Core promoter gene transcriptions: Difference between revisions
| Line 780: | Line 780: | ||
! Cores pn (2846-2811) !! Cores pn (2846-2811) !! Cores pn (2846-2811) !! Cores pn (2846-2811) | ! Cores pn (2846-2811) !! Cores pn (2846-2811) !! Cores pn (2846-2811) !! Cores pn (2846-2811) | ||
|- | |- | ||
| - || - || ciAAAACAA at 2842 || | | - || - || ciAAAACAA at 2842 || CAACC at 2844 | ||
|- | |- | ||
! Proximals nn (2811-2596) !! Proximals nn (2811-2596) !! Proximals nn (2811-2596) !! Proximals nn (2811-2596) | ! Proximals nn (2811-2596) !! Proximals nn (2811-2596) !! Proximals nn (2811-2596) !! Proximals nn (2811-2596) | ||
| Line 788: | Line 788: | ||
| - || - || TCGTACT at 2784 || - | | - || - || TCGTACT at 2784 || - | ||
|- | |- | ||
| - || - || TCGGACC at 2770 || | | - || - || TCGGACC at 2770 || GGACC at 2770 | ||
|- | |- | ||
| - || - || ciAGTACGG at 2753 || | | - || - || ciAGTACGG at 2753 || ciTGTCT at 2778 | ||
|- | |- | ||
| - || - || GTCACT at 2739 Ngoc (2017) || | | - || - || GTCACT at 2739 Ngoc (2017) || GGTCG at 2766 | ||
|- | |- | ||
| - || - || TTGGACC at 2720 || | | - || - || TTGGACC at 2720 || GGACC at 2720 | ||
|- | |- | ||
| - || - || | | - || - || - || ciCGACT at 2744 | ||
|- | |- | ||
| - || | | - || - || - || ciGAACT at 2714 | ||
|- | |- | ||
| - || - || | | - || - || - || ciCAACT at 2705 | ||
|- | |- | ||
| - || - || | | - || - || - || ciCGACT at 2696 | ||
|- | |- | ||
| - || - || TCACACC at 2605 || | | - || - || - || ciTGTCC at 2689 | ||
|- | |||
| - || - || GTCACA at 2656 Ngoc (2017) || GGTCG at 2681 | |||
|- | |||
| - || - || - || GGACAT at 2673 | |||
|- | |||
| - || ciATTTATA at 2638 || TCACACC at 2658 || GGACA at 2672 | |||
|- | |||
| - || - || CCACTTT at 2619 || GGTCA at 2654 | |||
|- | |||
| - || - || TTGTACC at 2614 || AGTCG at 2650 | |||
|- | |||
| - || - || - || GGTTGT at 2611 Juven-Gershon (2010) | |||
|- | |||
| - || - || TCACACC at 2605 || GGTTG at 2610 | |||
|- | |- | ||
| - || - || GTCACA at 2603 Ngoc (2017) || - | | - || - || GTCACA at 2603 Ngoc (2017) || - | ||
|- | |||
| - || - || - || GGTCA at 2601 | |||
|- | |||
! Proximals pn (2811-2596) !! Proximals pn (2811-2596) !! Proximals pn (2811-2596) !! Proximals pn (2811-2596) | |||
|- | |||
| - || - || - || ATGACT at 2786 Juven-Gershon (2010) | |||
|- | |||
| - || - || - || AGTTG at 2733 | |||
|- | |||
| - || - || - || ciGAACC at 2717 | |||
|- | |||
| - || - || - || AGTTG at 2704 | |||
|- | |||
| - || - || - || - | |||
|- | |||
| - || - || - || AGACC at 2598 | |||
|- | |- | ||
! Distal nn (2596-1) !! Distal nn (2596-1) !! Distal nn (2596-1) !! Distal nn (2596-1) | ! Distal nn (2596-1) !! Distal nn (2596-1) !! Distal nn (2596-1) !! Distal nn (2596-1) | ||
|- | |||
| - || - || - || ciCAACT at 2593 | |||
|- | |- | ||
| - || - || - || AGTCCT at 2588 Juven-Gershon (2010) | | - || - || - || AGTCCT at 2588 Juven-Gershon (2010) | ||
|- | |- | ||
| - || - || - || AGTCC at 2587 | | - || - || - || AGTCC at 2587 | ||
|- | |||
| - || - || - || ciGAACT at 2580 | |||
|- | |||
| - || - || - || ciCGTCC at 2568 | |||
|- | |||
| - || - || - || ciCGACT at 2562 | |||
|- | |- | ||
| - || - || - || GGTTG at 2547 | | - || - || - || GGTTG at 2547 | ||
| Line 819: | Line 857: | ||
|- | |- | ||
| - || - || - || GGTCC at 2519 | | - || - || - || GGTCC at 2519 | ||
|- | |||
| - || - || - || ciTGTCC at 2514 | |||
|- | |- | ||
| - || - || - || AGTTA at 2496 | | - || - || - || AGTTA at 2496 | ||
|- | |||
| - || - || - || ciTGTCT at 2443 | |||
|- | |- | ||
| - || - || - || GGACC at 2435 | | - || - || - || GGACC at 2435 | ||
|- | |- | ||
| - || - || - || GGTCG at 2431 | | - || - || - || GGTCG at 2431 | ||
|- | |||
| - || - || - || ciCGTCC at 2389 | |||
|- | |- | ||
| - || - || - || GGACC at 2385 | | - || - || - || GGACC at 2385 | ||
|- | |||
| - || - || - || ciGAACT at 2379 | |||
|- | |||
| - || - || - || ciCGTCC at 2367 | |||
|- | |||
| - || - || - || ciCGACT at 2361 | |||
|- | |- | ||
| - || - || - || GGTCG at 2346 | | - || - || - || GGTCG at 2346 | ||
|- | |||
| - || - || - || ciCGACC at 2326 | |||
|- | |- | ||
| - || - || - || GGACA at 2337 | | - || - || - || GGACA at 2337 | ||
| Line 839: | Line 891: | ||
|- | |- | ||
| - || - || - || AGTCC at 2250 | | - || - || - || AGTCC at 2250 | ||
|- | |||
| - || - || - || ciCGACT at 2226 | |||
|- | |- | ||
| - || - || - || GGTCA at 2211 | | - || - || - || GGTCA at 2211 | ||
| Line 847: | Line 901: | ||
|- | |- | ||
| - || - || - || AGTCC at 2134 | | - || - || - || AGTCC at 2134 | ||
|- | |||
| - || - || - || ciGAACT at 2127 | |||
|- | |||
| - || - || - || ciTGTCT at 2119 | |||
|- | |||
| - || - || - || ciCGACT at 2109 | |||
|- | |- | ||
| - || - || - || GGATC at 2093 | | - || - || - || GGATC at 2093 | ||
|- | |- | ||
| - || - || - || GGTCC at 2077 | | - || - || - || GGTCC at 2077 | ||
|- | |||
| - || - || - || ciTGTCT at 2017 | |||
|- | |- | ||
| - || - || - || AGACA at 2029 | | - || - || - || AGACA at 2029 | ||
|- | |||
| - || - || - || ciCGACC at 2069 | |||
|- | |- | ||
| - || - || - || GGACC at 2009 | | - || - || - || GGACC at 2009 | ||
|- | |- | ||
| - || - || - || GGTCG at 2005 | | - || - || - || GGTCG at 2005 | ||
|- | |||
| - || - || - || ciCGTCT at 1967 | |||
|- | |- | ||
| - || - || - || GGACC at 1959 | | - || - || - || GGACC at 1959 | ||
|- | |||
| - || - || - || ciCGTCC at 1941 | |||
|- | |||
| - || - || - || ciTGACT at 1935 | |||
|- | |||
| - || - || - || ciGAACC at 1927 | |||
|- | |- | ||
| - || - || - || GGTCG at 1920 | | - || - || - || GGTCG at 1920 | ||
|- | |- | ||
| - || - || - || GGACA at 1911 | | - || - || - || GGACA at 1911 | ||
|- | |||
| - || - || - || ciCGACC at 1891 | |||
|- | |- | ||
| - || - || - || AGATG at 1867 | | - || - || - || AGATG at 1867 | ||
|- | |||
| - || - || - || ciCAACT at 1853 | |||
|- | |- | ||
| - || - || - || GGACC at 1841 | | - || - || - || GGACC at 1841 | ||
|- | |||
| - || - || - || ciCGTCC at 1823 | |||
|- | |- | ||
| - || - || - || GGTTC at 1817 | | - || - || - || GGTTC at 1817 | ||
|- | |||
| - || - || - || ciCGACT at 1800 | |||
|- | |- | ||
| - || - || - || GGTCG at 1785 | | - || - || - || GGTCG at 1785 | ||
|- | |- | ||
| - || - || - || AGACA at 1776 | | - || - || - || AGACA at 1776 | ||
|- | |||
| - || - || - || ciCGACC at 1756 | |||
|- | |||
| - || - || - || ciCGACC at 1746 | |||
|- | |||
| - || - || - || ciTATCT at 1710 | |||
|- | |||
| - || - || - || ciGGTCT at 1670 | |||
|- | |||
| - || - || - || ciCATCT at 1653 | |||
|- | |||
| - || - || - || ciGAACC at 1649 | |||
|- | |||
| - || - || - || ciGGACT at 1623 | |||
|- | |||
| - || - || - || ciCGTCT at 1614 | |||
|- | |- | ||
| - || - || - || GGTCG at 1611 | | - || - || - || GGTCG at 1611 | ||
|- | |||
| - || - || - || ciTGTCT at 1567 | |||
|- | |- | ||
| - || - || - || GGTCA at 1532 | | - || - || - || GGTCA at 1532 | ||
|- | |- | ||
| - || - || - || AGATA at 1525 | | - || - || - || AGATA at 1525 | ||
|- | |||
| - || - || - || ciGGTCT at 1518 | |||
|- | |- | ||
| - || - || - || AGTTG at 1513 | | - || - || - || AGTTG at 1513 | ||
|- | |- | ||
| - || - || - || AGTCG at 1486 | | - || - || - || AGTCG at 1486 | ||
|- | |||
| - || - || - || ciCGACC at 1464 | |||
|- | |- | ||
| - || - || - || GGTCC at 1460 | | - || - || - || GGTCC at 1460 | ||
|- | |- | ||
| - || - || - || AGACA at 1452 | | - || - || - || AGACA at 1452 | ||
|- | |||
| - || - || - || ciGGTCT at 1411 | |||
|- | |- | ||
| - || - || - || AGTTG at 1406 | | - || - || - || AGTTG at 1406 | ||
| Line 893: | Line 997: | ||
|- | |- | ||
| - || - || - || GGTCA at 1352 | | - || - || - || GGTCA at 1352 | ||
|- | |||
| - || - || - || ciCGTCT at 1314 | |||
|- | |||
| - || - || - || ciGATCC at 1307 | |||
|- | |- | ||
| - || - || - || GGATC at 1306 | | - || - || - || GGATC at 1306 | ||
|- | |||
| - || - || - || ciGAACT at 1300 | |||
|- | |||
| - || - || - || ciCGTCC at 1288 | |||
|- | |||
| - || - || - || ciCGACT at 1282 | |||
|- | |- | ||
| - || - || - || AGTCC at 1275 | | - || - || - || AGTCC at 1275 | ||
| Line 909: | Line 1,023: | ||
|- | |- | ||
| - || - || - || GGTCG at 1194 | | - || - || - || GGTCG at 1194 | ||
|- | |||
| - || - || - || ciGGACT at 1173 | |||
|- | |- | ||
| - || - || - || GGTCG at 1140 | | - || - || - || GGTCG at 1140 | ||
|- | |- | ||
| - || - || - || GGACA at 1131 | | - || - || - || GGACA at 1131 | ||
|- | |||
| - || - || - || ciCGACC at 1111 | |||
|- | |- | ||
| - || - || - || AGACA at 1085 | | - || - || - || AGACA at 1085 | ||
|- | |||
| - || - || - || ciTGTCT at 1073 | |||
|- | |- | ||
| - || - || - || GGTCG at 1061 | | - || - || - || GGTCG at 1061 | ||
|- | |||
| - || - || - || ciTAACC at 1045 | |||
|- | |||
| - || - || - || ciCGTCT at 1023 | |||
|- | |- | ||
| - || - || - || GGACC at 1015 | | - || - || - || GGACC at 1015 | ||
|- | |||
| - || - || - || ciGAACT at 1009 | |||
|- | |||
| - || - || - || ciCGTCC at 997 | |||
|- | |||
| - || - || - || ciCGACT at 991 | |||
|- | |- | ||
| - || - || - || AGTCC at 984 | | - || - || - || AGTCC at 984 | ||
|- | |- | ||
| - || - || - || GGTCG at 976 | | - || - || - || GGTCG at 976 | ||
|- | |||
| - || - || - || ciCATCT at 970 | |||
|- | |- | ||
| - || - || - || GGACA at 967 | | - || - || - || GGACA at 967 | ||
| Line 929: | Line 1,061: | ||
|- | |- | ||
| - || - || - || AGACA at 919 | | - || - || - || AGACA at 919 | ||
|- | |||
| - || - || - || ciTGTCT at 907 | |||
|- | |- | ||
| - || - || - || GGACC at 899 | | - || - || - || GGACC at 899 | ||
| Line 937: | Line 1,071: | ||
|- | |- | ||
| - || - || - || GGTCC at 850 | | - || - || - || GGTCC at 850 | ||
|- | |||
| - || - || - || ciGAACT at 843 | |||
|- | |||
| - || - || - || ciCGTCC at 831 | |||
|- | |||
| - || - || - || ciCGACT at 825 | |||
|- | |- | ||
| - || - || - || GGTCG at 810 | | - || - || - || GGTCG at 810 | ||
|- | |- | ||
| - || - || - || GGACA at 801 | | - || - || - || GGACA at 801 | ||
|- | |||
| - || - || - || ciCGACC at 781 | |||
|- | |- | ||
| - || - || - || AGATG at 758 | | - || - || - || AGATG at 758 | ||
|- | |- | ||
| - || - || - || GGTCG at 737 | | - || - || - || GGTCG at 737 | ||
|- | |||
| - || - || - || ciGGACT at 732 | |||
|- | |- | ||
| - || - || - || GGTCG at 728 | | - || - || - || GGTCG at 728 | ||
|- | |- | ||
| - || - || - || AGTCC at 714 | | - || - || - || AGTCC at 714 | ||
|- | |||
| - || - || - || ciCGTCC at 697 | |||
|- | |- | ||
| - || - || - || GGTTC at 692 | | - || - || - || GGTTC at 692 | ||
| Line 957: | Line 1,103: | ||
|- | |- | ||
| - || - || - || GGTCC at 648 | | - || - || - || GGTCC at 648 | ||
|- | |||
| - || - || - || ciTAACC at 643 | |||
|- | |- | ||
| - || - || - || AGATG at 624 | | - || - || - || AGATG at 624 | ||
|- | |- | ||
| - || - || - || GGACC at 596 | | - || - || - || GGACC at 596 | ||
|- | |||
| - || - || - || ciTAACT at 585 | |||
|- | |- | ||
| - || - || - || AGTCC at 578 | | - || - || - || AGTCC at 578 | ||
|- | |||
| - || - || - || ciCGTCC at 565 | |||
|- | |||
| - || - || - || ciTGTCC at 561 | |||
|- | |- | ||
| - || - || - || GGTTC at 556 | | - || - || - || GGTTC at 556 | ||
| Line 977: | Line 1,131: | ||
|- | |- | ||
| - || - || - || AGTCC at 441 | | - || - || - || AGTCC at 441 | ||
|- | |||
| - || - || - || ciTGTCC at 424 | |||
|- | |- | ||
| - || - || - || GGTTC at 419 | | - || - || - || GGTTC at 419 | ||
| Line 983: | Line 1,139: | ||
|- | |- | ||
| - || - || - || GGACA at 394 | | - || - || - || GGACA at 394 | ||
|- | |||
| - || - || - || ciTATCT at 355 | |||
|- | |||
| - || - || - || ciGAACC at 328 | |||
|- | |||
| - || - || - || ciTGACT at 307 | |||
|- | |||
| - || - || - || ciTGTCT at 289 | |||
|- | |||
| - || - || - || ciCATCT at 284 | |||
|- | |- | ||
| - || - || - || GGTCC at 262 | | - || - || - || GGTCC at 262 | ||
|- | |- | ||
| - || - || - || AGATA at 234 | | - || - || - || AGATA at 234 | ||
|- | |||
| - || - || - || ciTGTCT at 168 | |||
|- | |||
| - || - || - || ciCGACT at 140 | |||
|- | |||
| - || - || - || ciTGACT at 130 | |||
|- | |||
| - || - || - || ciCATCC at 119 | |||
|- | |||
| - || - || - || ciTATCT at 100 | |||
|- | |||
| - || - || - || ciCAACT at 85 | |||
|- | |||
| - || - || - || GGTCG at 35 | |||
|- | |||
| - || - || - || ciTGACT at 17 | |||
|- | |||
| - || - || - || ciTGTCT at 13 | |||
|- | |||
! Distal pn (2596-1) !! Distal pn (2596-1) !! Distal pn (2596-1) !! Distal pn (2596-1) | |||
|- | |- | ||
| - || - || - || GGTCG at 35 | | - || - || - || GGTCG at 35 | ||
Revision as of 07:11, 9 July 2023
Editor-In-Chief: Henry A. Hoff
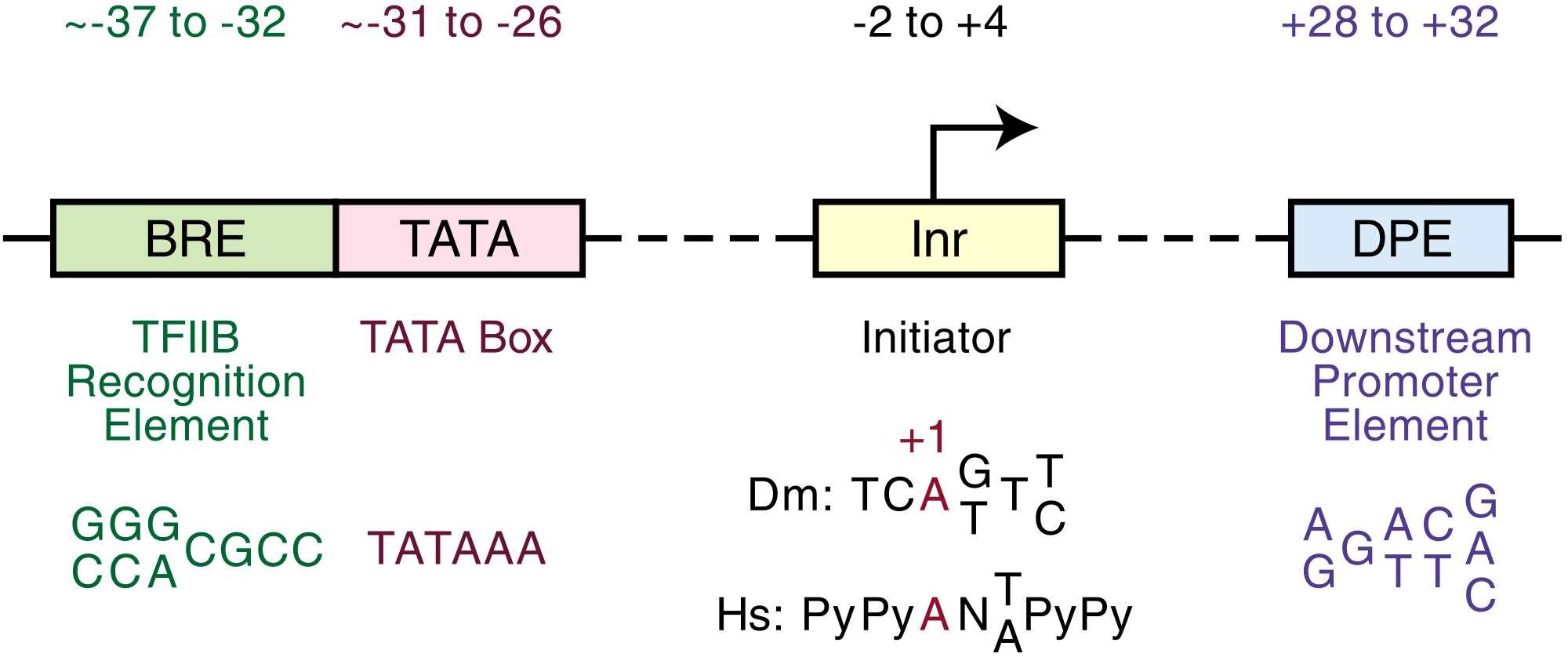
{{free media}}A core promoter is that portion of the proximal promoter that contains the transcription start sites.
Biochemical definition: the minimal stretch of DNA sequence that is sufficient to direct accurate initiation of transcription. An acceptable range of the length of a core promoter is typically 60 to 120 base pairs.
Genomics definition: short sequences surrounding the transcription start sites (TSSs).
It contains a binding site for RNA polymerase (RNA polymerase I, RNA polymerase II, or RNA polymerase III) holoenzymes.
A vast network of regulatory factors that contribute to the initiation of transcription by RNA polymerase ultimately target any specific gene’s core promoter.
The core promoter includes the transcription start site(s) (TSS).
That portion of the core promoter that is upstream of the TSS is also part of the proximal promoter.
The core promoter is approximately -34 bp upstream from the TSS. "Several factors have been identified that bind to core promoters (reviewed in Smale, 1997)"[2][3].
Genetics
Genetics involves the expression, transmission, and variation of inherited characteristics.
Gene transcriptions
DNA is a double helix of interlinked nucleotides surrounded by an epigenome. On the basis of biochemical signals, an enzyme, specifically a ribonucleic acid (RNA) polymerase, is chemically bonded to one of the strands (the template strand) of this double helix. The polymerase, once phosphorylated, begins to catalyze the formation of RNA using the template strand. Although the catalysis may have more than one beginning nucleotide (a start site) and more than one ending nucleotide (a stop site) along the DNA, each nucleotide sequence catalyzed that ultimately produces approximately the same RNA is part of a gene. The catalysis of each RNA representation from the template DNA is a transcription, specifically a gene transcription. The overall process is also referred to as gene transcription.
Promoters
Def. a "section of DNA that controls the initiation of RNA transcription as a product of a gene"[4] is called a promoter.
Proximal promoters
Def a section of promoter DNA which includes the transcription start sites that is neighboring the start sites is called a proximal promoter.
Cores
Def. a central or most important part of something is called a core.
Theoretical core promoters
Def. "the factors, including RNA polymerase II itself, that are minimally essential for transcription in vitro from an isolated core promoter" is called the basal machinery, or basal transcription machinery.[5]
Def. one or more sequence motifs containing the transcription start sites (TSSs), juxtaposed to the motif containing the TSSs, or in the proximal promoter that are only found in this core of motifs is called a core promoter.
Metal responsive elements
A metal responsive element (MRE), or TGC box, may occur in the core promoter of some human DNA genes.
"The metallothionein (MT) genes provide a good example of eucaryotic promoter architecture. MT genes specify the synthesis of low-molecular-weight metal-binding proteins. They are transcriptionally regulated by the metal ions cadmium and zinc (11), glucocorticoid hormones (18), interferon (14), interleukin-1 (22), and tumor promoters (2). The metal ion regulation of MTs is conferred by a short sequence element called the metal-responsive element (MRE [21]) or TGC box (31, 34), which functions as a metal ion-dependent enhancer."[6]
GC boxes
Def. a "sequence of contiguous guanine, guanine, guanine, cytosine, and guanine, in that order, along a DNA strand"[7] is called a GC box.
"[A] GC box is a distinct pattern of nucleotides found in the promoter region of some eukaryotic genes upstream of the TATA box and approximately 110 bases upstream from the transcription initiation site. It has a consensus sequence GGGCGG which is position dependent and orientation independent. The GC elements are bound by transcription factors and have similar functions to enhancers.[8]"[9]
"A large subclass of polymerase II promoters lacks both TATAA and CCAAT sequence motifs but contains multiple GC boxes. This promoter class includes several housekeeping genes (e.g., the genes encoding dihydrofolate reductase [DHFR] ..., hydroxymethylglutaryl coenzyme A reductase [39], hypoxanthine guanine phosphoribosyltransferase [33], and adenosine deaminase [46]) [and] nonhousekeeping genes (e.g., the transforming growth factor alpha [9, 23], rat malic enzyme [36], human c-Ha-ras [21], epidermal growth factor receptor [22], and nerve growth factor receptor [42] genes)."[10]
"[A] GC box-binding factor is required for transcription and ... a truncated promoter containing one GC box is transcriptionally inactive (44). ... the DNA-protein interactions occurring at the GC boxes in the DHFR promoter are functionally distinct and that factors binding to the GC boxes must interact in a position-dependent manner."[10]
"In promoters containing multiple GC boxes but lacking the TATAA box, transcription start sites may be single and specific, as observed in the nerve growth factor receptor gene (42) and the cellular retinol-binding protein gene (37), or there may be multiple heterogeneous start sites, such as those found in the c-myb (4), insulin receptor (45), and Ha-ras (21) genes. ... GC boxes are responsible for directing transcription from the major and the minor start sites. ... All TATAA-less promoters have at least two GC boxes"[10].
"A GC box sequence, one of the most common regulatory DNA elements of eukaryotic genes, is recognized by the Spl transcription factor; its consensus sequence is represented as 5'-G/T G/A GGCG G/T G/A G/A C/T-3' [or 5′-KRGGCGKRRY-3′] (Briggs et al., 1986)."[11]
HY boxes
A core responsive element is the hypertrophy region HY box between -89 and -60 nucleotides (nts) upstream from the transcription start site.[12]
CAAT boxes
"[A] CCAAT box (also sometimes abbreviated a CAAT box or CAT box) is a distinct pattern of nucleotides with GGCCAATCT consensus sequence that occur upstream by 75-80 bases to the initial transcription site. The CAAT box signals the binding site for the RNA transcription factor, and is typically accompanied by a conserved consensus sequence. It is an invariant DNA sequence at about minus 70 base pairs from the origin of transcription in many eukaryotic promoters. Genes that have this element seem to require it for the gene to be transcribed in sufficient quantities. It is frequently absent from genes that encode proteins used in virtually all cells. This box along with the GC box is known for binding general transcription factors. CAAT and GC are primarily located in the region from 100-150bp upstream from the TATA box. Both of these consensus sequences belong to the regulatory promoter. Full gene expression occurs when transcription activator proteins bind to each module within the regulatory promoter. Protein specific binding is required for the CCAAT box activation. These proteins are known as CCAAT box binding proteins/CCAAT box binding factors. A CCAAT box is a feature frequently found before eukaryote coding regions".[13]
B recognition elements
"The B recognition element (BRE) is a DNA sequence found in the promoter region of most genes in eukaryotes and Archaea.[14][15] The BRE is a cis-regulatory element that is found immediately upstream of the TATA box, and consists of 7 nucleotides."[16]
"The Transcription Factor IIB (TFIIB) recognizes this sequence in the DNA, and binds to it. The fourth and fifth alpha helices of TFIIB intercalate with the major groove of the DNA at the BRE. TFIIB is one part of the preinitiation complex that helps RNA Polymerase II bind to the DNA."[16]
The consensus sequence is 5’-G/C G/C G/A C G C C-3’.[17]
The general consensus sequence using degenerate nucleotides is 5’-SSRCGCC-3’, where S = G or C and R = A or G.[18]
"The position in nucleotides (nt) relative to the transcription start site (TSS, +1)" is -35 for the BRE. Of human promoters, some "22-25% [are] BRE containing promoters ... the functional consensus sequences for BRE ... motif [is] still poorly defined."[18]
EIF4E basal elements
The EIF4E basal element, also eIF4E, (4EBE) is a basal promoter element for the eukaryotic translation initiation factor 4E. "Interactions between 4EBE and upstream activator sites are position, distance, and sequence dependent."[19]
TATA boxes
Def. a "DNA sequence (cis-regulatory element) found in the promoter region of genes in archaea and eukaryotes"[20] is called a TATA box.
The TATA box can be an AT-rich sequence "located at a fixed distance upstream of the transcription start site"[5].
TBP-like factors
Notation: let the symbol TLF designate a TATA binding protein-like factor.
The human gene TBPL1 (TBP-like 1, also TLF and TRF2[5]), GeneID: 9519, encodes a protein that "does not bind to the TATA box and initiates transcription from TATA-less promoters."[21]
Downstream TFIIB recognition
The downstream TFIIB recognition element (dBRE) has a consensus sequence in the transcription direction on the template strand of 3'-RTDKKKK-5', using degenerate nucleotides, or 3'-A/G-T-A/G/T-G/T-G/T-G/T-G/T-5'.[22]
dBRE is cis-TATA box, between the TATA box and the Inr or transcription start site (TSS) and trans-TSS.[22]
Initiator elements
For RNA polymerase II holoenzyme to transcribe a gene, the gene's promoter must be located. After the promoter is located, the transcription start site (TSS) is pinpointed by using nucleotide sequences that include the TSS or perhaps allow distance measurement to the TSS. Within the promoter, most human genes lack a TATA box and have an initiator element (Inr) or downstream promoter element instead.
"RNA pol II itself recognizes features of the Inr which might assist the correct positioning of the polymerase on the promoter (Carcamo et al., 1991; Weis and Reinberg, 1997)."[2][23][24]
Transcription start sites
The transcription start site (TSS) is the location on the DNA template strand where transcription begins at the 3'-end of a gene.[25] This location corresponds to the 5'-end of the mRNA which by convention is used to designate DNA locations.[25] For example, the 5'-TATA-box-3' designation refers to the directionality of the mRNA and corresponds to the 3'-TATA-box-5' designation for nucleotides on the template strand.[25] The template strand is the DNA strand being transcribed by RNA polymerase.[25]
Downstream core elements
"[N]onredundant human promoter sequences 600 bp long (−499 to +100 bp around the TSS) [are available] from [the] Eukaryotic Promoter Database (EPD) release 75 (4, 68) (http://www.epd.isb-sib.ch/), and ... promoters sequences 1,200 bp long (−1,000 to +200 bp) [are available] from the Database of Transcriptional Start Sites (DBTSS) (59, 74, 75) (http://dbtss.hgc.jp/index.html)"[26].
The downstream core element (DCE) is a transcription core promoter sequence that is within the transcribed portion of a gene.
The consensus sequence for the DCE is CTTC...CTGT...AGC.[26] These three consensus elements are referred to as subelements: "SI is CTTC, SII is CTGT, and SIII is AGC."[26]
The number of nucleotides between each subelement can apparently vary down to none.
A core promoter that contains all three subelements may be much less common than one containing only one or two.[26] "SI resides approximately from +6 to +11, SII from +16 to +21, and SIII from +30 to +34."[26]
SI as 3'-CTTC-5' can occur as 3 of 4 (CTT, TTC) or 4 of 4 (CTTC). SII as 3'-CTGT-5' can also occur as 3 of 4 (CTG, TGT) or 4 of 4 (CTGT). SIII as AGC is not known to vary.
DCE SIII can function independently of SI and SII.[26]
Transcription factor II D (TFIID), a transcription factor that is part of the RNA polymerase II holoenzyme, interacts with promoters containing only SIII of the DCE suggesting a critical spacing parameter between SIII and the TATA box, initiator element, or some combination of the two.[26] TFIID probably serves as a core promoter recognition complex.[26]
TAF1 interacts with the DCE in a sequence-dependent manner.[26]
The differences between core promoters with downstream elements may be explained by
- "TATA- and DPE-dependent promoters are specific for particular enhancers"[26],
- "preferences of activators for specific core promoter architectures"[26], and
- "the presence of a DCE or [downstream core promoter element (DPE)] might be indicative of an architecture designed for specific regulatory networks, such as the regulation of housekeeping promoters versus tissue-specific promoters (or other highly regulated promoters) or the regulation of subsets of viral promoters."[26]
Motif ten elements
The motif ten element (MTE) is a downstream core promoter element that "promotes transcription by RNA polymerase II when it is located precisely at positions +18 to +27 relative to A+1 in the initiator (Inr) element."[27]
The motif 10 consensus sequence is CSARCSSAACGS [5'-C-C/G-A-A/G-C-C/G-C/G-A-A-C-G-C/G-3'].[27] By convention, the consensus sequence 5'-C-C/G-A-A/G-C-C/G-C/G-A-A-C-G-C/G-3' is stated as it would be translated into mRNA. In the direction of transcription on the template strand this consensus sequence becomes 3'-C-C/G-A-A/G-C-C/G-C/G-A-A-C-G-C/G-5'.
Downstream promoter elements
"The downstream promoter element (DPE) is a core promoter element ... present in other species including humans and excluding Saccharomyces cerevisiae.[28] Like all core promoters, the DPE plays an important role in the initiation of gene transcription by RNA polymerase II."[29]
The core sequence of the DPE is located precisely +28 to +32 nts relative to the A+1 nt in the Inr.[17]
Hypotheses
- Each portion of a DNA that becomes active has a core promoter.
- The "minimal portion of the promoter required to properly initiate transcription".[30]
Comparisons of negative direction promoter elements
| Butler (2002) | Watson (2014) | Juven-Gershon (2008) | Butler (2002) |
|---|---|---|---|
| ~-37 to -32 BREu SSRCGCC | ~-31 to -26 TATAWAW | -2 to +4 Inr YYRNWYY | +28 to +32 DPE RGWYV |
| UTR nn(4560-2846) | UTR nn(4560-2846) | UTR nn(4560-2846) | UTR nn(4560-2846) |
| - | - | TTACTCC at 4557 | - |
| - | - | ciAGTGTAA at 4533 | - |
| - | - | - | GGACC at 4546 |
| - | - | - | ciTGTCT at 4518 |
| - | - | - | GGACC at 4494 |
| - | - | - | AGTCG at 4489 |
| - | - | - | GGTCG at 4480 |
| - | - | - | ciGGTCT at 4448 |
| - | - | - | AGTCC at 4436 |
| - | - | - | AGATG at 4430 |
| - | - | - | GGTCA at 4415 |
| - | - | TCACACT at 4361 | - |
| - | - | GTCACA at 4359 Ngoc (2017) | - |
| - | - | CCCACT at 4353 Ngoc (2017) | GGACA at 4369 |
| - | - | TCGGACC at 4349 | - |
| - | - | GTCACT at 4319 Ngoc (2017) | GGACC at 4349 |
| - | - | - | GGTCG at 4345 |
| - | - | CCAGTTT at 4309 | - |
| - | - | TCCAGT at 4307 Ngoc (2017) | - |
| - | - | TCGGACC at 4300 | GGACC at 4300 |
| - | - | - | ciGAACT at 4294 |
| - | - | ciGGTCCGA at 4255 | ciTGTCC at 4282 |
| - | - | - | ciCGACT at 4276 |
| - | - | CTGCACC at 4238 | ciGAACC at 4268 |
| - | - | TCGGTCT at 4233 | GGTCG at 4261 |
| - | - | - | GGTCC at 4253 |
| - | - | TCACTCT at 4202 | ciGGTCT at 4233 |
| - | - | GTCACT at 4200 Ngoc (2017) | - |
| - | - | TCGAACC at 4188 | AGATG at 4212 |
| - | - | - | GGACA at 4208 |
| - | - | - | ciGAACC at 4188 |
| - | - | - | AGTTC at 4178 |
| - | - | CCGGTCC at 4170 | GGTCC at 4170 |
| - | - | ciAGTACGG at 4118 | ciCGACT at 4145 |
| - | - | CCGTACC at 4107 | AGTCC at 4138 |
| - | - | CCGGTCC at 4102 | GGTCG at 4130 |
| - | - | TTACACT at 4092 | GGACA at 4121 |
| - | - | - | GGTCC at 4102 |
| - | - | - | ciCAACC at 4097 |
| - | - | TCACTCT at 4051 | ciTATCT at 4079 |
| - | - | TTGTATC at 4046 | - |
| - | - | TCGGACC at 4037 | AGATG at 4062 |
| - | - | - | GGACC at 4037 |
| - | - | - | GGTCG at 4033 |
| - | - | - | AGTTC at 4027 |
| - | - | - | GGTTC at 4019 |
| - | - | - | ciGAACT at 4012 |
| - | - | ciAGTGTGG at 3967 | ciCGACT at 3994 |
| - | - | CCGGTCC at 3951 | GGTTG at 3979 |
| - | - | TTCACA at 3939 Ngoc (2017) | GGACA at 3970 |
| - | - | CTACTTT at 3922 | GGTCC at 3951 |
| - | - | CTACTTT at 3922 | ciCAACC at 3946 |
| - | - | - | ciCAACC at 3942 |
| - | - | - | ciGGACT at 3932 |
| - | - | TCATTCT at 3893 | ciTGTCT at 3917 |
| - | - | TCATTC at 3892 (Butler 2002) | ciTGTCT at 3917 |
| - | - | CTCATT at 3891 Ngoc (2017) | ciTGTCT at 3917 |
| - | - | - | GGACC at 3906 |
| - | - | ciGGTCCGG at 3873 | GGTCC at 3885 |
| - | - | CTGGTCC at 3871 | GGTCC at 3871 |
| - | - | ciGGTATGG at 3858 | ciCGACC at 3864 |
| - | - | CTCATA at 3829 Ngoc (2017) | GGACG at 3861 |
| - | - | TCCACT at 3825 Ngoc (2017) | AGTTC at 3844 |
| - | - | CTACACC at 3810 | AGACC at 3835 |
| - | - | - | ciCATCT at 3820 |
| - | - | - | ciCAACT at 3805 |
| - | - | - | ciGAACC at 3793 |
| - | - | CTGTTCT at 3759 | ciGGACT at 3781 |
| - | - | - | ciTGACC at 3749 |
| - | - | - | GGACC at 3744 |
| - | - | - | GGTCGTG at 3733 Kadonaga (2002) |
| - | - | - | GGTCG at 3731 |
| - | - | - | ciCGACC at 3719 |
| - | - | - | AGACG at 3706 |
| - | - | - | GGTCG at 3701 |
| - | - | - | GGTCG at 3682 |
| - | - | - | ciTGTCT at 3672 |
| - | - | - | ciCGACT at 3649 |
| - | - | - | AGTCTC at 3645 Matsumoto (2020) |
| - | - | - | ciCAACC at 3606 |
| - | - | - | ciCGTCT at 3589 |
| - | - | - | GGTCC at 3585 |
| - | - | - | GGACG at 3579 |
| - | - | - | ciGAACT at 3571 |
| - | - | - | GGTCC at 3564 |
| - | - | - | AGACA at 3556 |
| - | - | - | ciTGACT at 3542 |
| - | - | - | ciCAACT at 3533 |
| - | - | - | ciTAACC at 3529 |
| - | - | ciGGTCTAG at 3488 | AGTTG at 3523 |
| - | - | TTGGTCT at 3486 | ciCAACT at 3505 |
| - | - | TCATTT at 3481 (Butler 2002) | ciCAACT at 3505 |
| - | - | GTCATT at 3480 Ngoc (2017) | ciCAACT at 3505 |
| - | - | TTGATCT at 3463 | ciGGTCT at 3486 |
| - | - | CCGTATC at 3446 | ciGATCT at 3463 |
| - | - | TTCACT at 3410 Ngoc (2017) | ciTATCC at 3447 |
| - | - | CCGAACT at 3401 | AGTTG at 3431 |
| - | - | ciAGTCCGA at 3398 | ciTATCT at 3422 |
| - | - | TCGTTCT at 3374 | ciGAACT at 3401 |
| - | - | TTGTTCT at 3340 | AGTCC at 3396 |
| - | - | TCGTTTT at 3313 | ciTAACT at 3358 |
| - | - | TTGTTCT at 3307 | AGACA at 3319 |
| - | - | TCGGACC at 3298 | GGACC at 3298 |
| - | - | TCGGTTC at 3273 | GGACC at 3298 |
| - | - | ciAGTGCGG at 3281 | GGTCG at 3294 |
| - | - | TCGGTTC at 3273 | GGTTC at 3273 |
| - | - | - | ciCATCT at 3256 |
| - | - | - | GGTCC at 3249 |
| - | - | - | ciCiGAACT at 3242 |
| - | - | - | ciCGACT at 3224 |
| - | - | - | AGTCC at 3217 |
| - | - | CCACACC at 3186 | GGTCG at 3209 |
| - | - | CCCACA at 3184 Ngoc (2017) | AGTCG at 3204 |
| - | - | - | GGACA at 3200 |
| - | - | TTGTATT at 3169 | ciCGACC at 3180 |
| - | - | CCACTTT at 3146 | - |
| - | - | TCCACT at 3144 Ngoc (2017) | - |
| - | - | TTGTTCC at 3141 | - |
| - | - | ciGGACCGG at 3130 | - |
| - | - | TCGGACC at 3128 | AGATG at 3158 |
| - | - | - | GGTTG at 3137 |
| - | - | TCGGACC at 3128 | GGACC at 3128 |
| - | - | - | GGTCG at 3124 |
| - | - | - | ciCAACC at 3116 |
| - | - | - | AGTCC at 3110 |
| - | - | - | ciGAACT at 3103 |
| - | - | - | ciCGACT at 3085 |
| - | - | - | GGTCGTG at 3072 Kadonaga (2002) |
| CCGCACC at 3047 | - | CCGCACC at 3047 | GGTCG at 3070 |
| - | - | ciGATTCGA at 3033 | GGACA at 3061 |
| - | - | TTGATTC at 3031 | GGACA at 3061 |
| - | - | - | ciCGACC at 3041 |
| - | - | CCGATTT at 3009 | ciCGACC at 3035 |
| - | - | - | GGATA at 2996 |
| - | - | - | AGATG at 2988 |
| - | - | TTGATTC at 2914 | ciCiGAACC at 2921 |
| - | - | ciAAAGTAG at 2887 | ciCiGAACC at 2921 |
| - | TATATAT at 2872 | TTCACA at 2860 Ngoc (2017) | ciTATCT at 2903 |
| - | ciTTATATA at 2871 | - | ciTGTCT at 2878 |
| - | ciTTTTATA at 2869 | - | ciTGTCT at 2878 |
| - | TATAAAA at 2853 | - | ciTGTCT at 2878 |
| - | - | - | GGTTA at 2848 |
| Cores nn (2846-2811) | Cores nn (2846-2811) | Cores nn (2846-2811) | Cores nn (2846-2811) |
| Cores pn (2846-2811) | Cores pn (2846-2811) | Cores pn (2846-2811) | Cores pn (2846-2811) |
| - | - | ciAAAACAA at 2842 | CAACC at 2844 |
| Proximals nn (2811-2596) | Proximals nn (2811-2596) | Proximals nn (2811-2596) | Proximals nn (2811-2596) |
| - | - | ciACTGAG at 2787 Ngoc (2017) | - |
| - | - | TCGTACT at 2784 | - |
| - | - | TCGGACC at 2770 | GGACC at 2770 |
| - | - | ciAGTACGG at 2753 | ciTGTCT at 2778 |
| - | - | GTCACT at 2739 Ngoc (2017) | GGTCG at 2766 |
| - | - | TTGGACC at 2720 | GGACC at 2720 |
| - | - | - | ciCGACT at 2744 |
| - | - | - | ciGAACT at 2714 |
| - | - | - | ciCAACT at 2705 |
| - | - | - | ciCGACT at 2696 |
| - | - | - | ciTGTCC at 2689 |
| - | - | GTCACA at 2656 Ngoc (2017) | GGTCG at 2681 |
| - | - | - | GGACAT at 2673 |
| - | ciATTTATA at 2638 | TCACACC at 2658 | GGACA at 2672 |
| - | - | CCACTTT at 2619 | GGTCA at 2654 |
| - | - | TTGTACC at 2614 | AGTCG at 2650 |
| - | - | - | GGTTGT at 2611 Juven-Gershon (2010) |
| - | - | TCACACC at 2605 | GGTTG at 2610 |
| - | - | GTCACA at 2603 Ngoc (2017) | - |
| - | - | - | GGTCA at 2601 |
| Proximals pn (2811-2596) | Proximals pn (2811-2596) | Proximals pn (2811-2596) | Proximals pn (2811-2596) |
| - | - | - | ATGACT at 2786 Juven-Gershon (2010) |
| - | - | - | AGTTG at 2733 |
| - | - | - | ciGAACC at 2717 |
| - | - | - | AGTTG at 2704 |
| - | - | - | - |
| - | - | - | AGACC at 2598 |
| Distal nn (2596-1) | Distal nn (2596-1) | Distal nn (2596-1) | Distal nn (2596-1) |
| - | - | - | ciCAACT at 2593 |
| - | - | - | AGTCCT at 2588 Juven-Gershon (2010) |
| - | - | - | AGTCC at 2587 |
| - | - | - | ciGAACT at 2580 |
| - | - | - | ciCGTCC at 2568 |
| - | - | - | ciCGACT at 2562 |
| - | - | - | GGTTG at 2547 |
| - | - | - | GGACA at 2538 |
| - | - | - | GGTCC at 2519 |
| - | - | - | ciTGTCC at 2514 |
| - | - | - | AGTTA at 2496 |
| - | - | - | ciTGTCT at 2443 |
| - | - | - | GGACC at 2435 |
| - | - | - | GGTCG at 2431 |
| - | - | - | ciCGTCC at 2389 |
| - | - | - | GGACC at 2385 |
| - | - | - | ciGAACT at 2379 |
| - | - | - | ciCGTCC at 2367 |
| - | - | - | ciCGACT at 2361 |
| - | - | - | GGTCG at 2346 |
| - | - | - | ciCGACC at 2326 |
| - | - | - | GGACA at 2337 |
| - | - | - | AGATG at 2294 |
| - | - | - | GGACC at 2268 |
| - | - | - | GGTCG at 2264 |
| - | - | - | AGTCC at 2250 |
| - | - | - | ciCGACT at 2226 |
| - | - | - | GGTCA at 2211 |
| - | - | - | AGATG at 2169 |
| - | - | - | GGTTG at 2148 |
| - | - | - | AGTCC at 2134 |
| - | - | - | ciGAACT at 2127 |
| - | - | - | ciTGTCT at 2119 |
| - | - | - | ciCGACT at 2109 |
| - | - | - | GGATC at 2093 |
| - | - | - | GGTCC at 2077 |
| - | - | - | ciTGTCT at 2017 |
| - | - | - | AGACA at 2029 |
| - | - | - | ciCGACC at 2069 |
| - | - | - | GGACC at 2009 |
| - | - | - | GGTCG at 2005 |
| - | - | - | ciCGTCT at 1967 |
| - | - | - | GGACC at 1959 |
| - | - | - | ciCGTCC at 1941 |
| - | - | - | ciTGACT at 1935 |
| - | - | - | ciGAACC at 1927 |
| - | - | - | GGTCG at 1920 |
| - | - | - | GGACA at 1911 |
| - | - | - | ciCGACC at 1891 |
| - | - | - | AGATG at 1867 |
| - | - | - | ciCAACT at 1853 |
| - | - | - | GGACC at 1841 |
| - | - | - | ciCGTCC at 1823 |
| - | - | - | GGTTC at 1817 |
| - | - | - | ciCGACT at 1800 |
| - | - | - | GGTCG at 1785 |
| - | - | - | AGACA at 1776 |
| - | - | - | ciCGACC at 1756 |
| - | - | - | ciCGACC at 1746 |
| - | - | - | ciTATCT at 1710 |
| - | - | - | ciGGTCT at 1670 |
| - | - | - | ciCATCT at 1653 |
| - | - | - | ciGAACC at 1649 |
| - | - | - | ciGGACT at 1623 |
| - | - | - | ciCGTCT at 1614 |
| - | - | - | GGTCG at 1611 |
| - | - | - | ciTGTCT at 1567 |
| - | - | - | GGTCA at 1532 |
| - | - | - | AGATA at 1525 |
| - | - | - | ciGGTCT at 1518 |
| - | - | - | AGTTG at 1513 |
| - | - | - | AGTCG at 1486 |
| - | - | - | ciCGACC at 1464 |
| - | - | - | GGTCC at 1460 |
| - | - | - | AGACA at 1452 |
| - | - | - | ciGGTCT at 1411 |
| - | - | - | AGTTG at 1406 |
| - | - | - | AGACC at 1356 |
| - | - | - | GGTCA at 1352 |
| - | - | - | ciCGTCT at 1314 |
| - | - | - | ciGATCC at 1307 |
| - | - | - | GGATC at 1306 |
| - | - | - | ciGAACT at 1300 |
| - | - | - | ciCGTCC at 1288 |
| - | - | - | ciCGACT at 1282 |
| - | - | - | AGTCC at 1275 |
| - | - | - | GGTCG at 1267 |
| - | - | - | GGACA at 1258 |
| - | - | - | AGATG at 1224 |
| - | - | - | GGTTG at 1203 |
| - | - | - | GGACC at 1198 |
| - | - | - | GGTCG at 1194 |
| - | - | - | ciGGACT at 1173 |
| - | - | - | GGTCG at 1140 |
| - | - | - | GGACA at 1131 |
| - | - | - | ciCGACC at 1111 |
| - | - | - | AGACA at 1085 |
| - | - | - | ciTGTCT at 1073 |
| - | - | - | GGTCG at 1061 |
| - | - | - | ciTAACC at 1045 |
| - | - | - | ciCGTCT at 1023 |
| - | - | - | GGACC at 1015 |
| - | - | - | ciGAACT at 1009 |
| - | - | - | ciCGTCC at 997 |
| - | - | - | ciCGACT at 991 |
| - | - | - | AGTCC at 984 |
| - | - | - | GGTCG at 976 |
| - | - | - | ciCATCT at 970 |
| - | - | - | GGACA at 967 |
| - | - | - | GGTCC at 948 |
| - | - | - | AGACA at 919 |
| - | - | - | ciTGTCT at 907 |
| - | - | - | GGACC at 899 |
| - | - | - | GGTCG at 895 |
| - | - | - | GGTTC at 874 |
| - | - | - | GGTCC at 850 |
| - | - | - | ciGAACT at 843 |
| - | - | - | ciCGTCC at 831 |
| - | - | - | ciCGACT at 825 |
| - | - | - | GGTCG at 810 |
| - | - | - | GGACA at 801 |
| - | - | - | ciCGACC at 781 |
| - | - | - | AGATG at 758 |
| - | - | - | GGTCG at 737 |
| - | - | - | ciGGACT at 732 |
| - | - | - | GGTCG at 728 |
| - | - | - | AGTCC at 714 |
| - | - | - | ciCGTCC at 697 |
| - | - | - | GGTTC at 692 |
| - | - | - | GGTCG at 676 |
| - | - | - | GGACA at 667 |
| - | - | - | GGTCC at 648 |
| - | - | - | ciTAACC at 643 |
| - | - | - | AGATG at 624 |
| - | - | - | GGACC at 596 |
| - | - | - | ciTAACT at 585 |
| - | - | - | AGTCC at 578 |
| - | - | - | ciCGTCC at 565 |
| - | - | - | ciTGTCC at 561 |
| - | - | - | GGTTC at 556 |
| - | - | - | GGTCG at 540 |
| - | - | - | GGACC at 508 |
| - | - | - | GGTCG at 504 |
| - | - | - | AGATG at 481 |
| - | - | - | GGACC at 459 |
| - | - | - | AGTCC at 441 |
| - | - | - | ciTGTCC at 424 |
| - | - | - | GGTTC at 419 |
| - | - | - | GGTCG at 403 |
| - | - | - | GGACA at 394 |
| - | - | - | ciTATCT at 355 |
| - | - | - | ciGAACC at 328 |
| - | - | - | ciTGACT at 307 |
| - | - | - | ciTGTCT at 289 |
| - | - | - | ciCATCT at 284 |
| - | - | - | GGTCC at 262 |
| - | - | - | AGATA at 234 |
| - | - | - | ciTGTCT at 168 |
| - | - | - | ciCGACT at 140 |
| - | - | - | ciTGACT at 130 |
| - | - | - | ciCATCC at 119 |
| - | - | - | ciTATCT at 100 |
| - | - | - | ciCAACT at 85 |
| - | - | - | GGTCG at 35 |
| - | - | - | ciTGACT at 17 |
| - | - | - | ciTGTCT at 13 |
| Distal pn (2596-1) | Distal pn (2596-1) | Distal pn (2596-1) | Distal pn (2596-1) |
| - | - | - | GGTCG at 35 |
Acknowledgements
The content on this page was first contributed by: Henry A. Hoff.
Initial content for this page in some instances came from Wikiversity.
See also
References
- ↑ Jennifer E.F. Butler, James T. Kadonaga (October 15, 2002). "The RNA polymerase II core promoter: a key component in the regulation of gene expression". Genes & Development. 16 (20): 2583–292. doi:10.1101/gad.1026202. PMID 12381658.
- ↑ 2.0 2.1 Gillian E. Chalkley and C. Peter Verrijzer (1999). "DNA binding site selection by RNA polymerase II TAFs: a TAFII250-TAFII150 complex recognizes the Initiator". The EMBO Journal. 18 (17): 4835–45. PMID 10469661. Retrieved 2012-04-26. Unknown parameter
|month=ignored (help); Unknown parameter|pdf=ignored (help) - ↑ S. T. Smale (1997). "Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes". Biochim. Biophys. Acta. 1351: 73–88.
|access-date=requires|url=(help) - ↑ Ceyockey (28 January 2005). promoter. San Francisco, California: Wikimedia Foundation, Inc. Retrieved 2012-09-29.
- ↑ 5.0 5.1 5.2 Stephen T. Smale and James T. Kadonaga (2003). "The RNA Polymerase II Core Promoter" (PDF). Annual Review of Biochemistry. 72 (1): 449–79. doi:10.1146/annurev.biochem.72.121801.161520. PMID 12651739. Retrieved 2012-05-07. Unknown parameter
|month=ignored (help) - ↑ Robert D. Andersen, Susan J. Taplitz, Sandy Wong, Greg Bristol, Bill Larkin, and Harvey R. Herschman (1987). "Metal-Dependent Binding of a Factor In Vivo to the Metal-Responsive Elements of the Metallothionein 1 Gene Promoter" (PDF). Molecular and Cellular Biology. 7 (10): 3574–81. doi:10.1128/MCB.7.10.3574 Check
|doi=value (help). Retrieved 2013-04-15. Unknown parameter|month=ignored (help); zero width space character in|doi=at position 9 (help) - ↑ Msh210 (23 February 2010). "GC box". San Francisco, California: Wikimedia Foundation, Inc. Retrieved 2013-01-27.
- ↑ Klug WS, Cummings MR, Spencer CA, Palladina, MA (2009). Concepts of Genetics: Ninth Edition. San Francisco: Pearson Benjamin Cummings. pp. 463–464. ISBN 978-0-321-54098-0.
- ↑ "GC box, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. June 23, 2012. Retrieved 2013-01-27.
- ↑ 10.0 10.1 10.2 Michael C. Blake, Robert C. Jambou, Andrew G. Swick, Jeanne W. Kahn, and Jane Clifford Azizkhan (1990). "Transcriptional Initiation Is Controlled by Upstream GC-Box Interactions in a TATAA-Less Promoter". Molecular and Cellular Biology. 10 (12): 6632–41. doi:10.1128/MCB.10.12.6632 Check
|doi=value (help). PMID 2247077. Retrieved 2013-01-27. Unknown parameter|month=ignored (help); Unknown parameter|pdf=ignored (help); zero width space character in|doi=at position 9 (help) - ↑ H Imataka, K Sogawa, KI Yasumoto, Y Kikuchi, K Sasano, A Kobayashi, M Hayami, and Y Fujii-Kuriyama (1992). "Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene". The EMBO Journal. 11 (10): 3663–71. PMID 1356762. Retrieved 2013-01-27. Unknown parameter
|month=ignored (help); Unknown parameter|pdf=ignored (help) - ↑ Akiro Higashikawa, Taku Saito, Toshiyuki Ikeda, Satoru Kamekura, Naohiro Kawamura, Akinori Kan, Yasushi Oshima, Shinsuke Ohba, Naoshi Ogata, Katsushi Takeshita, Kozo Nakamura, Ung-Il Chung, Hiroshi Kawaguchi (2009). "Identification of the core element responsive to runt-related transcription factor 2 in the promoter of human type x collagen gene". Arthritis & Rheumatism. 60 (1): 166–78. doi:10.1002/art.24243. PMID 19116917. Retrieved 2013-06-18. Unknown parameter
|month=ignored (help) - ↑ "CAAT box". San Francisco, California: Wikimedia Foundation, Inc. April 8, 2013. Retrieved 2013-04-14.
- ↑ Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH (1998). "New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB". Genes & Development. 12 (1): 34–44. doi:10.1101/gad.12.1.34. PMC 316406. PMID 9420329.
- ↑ Littlefield O, Korkhin Y, Sigler PB (1999). "The structural basis for the oriented assembly of a TBP/TFB/promoter complex". Proceedings of the National Academy of Sciences of the USA. 96 (24): 13668–73. doi:10.1073/pnas.96.24.13668. PMC 24122. PMID 10570130.
- ↑ 16.0 16.1 "B recognition element, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. January 30, 2013. Retrieved 2013-01-30.
- ↑ 17.0 17.1 Alan K. Kutach, James T. Kadonaga (2000). "The Downstream Promoter Element DPE Appears To Be as Widely Used as the TATA Box in Drosophila Core Promoters" (PDF). Molecular and Cellular Biology. 20 (13): 4754–64. PMID 10848601. Retrieved 2012-07-15. Unknown parameter
|month=ignored (help) - ↑ 18.0 18.1 Chuhu Yang, Eugene Bolotin, Tao Jiang, Frances M. Sladek, Ernest Martinez. (2007). "Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters". Gene. 389 (1): 52–65. doi:10.1016/j.gene.2006.09.029. PMID 17123746. Unknown parameter
|month=ignored (help) - ↑ Mary Lynch, Li Chen, Michael J. Ravitz, Sapna Mehtani, Kevin Korenblat, Michael J. Pazin and Emmett V. Schmidt (2005). "hnRNP K Binds a Core Polypyrimidine Element in the Eukaryotic Translation Initiation Factor 4E (eIF4E) Promoter, and Its Regulation of eIF4E Contributes to Neoplastic Transformation". Molecular and Cellular Biology. 25 (15): 6436–53. doi:10.1128/MCB.25.15.6436-6453.2005 Check
|doi=value (help). Retrieved 2013-03-17. Unknown parameter|month=ignored (help); Unknown parameter|pdf=ignored (help); zero width space character in|doi=at position 9 (help) - ↑ "TATA box, In: Wiktionary". San Francisco, California: Wikimedia Foundation, Inc. June 17, 2013. Retrieved 2014-05-07.
- ↑ National Center for Biotechnology Information (April 28, 2012). "TBPL1 TBP-like 1 [ Homo sapiens ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: U.S. National Library of Medicine. Retrieved 2012-04-30.
- ↑ 22.0 22.1 Wensheng Deng, Stefan G.E. Roberts (2005). "A core promoter element downstream of the TATA box that is recognized by TFIIB". Genes & Development. 19 (20): 2418–23. doi:10.1101/gad.342405. PMID 16230532. Unknown parameter
|month=ignored (help); Unknown parameter|pdf=ignored (help) - ↑ J. Carcamo, L. Buckbinder and D. Reinberg (1991). "The initiator directs the assembly of a transcription factor IID-dependent transcription complex". Proc. Natl. Acad. Sci, USA. 88: 8052–6.
|access-date=requires|url=(help) - ↑ L. Weis and D. Reinberg (1997). "Accurate positioning of RNA polymerase II on a natural TATA-less promoter is independent of TATA-binding protein associated factors and initiator-binding proteins". Mol. Cell. Biol. 17: 2973–84.
|access-date=requires|url=(help) - ↑ 25.0 25.1 25.2 25.3 Marketa J. Zvelebil, Jeremy O. Baum (2008). Dom Holdsworth, ed. Understanding bioinformatics. New York: Garland Science. p. 772. ISBN 978-0815340249.
- ↑ 26.00 26.01 26.02 26.03 26.04 26.05 26.06 26.07 26.08 26.09 26.10 26.11 Dong-Hoon Lee, Naum Gershenzon, Malavika Gupta, Ilya P. Ioshikhes, Danny Reinberg and Brian A. Lewis (2005). "Functional Characterization of Core Promoter Elements: the Downstream Core Element Is Recognized by TAF1". Molecular and Cellular Biology. 25 (21): 9674–86. doi:10.1128/MCB.25.21.9674-9686.2005. PMID 16227614. Retrieved 2010-10-23. Unknown parameter
|month=ignored (help) - ↑ 27.0 27.1 Chin Yan Lim, Buyung Santoso, Thomas Boulay, Emily Dong, Uwe Ohler, and James T. Kadonaga (2004). "The MTE, a new core promoter element for transcription by RNA polymerase II". Genes & Development. 18 (13): 1606–17. doi:10.1101/gad.1193404. PMID 15231738. Retrieved 2013-02-10. Unknown parameter
|month=ignored (help); Unknown parameter|pdf=ignored (help) - ↑
Tamar Juven-Gershon, James T. Kadonaga (2010). "Regulation of Gene Expression via the Core Promoter and the Basal Transcriptional Machinery". Developmental Biology. 339 (2): 225–9. doi:10.1016/j.ydbio.2009.08.009. PMC 2830304. PMID 19682982. Unknown parameter
|month=ignored (help) - ↑ "Downstream promoter element". San Francisco, California: Wikimedia Foundation, Inc. May 6, 2012. Retrieved 2012-05-20.
- ↑ Cquan (2 October 2006). "Promoter (genetics)". San Francisco, California: Wikimedia Foundation, Inc. Retrieved 2016-01-09.
Further reading
- Stephen T. Smale and James T. Kadonaga (2003). "The RNA Polymerase II Core Promoter". Annual Review of Biochemistry. 72 (1): 449–79. doi:10.1146/annurev.biochem.72.121801.161520. PMID 12651739. Retrieved 2012-05-07. Unknown parameter
|month=ignored (help); Unknown parameter|pdf=ignored (help)
External links
- GenomeNet KEGG database
- Home - Gene - NCBI
- NCBI All Databases Search
- Office of Scientific & Technical Information
- PubChem Public Chemical Database
- Scirus for scientific information only advanced search
{{Phosphate biochemistry}}{{Terminology resources}}Template:Sisterlinks