The PCR is carried out in small reaction tubes (0.2-0.5 ml volumes), containing a reaction volume typically of 15-100 μl, that are inserted into a thermal cycler. This is a machine that heats and cools the reaction tubes within it to the precise temperature required for each step of the reaction. The tubes are generally made with special thin walls that have a favorable thermal conductivity to allow for rapid thermal equilibration. Most contemporary thermal cyclers also have heated lids to prevent condensation on the top of the reaction tube caps. Alternatively, a layer of oil may be placed on top of the reaction mixture, or a ball of wax inside the tube, to prevent evaporation.
Procedure
The PCR usually consists of a series of 20 to 35 cycles. Most commonly, PCR is carried out in three steps (Fig. 2), often preceded by one temperature hold at the start and followed by one hold at the end.
- Prior to the first cycle, during an initialization step, the PCR reaction is often heated to a temperature of 94-96°C (or 98°C if extremely thermostable polymerases are used), and this temperature is then held for 1-9 minutes. This first hold is employed to ensure that most of the DNA template and primers are denatured, i.e., that the DNA is melted by disrupting the hydrogen bonds between complementary bases of the DNA strands, yielding two single strands of DNA. Also, some PCR polymerases require this step for activation (see hot-start PCR).[5] Following this hold, cycling begins, with one step at 94-98°C for 20-30 seconds (denaturation step).
- The denaturation is followed by the annealing step. In this step the reaction temperature is lowered so that the primers can anneal to the single-stranded DNA template. Brownian motion causes the primers to move around, and DNA-DNA hydrogen bonds are constantly formed and broken between primer and template. Stable bonds are only formed when the primer sequence very closely matches the template sequence, and to this short section of double-stranded DNA the polymerase attaches and begins DNA synthesis. The temperature at this step depends on the melting temperature of the primers, and is usually between 50-64°C for 20-40 seconds. Once the primers have annealed, some extension occurs, so that the primers do not melt off of the template in the next step.
- The annealing step is followed by an extension/elongation step during which the DNA polymerase synthesizes new DNA strands complementary to the DNA template strands. The temperature at this step depends on the DNA polymerase used. Taq polymerase has a temperature optimum of 70-74°C; thus, in most cases a temperature of 72°C is used. The hydrogen bonds between the extended primer and the DNA template are now strong enough to withstand forces breaking these attractions at the higher temperature. Primers that have annealed to DNA regions with mismatching bases dissociate from the template and are not extended. The DNA polymerase condenses the 5'-phosphate group of the dNTPs with the 3'-hydroxyl group at the end of the nascent (extending) DNA strand, i.e., the polymerase adds dNTP's that are complementary to the template in 5' to 3' direction, thus reading the template in 3' to 5' direction. The extension time depends both on the DNA polymerase used and on the length of the DNA fragment to be amplified. As a rule-of-thumb, at its optimum temperature, the DNA polymerase will polymerize a thousand bases in one minute. A final elongation step of 5-15 minutes (depending on the length of the DNA template) after the last cycle may be used to ensure that any remaining single-stranded DNA is fully extended. A final hold of 4-15°C for an indefinite time may be employed for short-term storage of the reaction, e.g., if reactions are run overnight.
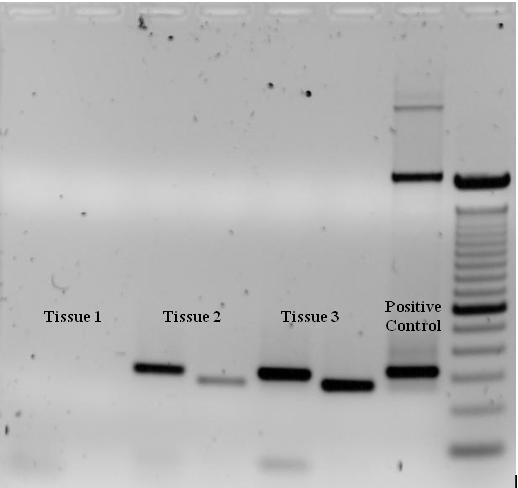
To check whether the PCR generated the anticipated DNA fragment (also sometimes referred to as amplimer), agarose gel electrophoresis is commonly employed for size separation of the PCR products. The size(s) of PCR products is thereby determined by comparison with a DNA ladder, which contains DNA fragments of known size, run on the gel alongside the PCR products (see Fig. 3).
PCR optimization
In practice, PCR can fail for various reasons, in part due to its sensitivity to contamination causing amplification of spurious DNA products. Because of this, a number of techniques and procedures have been developed for optimizing PCR conditions. Contamination with extraneous DNA is addressed with lab protocols and procedures that separate pre-PCR reactions from potential DNA contaminants.[3] This usually involves spatial separation of PCR-setup areas from areas for analysis or purification of PCR products, and thoroughly cleaning the work surface between reaction setups. Primer-design techniques are important in improving PCR product yield and in avoiding the formation of spurious products, and the usage of alternate buffer components or polymerase enzymes can help with amplification of long or otherwise problematic regions of DNA.
Uses of PCR
Isolation of genomic material
Because it selectively amplifies a specific region of DNA, PCR can be used to isolate desired sections of DNA sequence from whole genomic material. Many techniques (such as Southern and northern blotting and DNA cloning) require a pool of DNA molecules isolated from a particular DNA region — although PCR is not always a necessary prerequisite, its usage has greatly augmented these methods. Because PCR also amplifies the isolated region, the techniques are endowed with high amounts of pure starting material and become applicable to samples otherwise too small for analysis.
- Sequencing and the detection of genetic diseases
Once a sample is created where all molecules derive from a single region of DNA, it is possible to go forward with DNA sequencing to determine the unknown sequence of nucleotides that spans the fragment between the two primers. One of the PCR primers is commonly used as the anchor for Sanger sequencing, which is currently the most common method for DNA sequencing. This method is commonly used to diagnose genetic disorders; a doctor can confirm a diagnosis by observing DNA sequence differences that are known to be associated with a disorder.
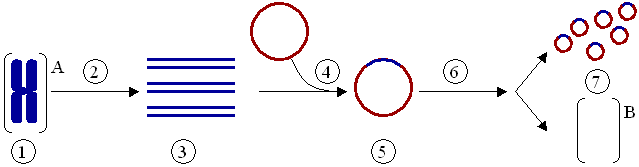
(1) Chromosomal DNA of organism A. (2) PCR. (3) Multiple copies of a single gene from organism A. (4) Insertion of the gene into a plasmid. (5) Plasmid with gene from organism A. (6) Insertion of the plasmid in organism B. (7) Multiplication or expression of the gene, originally from organism A, occurring in organism B.
- Recombinant DNA techniques
Isolating a DNA sequence also enables recombinant DNA technologies, which involve the insertion of a given DNA sequence into the genetic material of another organism (now termed a genetically modified organism (GMO)). PCR is often used to amplify a gene, which can then be inserted into a target genome through homologous recombination or, more commonly, inserted into a vector that carries the DNA into the target organism, such as a plasmid.
- Genetic fingerprinting and paternity testing
Genetic fingerprinting is a forensic technique used to identify a person by comparing his or her DNA with the DNA in a given sample. PCR can be used to amplify a specific set regions which vary in length from person to person. The combination of all these variations creates "genetic fingerprint" – when these fragments are run out on an agarose gel they produce a specific pattern of bands. With the use of PCR, in theory, only a single DNA strand is needed, providing very high sensitivity to this technique, although such sensitive amplification increases the risk of confounding results due to possible contamination with, and amplification of, DNA from extraneous sources. There are different PCR-based methods for fingerprinting, summarized in Genetic fingerprinting. The overall pattern of PCR-generated DNA fragments after gel electrophoresis and visualization by ethidium bromide staining (or hybridization with a DNA probe after Southern blotting), can be considered a DNA fingerprint analogous to the fingerprint pattern unique to each individual.
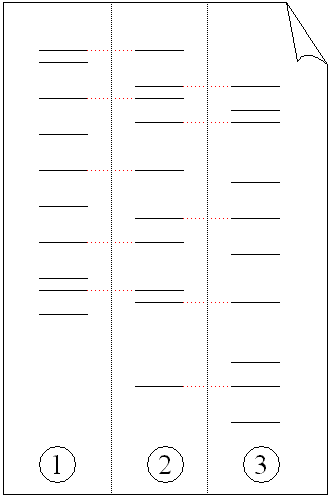
Although these resulting 'fingerprints' are unique , genetic relationships, for example, parent-child or siblings, can be determined from two or more genetic fingerprints, which can be used for paternity tests (Fig. 4). A variation of this technique can also be used to determine evolutionary relationships between organisms.
Amplification of DNA & DNA quantitation
Because PCR amplifies the regions of DNA that it targets, PCR can be used to analyze extremely small amounts of sample. This is often critical for forensic analysis, when only a trace amount of DNA is available as evidence. The amount of time it takes to amplify DNA to a given level is related to the original amount in the sample, which means that PCR can also be used to quantify the presence of particular DNA fragments.
- Analysis of ancient DNA
Using PCR, it becomes possible to analyze DNA that is thousands of years old. PCR techniques have been successfully used on animals, such as a forty-thousand-year-old mammoth, and also on human DNA, in applications ranging from the analysis of Egyptian mummies to the identification of a Russian Tsar[6].
- Detection of viral DNA
Viral DNA can likewise be detected by PCR amplification. Primers need to be specific to sequences in the DNA of a given virus, and the PCR can be used for diagnostic analyses or DNA sequencing of parts or all of the viral genome. Because of the high sensitivity of PCR, virus detection may be possible soon after infection and even before the onset of disease symptoms. Such early detection may give physicians a significant lead in treatment. The the amount of virus ("viral load") in a patient can be quantified by PCR-based DNA quantitation techniques (see below).
- DNA quantitation and comparison of gene expression
Because the amount of product produced by PCR roughly correlates to the amount of starting material, PCR can be used to estimate the amount of a given sequence that is present in a sample – a technique especially useful for determining gene expression levels. In cells, each gene is expressed through the production of messenger RNA (mRNA), which is then used to create a protein corresponding to the gene. The amount of mRNA in the cell for a given gene reflects how active that gene is. By using reverse transcription to produce DNA complementary to the mRNA (called cDNA) and subsequently using PCR to amplify these molecules, the amount of DNA produced for each gives a rough measure of the underlying expression for that gene.
More accurate measurements are possible when the amount of double-stranded DNA is measured after each round of PCR amplification, a technique known as "quantitative real-time PCR". A dye that fluoresces in the presence of double-stranded DNA is added to the mixture and, as the fluorescence increases, the timing as well as amount of amplification can be measured.
History
A 1971 paper in the Journal of Molecular Biology by Kleppe and co-workers first described a method using an enzymatic assay to replicate a short DNA template with primers in vitro. [7] However, this early manifestation of the basic PCR principle did not receive much attention, and the invention of the polymerase chain reaction in 1983 is generally credited to Kary Mullis [8][9]. He was awarded the Nobel Prize in Chemistry in 1993 for his invention, seven years after he and his colleagues at Cetus first put his proposal to practice. However, some controversies have remained about the intellectual and practical contributions of other scientists to Mullis' work, and whether he had been the sole inventor of the PCR principle. (see main article:Kary Mullis)
At the time he developed PCR in 1983, Mullis was working in Emeryville, California for Cetus Corporation, one of the first biotechnology companies. There, he was responsible for synthesizing short chains of DNA. Mullis has written that he conceived of PCR while cruising along the Pacific Coast Highway one night in his car[8]. He was playing in his mind with a new way of analyzing changes (mutations) in DNA when he realized that he had instead invented a method of amplifying any DNA region through repeated cycles of duplication driven by an enzyme called DNA polymerase. Mullis credits the psychedelic drug LSD for his invention of the technique. [2](Video)
In Scientific American, Mullis summarized the accomplishment: "Beginning with a single molecule of the genetic material DNA, the PCR can generate 100 billion similar molecules in an afternoon. The reaction is easy to execute. It requires no more than a test tube, a few simple reagents, and a source of heat."[10]
DNA polymerase occurs naturally in living organisms. In cells it functions to duplicate DNA when cells divide in mitosis and meiosis. Polymerase works by binding to a single DNA strand and creating the complementary strand. In the first of many original processes, the enzyme was used in vitro (in a controlled environment outside an organism). The double-stranded DNA was separated into two single strands by heating it to 94°C (201°F). At this temperature, however, the DNA polymerase used at the time were destroyed, so the enzyme had to be replenished after the heating stage of each cycle. The original procedure was very inefficient, since it required a great deal of time, large amounts of DNA polymerase, and continual attention throughout the process.
In 1986, this original PCR process was greatly improved by the use of DNA polymerase taken from thermophilic bacteria grown in geysers at a temperature of over 110°C (230°F). The DNA polymerase taken from these organisms is stable at high temperatures and, when used in PCR, does not break down when the mixture was heated to separate the DNA strands. Since there was no longer a need to add new DNA polymerase for each cycle, the process of copying a given DNA strand could be simplified and automated.
One of the first thermostable DNA polymerases was obtained from Thermus aquaticus and was called "Taq." Taq polymerase is widely used in current PCR practice. A disadvantage of Taq is that it sometimes makes mistakes when copying DNA, leading to mutations (errors) in the DNA sequence, since it lacks 3'→5' proofreading exonuclease activity. Polymerases such as Pwo or Pfu, obtained from Archaea, have proofreading mechanisms (mechanisms that check for errors) and can significantly reduce the number of mutations that occur in the copied DNA sequence. However these enzymes polymerise DNA at a much slower rate than Taq. Combinations of both Taq and Pfu are available nowadays that provide both high processivity (fast polymerisation) and high fidelity (accurate duplication of DNA).
PCR has been performed on DNA larger than 10 kilobases, but the average PCR is only several hundred to a few thousand bases of DNA. The problem with long PCR is that there is a balance between accuracy and processivity of the enzyme. Usually, the longer the fragment, the greater the probability of errors.
Patent wars
The PCR technique was patented by Cetus Corporation, where Mullis worked when he invented the technique in 1983. The Taq polymerase enzyme was also covered by patents. There have been several high-profile lawsuits related to the technique, including an unsuccessful lawsuit brought by DuPont. The pharmaceutical company Hoffmann-La Roche purchased the rights to the patents in 1992 and currently holds those that are still protected.
A related patent battle over the Taq polymerase enzyme is still ongoing in several jurisdictions around the world between Roche and Promega. Interestingly, it seems possible that the legal arguments will extend beyond the life of the original PCR and Taq polymerase patents, which expired on March 28, 2005.[11]
Variations on the basic PCR technique
- Allele-specific PCR: AS-PCR is used to determine the genotype of single-nucleotide polymorphisms (SNPs) (single base differences in DNA) by using primers whose ends overlap the SNP and differ by that single base. PCR amplification is less efficient in the presence of a mismatch, so the differences in amplification resulting from different primers can be used to quickly determine which primer matches the sample genotype.
- Assembly PCR: Assembly PCR is the completely artificial synthesis of long gene products by performing PCR on a pool of long oligonucleotides with short overlapping segments. The oligonucleotides alternate between sense and antisense directions, and the overlapping segments serve to order the PCR fragments so that they selectively produce their final product.
- Asymmetric PCR: Asymmetric PCR is used to preferentially amplify one strand of the original DNA more than the other. It finds use in some types of sequencing and hybridization probing where having only one of the two complementary stands is required. PCR is carried out as usual, but with a great excess of the primers for the chosen strand. Due to the slow (arithmetic) amplification later in the reaction after the limiting primer has been used up, extra cycles of PCR are required. A recent modification on this process, known as Linear-After-The-Exponential-PCR (LATE-PCR), uses a limiting primer with a higher melting temperature (Tm) than the excess primer to maintain reaction efficiency as the limiting primer concentration decreases mid-reaction.
- Colony PCR: Bacterial clones (E.coli) can be rapidly screened for correct DNA vector constructs. Selected bacterial colonies are picked with a sterile toothpick from an agarose plate and dabbed into the master mix or sterile water. Primers (and the master mix) are added, and the PCR is started with an extended time at 95˚C when standard polymerase is used or with a shortened denaturation step at 100˚C and special chimeric DNA polymerase.[12]
- Helicase-dependent amplification: similar to traditional PCR, but maintains a constant temperature rather than cycling through denaturation and annealing/extension cycles. Helicase, an enzyme that unwinds DNA, is used in place of thermal denaturation.[13]
- Hot-start PCR: a technique that reduces non-specific amplification during the initial set up stages of the PCR. The technique may be performed manually by simply heating the reaction components briefly at the melting temperature (e.g., 95˚C) before adding the polymerase.[14] Specialized enzyme systems have been developed that inhibit the polymerase's activity at ambient temperature, either by the binding of an antibody[5] or by the presence of covalently bound inhibitors that only dissociate after a high-temperature activation step. Hot-start/cold-finish PCR is achieved with new hybrid polymerases that are inactive at ambient temperature and are instantly activated at elongation temperature.
- Intersequence-specific (ISSR) PCR: a PCR method for DNA fingerprinting that amplifies regions between some simple sequence repeats to produce a unique fingerprint of amplified fragment lengths.[15]
- Inverse PCR: a method used to allow PCR when only one internal sequence is known. This is especially useful in identifying flanking sequences to various genomic inserts. This involves a series of DNA digestions and self ligation, resulting in known sequences at either end of the unknown sequence.
- Ligation-mediated PCR: Ligation-mediated PCR (LM-PCR) can be used to detect vector insertion sites into a genome.
- Methylation-specific PCR (MSP): The MSP method was developed by Stephen Baylin and Jim Herman at the Johns Hopkins School of Medicine, [16] and is used to detect methylation of CpG islands in genomic DNA. DNA is first treated with sodium bisulfite, which converts unmethylated cytosine bases to uracil, which is recognized by PCR primers as thymine. Two PCR reactions are then carried out on the modified DNA, using primer sets identical except at any CpG islands within the primer sequences. At these points, one primer set recognizes DNA with cytosines to amplify methylated DNA, and one set recognizes DNA with uracil or thymine to amplify unmethylated DNA. MSP using qPCR can also be performed to obtain quantitative rather than qualitative information about methylation.
- Multiplex Ligation-dependent Probe Amplification (MLPA): permits multiple targets to be amplified with only a single primer pair, thus avoiding the resolution limitations of multiplex PCR (see below).
- Multiplex-PCR: The use of multiple, unique primer sets within a single PCR reaction to produce amplicons of varying sizes specific to different DNA sequences. By targeting multiple genes at once, additional information may be gained from a single test run that otherwise would require several times the reagents and more time to perform. Annealing temperatures for each of the primer sets must be optimized to work correctly within a single reaction, and amplicon sizes, i.e., their base pair length, should be different enough to form distinct bands when visualized by gel electrophoresis.
- Nested PCR: increases the specificity of DNA amplification, by reducing background due to non-specific amplification of DNA. Two sets of primers are being used in two successive PCR reactions. In the first reaction, one pair of primers is used to generate DNA products, which besides the intended target, may still consist of non-specifically amplified DNA fragments. The product(s) (sometimes after gel purification after electrophoresis of the PCR product) are then used in a second PCR reaction with a set of primers whose binding sites are completely or partially different from the primer pair used in the first reaction, but are completely within the DNA target fragment. Nested PCR is often more successful in specifically amplifying long DNA fragments than conventional PCR, but it requires more detailed knowledge of the target sequences.
- Quantitative PCR (Q-PCR): is used to measure the quantity of a PCR product (preferably real-time). It is the method of choice to quantitatively measure starting amounts of DNA, cDNA or RNA. Q-PCR is commonly used to determine whether a DNA sequence is present in a sample and the number of its copies in the sample. The method with currently the highest level of accuracy is Quantitative real-time PCR. It is often confusingly known as RT-PCR (Real Time PCR) or RQ-PCR. QRT-PCR or RTQ-PCR are more appropriate contractions. RT-PCR commonly refers to reverse transcription PCR (see below), which is often used in conjunction with Q-PCR. QRT-PCR methods use fluorescent dyes, such as Sybr Green, or fluorophore-containing DNA probes, such as TaqMan, to measure the amount of amplified product in real time.
- RT-PCR: (Reverse Transcription PCR) is a method used to amplify, isolate or identify a known sequence from a cellular or tissue RNA. The PCR is preceded by a reaction using reverse transcriptase to convert RNA to cDNA. RT-PCR is widely used in expression profiling, to determine the expression of a gene or to identify the sequence of an RNA transcript, including transcription start and termination sites and, if the genomic DNA sequence of a gene is known, to map the location of exons and introns in the gene. The 5' end of a gene (corresponding to the transcription start site) is typically identified by a RT-PCR method, named RACE-PCR, short for Rapid Amplification of cDNA Ends.
- TAIL-PCR: thermal asymmetric interlaced PCR is used to isolate unknown sequence flanking a known sequence. Within the known sequence TAIL-PCR uses a nested pair of primers with differing annealing temperatures; a degenerate primer is used to amplify in the other direction from the unknown sequence.[17]
- Touchdown PCR: a variant of PCR that aims to reduce nonspecific background by gradually lowering the annealing temperature as PCR cycling progresses. The annealing temperature at the initial cycles is usually a few degrees above the Tm of the primers used, while at the later cycles, it is a few degrees below the primer Tm. The higher temperatures give greater specificity for primer binding, and the lower temperatures permit more efficient amplification from the specific products formed during the initial cycles.
- PAN-AC: Another isothermal amplification reaction. The authors claim that it can be used in living cells.[18][19].
References
- ↑ The history of PCR: Smithsonian Institution Archives, Institutional History Division. Retrieved 24 June 2006.
- ↑ Cheng S, Fockler C, Barnes WM, Higuchi R (1994). "Effective amplification of long targets from cloned inserts and human genomic DNA". Proc Natl Acad Sci. 91: 5695–5699. PMID 8202550.
- ↑ 3.0 3.1 Joseph Sambrook and David W. Russel (2001). Molecular Cloning: A Laboratory Manual (3rd ed. ed.). Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press. ISBN 0-87969-576-5. Chapter 8: In vitro Amplification of DNA by the Polymerase Chain Reaction
- ↑ Pavlov AR, Pavlova NV, Kozyavkin SA, Slesarev AI (2004). "Recent developments in the optimization of thermostable DNA polymerases for efficient applications". Trends Biotechnol. 22: 253–260. PMID 15109812.
- ↑ 5.0 5.1 D.J. Sharkey, E.R. Scalice, K.G. Christy Jr., S.M. Atwood, and J.L. Daiss (1994). "Antibodies as Thermolabile Switches: High Temperature Triggering for the Polymerase Chain Reaction". Bio/Technology. 12: 506–509.
- ↑ http://photoscience.la.asu.edu/photosyn/courses/BIO_343/lecture/DNAtech.html
- ↑ Kleppe K, Ohtsuka E, Kleppe R, Molineux I, Khorana HG (1971). "Studies on polynucleotides. XCVI. Repair replications of short synthetic DNA's as catalyzed by DNA polymerases". J. Mol. Biol. 56: 341–361.
- ↑ 8.0 8.1 Mullis, Kary (1998). Dancing Naked in the Mind Field. New York: Pantheon Books. ISBN 0-679-44255-3.
- ↑ Rabinow, Paul (1996). Making PCR: A Story of Biotechnology. Chicago: University of Chicago Press. ISBN 0-226-70146-8.
- ↑ Mullis, Kary (1990). "The unusual origin of the polymerase chain reaction". Scientific American. 262 (4): 56–61, 64–5.
- ↑ Advice on How to Survive the Taq Wars ¶2: GEN Genetic Engineering News Biobusiness Channel: Article. May 1 2006 (Vol. 26, No. 9).
- ↑ Pavlov AR, Pavlova NV, Kozyavkin SA, Slesarev AI (2006). "Thermostable DNA Polymerases for a Wide Spectrum of Applications: Comparison of a Robust Hybrid TopoTaq to other enzymes". In Kieleczawa J. DNA Sequencing II: Optimizing Preparation and Cleanup. Jones and Bartlett. pp. pp. 241-257. ISBN 0-7637338-3-0.
- ↑ Myriam Vincent, Yan Xu and Huimin Kong (2004). "Helicase-dependent isothermal DNA amplification". EMBO reports. 5 (8): 795–800.
- ↑ Q. Chou, M. Russell, D.E. Birch, J. Raymond and W. Bloch (1992). "Prevention of pre-PCR mis-priming and primer dimerization improves low-copy-number amplifications". Nucleic Acids Research. 20: 1717–1723.
- ↑ E. Zietkiewicz, A. Rafalski, and D. Labuda (1994). "Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification". Genomics. 20 (2): 176–83.
- ↑ Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB (1996). "Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands". Proc Natl Acad Sci U S A. 93 (13): 9821–9826. PMID 8790415.
- ↑ Y.G. Liu and R. F. Whittier (1995). "Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking". Genomics. 25 (3): 674–81.
- ↑ David, F.Turlotte, E., (1998). "An Isothermal Amplification Method". C.R.Acad. Sci Paris, Life Science. 321 (1): 909–914.
- ↑ http://www.lab-rech-associatives.com/pdf/Utiliser%20la%20Topologie%20de%20l'ADN.pdf
Resources
- PCR at Home - Amateur Scientist article in the July 2000 issue of Scientific American on performing PCR reactions with low-cost household materials.
- US Patent for PCR
- Narrated animation and step-through animation of PCR - From the educational multimedia company Sumanas. Adobe Flash required.
- Step-through animation of PCR - From Cold Spring Harbor's Dolan DNA Learning Center. Adobe Flash required.
ar:تفاعل البلمرة المتسلسل az:Polimeraz zəncir reaksiyası bg:Полимеразна верижна реакция ca:Reacció en cadena per la polimerasa cs:Polymerázová řetězová reakce da:PCR de:Polymerase-Kettenreaktion el:Αλυσιδωτή αντίδραση πολυμεράσης eo:PĈR ko:중합 효소 연쇄 반응 hr:PCR id:Reaksi berantai polimerase it:Reazione a catena della polimerasi he:PCR lt:PGR hu:PCR nl:Polymerase-kettingreactie no:PCR sl:Verižna reakcija s polimerazo fi:Polymeraasiketjureaktio sv:PCR uk:Полімеразна ланцюгова реакція ur:پی سی آر