Epigenomes
Editor-In-Chief: Henry A. Hoff
{{free media}}{{free media}}Inside each eukaryote nucleus is genetic material (DNA) surrounded by protective and regulatory proteins. These protective and regulatory proteins and the dynamic changes to them that occur during the course of a eukaryote's existence are the epigenome.
An epigenome consists of a record of the chemical changes to the DNA and histone proteins of an organism that can be passed down to an organism's offspring via transgenerational epigenetic inheritance, where changes to the epigenome can result in changes to the structure of chromatin and changes to the function of the genome.[1]
Unlike the underlying genome which is largely static within an individual, the epigenome can be dynamically altered by environmental conditions.[2]
Evolution
Evolution, the accumulation of change, while broadly applicable to anything which accumulates changes, is often thought of as gradual change or a series of changes, such as changes in the genetic composition of a population over successive generations.
Lamarckism
Lamarckism (or Lamarckian inheritance) is the idea that an organism can pass on characteristics that it acquired during its lifetime to its offspring (also known as heritability of acquired characteristics or soft inheritance). It is named after the French biologist Jean-Baptiste Lamarck (1744–1829), who incorporated the action of soft inheritance into his evolutionary theories.
After Erasmus Darwin wrote Zoonomia suggesting "that all warm-blooded animals have arisen ... with the power of acquiring new parts" in response to stimuli, with each round of "improvements" being inherited by successive generations",[3] Jean-Baptiste Lamarck repeated in his Philosophie Zoologique of 1809 the folk wisdom that characteristics which were "needed" were acquired (or diminished) during the lifetime of an organism then passed on to the offspring.
Neo-Lamarckism is a theory of inheritance based on a modification and extension of Lamarckism, essentially maintaining the principle that genetic changes can be influenced and directed by environmental factors.
Epigenetics
{{free media}}Epigenetics is the study of genome or epigenome changes resulting from external rather than genetic influences.
"Epigenetic mechanisms are affected by several factors and processes including development in utero and in childhood, environmental chemicals, drugs and pharmaceuticals, aging, and diet. DNA methylation is what occurs when methyl groups, an epigenetic factor found in some dietary sources, can tag DNA and activate or repress genes. Histones are proteins around which DNA can wind for compaction and gene regulation. Histone modification occurs when the binding of epigenetic factors to histone "tails"; alters the extent to which DNA is wrapped around histones and the availability of genes in the DNA to be activated. All of these factors and processes can have an effect on people's health and influence their health possibly resulting in cancer, autoimmune disease, mental disorders, or diabetes among other illnesses."[4]
Epigenomic theory
Def. a chemical entity anterior to, after, at, besides, near to, on, outer to, over, related to, or upon another chemical is called an epi (or epi-) chemical.
Def. the "complete genetic information ... of an organism"[5] is called a genome.
Here's a theoretical definition:
Def. a chemical entity anterior to, after, at, besides, near to, on, outer to, over, related to, or upon the complete genetic information of an organism is called an epi (or epi-) genome, or epigenome.
Genomes
The genome is the entirety of an organism's hereditary information. In humans, it is encoded in DNA. The genome includes both the genes and the non-coding sequences of the DNA.[6]
Homo sapiens estimated genome size [is] 3.2 billion bp.[7]
Genetic information is encoded as a sequence of nucleobases: adenine (A), cytosine (C), guanine (G), and thymine (T).
Deoxyribonucleic acid molecules

Deoxyribonucleic acid (DNA) is composed of nucleobases (the sequence of which is the genome), deoxyribose (a sugar), and phosphate groups. Each nucleobase is attached to one deoxyribose molecule and one (PO4) phosphate molecule to form a chain of nucleotides (nucleobase + deoxyribose + phosphate) for a haploid genome. A linking of nucleobases may occur without the phosphate or the deoxyribose. The phosphate and the sugar are part of the epigenome.
DNA often occurs as a double helix. The linking between one nitrogenous nucleobase of a DNA molecule and another nitrogenous nucleobase of a second DNA molecule is via hydrogen bonds. Each hydrogen bond (the electromagnetic attractive interaction of a hydrogen atom and an electronegative atom, such as nitrogen or oxygen of a nucleobase) is part of the epigenome.
The structure a DNA molecule shown in the top image on the left depends on its environment. In aqueous environments, including the majority of DNA in a cell, B-DNA is the most common structure. The A-DNA structure dominates in dehydrated samples and is similar to the double-stranded RNA and DNA/RNA hybrids. Z-DNA is a rarer structure found in DNA bound to certain proteins.
Nucleosomes
DNA packaging in eukaryotes consists of "DNA wound in sequence around four histone protein cores.[10]
Nucleosomes form the fundamental repeating units of eukaryotic chromatin.[11]
The nucleosome core particle consists of approximately 147 base pairs of DNA wrapped in 1.67 left-handed superhelical turns around a histone octamer consisting of 2 copies each of the core histones H2A, H2B, H3, and H4.[12]
Core particles are connected by stretches of "linker DNA", which can be up to about 80 bp long.
Histones
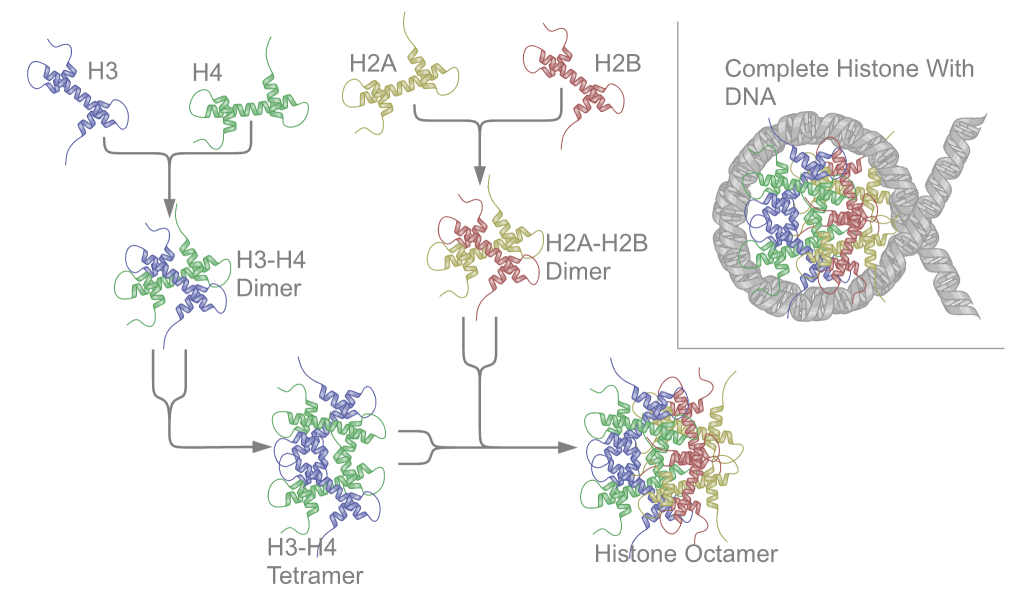
Histone deacetylases (HDAC) ([Enzyme Commission number] EC number 3.5.1) are a class of enzymes that remove acetyl groups (O=C-CH3) from an ε-N-acetyl lysine amino acid on a histone, allowing the histones to wrap the DNA more tightly.
Histone deacetylase action is opposite to that of histone acetyltransferase.
Chromatin
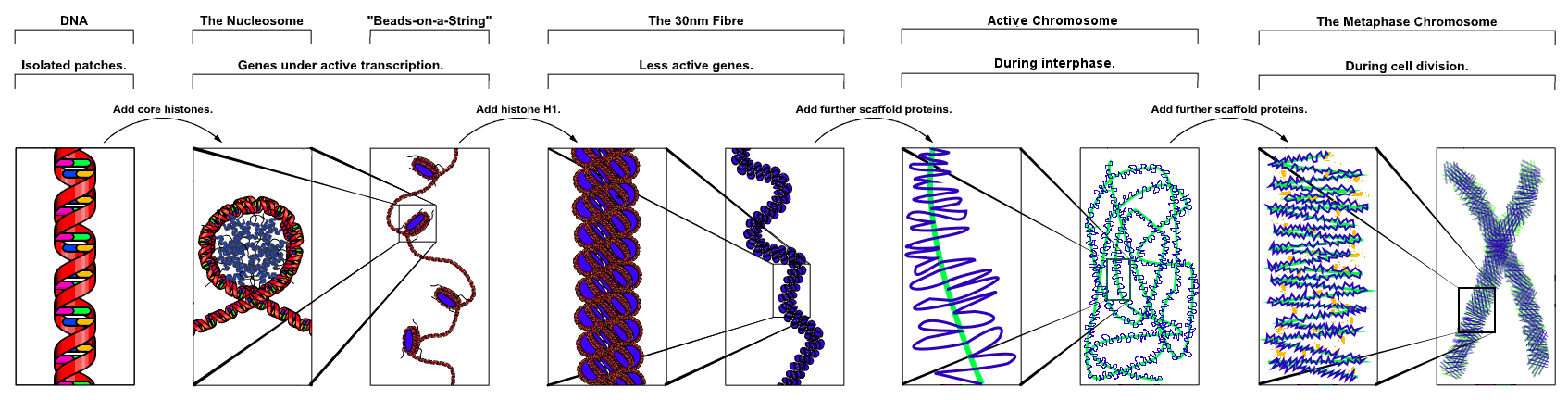
Chromatin, or the Chromatin network, is a complex of macromolecules found in cells, consisting of DNA, protein, and RNA.[13]
DNA which codes genes that are actively transcribed ("turned on") is more loosely packaged and associated with RNA polymerases (referred to as euchromatin) while that DNA which codes inactive genes ("turned off") is more condensed and associated with structural proteins (heterochromatin).[14][15]
Polycomb-group proteins play a role in regulating genes through modulation of chromatin structure.[16]
Euchromatin
Def. "uncoiled dispersed threads of chromosomal material that occurs during interphase"[17] is called euchromatin.
The structure of euchromatin is reminiscent of an unfolded set of beads along a string, wherein those beads represent nucleosomes.
The presence of methylated lysine 4 on the histone tails may act as a general marker for euchromatin.
One example of constitutive euchromatin that is 'always turned on' is housekeeping genes, which code for the proteins needed for basic functions of cell survival.
Heterochromatin
Heterochromatin mainly consists of genetically inactive satellite sequences,[18] and many genes are repressed to various extents, although some cannot be expressed in euchromatin at all.[19] Both centromeres and telomeres are heterochromatic, as is the Barr body of the second, inactivated X-chromosome in a female.
Constitutive heterochromatin
Sections of DNA that occur particularly at the centromeres and telomeres often consisting of repetitive DNA that is largely transcriptionally silent are constitutive heterochromatin.
Regions of DNA that exist as constitutive heterochromatin are the same for all cells of a given species.
All human chromosomes 1, 9, 16, and the Y-chromosome contain large regions of constitutive heterochromatin. In most organisms, constitutive heterochromatin occurs around the chromosome centromere and near telomeres.
Facultative heterochromatin
Genes that are silenced through a mechanism such as histone methylation or siRNA through RNAi produce facultative heterochromatin.
The regions of DNA packaged in facultative heterochromatin are not consistent between the cell types within a species, and thus a sequence in one cell that is packaged in facultative heterochromatin (and the genes within poorly expressed) may be packaged in euchromatin in another cell (and the genes within no longer silenced).
An example of facultative heterochromatin is X-chromosome inactivation in female mammals such as the cat in the image on the right: one X chromosome is packaged as facultative heterochromatin and silenced, while the other X chromosome is packaged as euchromatin and expressed. The black and orange alleles of a fur coloration gene reside on the X chromosome. For any given patch of fur, the inactivation of an X chromosome that carries one gene results in the fur color of the other, active gene.
Centric heterochromatin
Centric heterochromatin, a variety of heterochromatin, is a tightly packed form of DNA that is a constituent in the formation of active centromeres in most higher-order organisms; the domain exists on both mitotic and interphase chromosomes.[21]
Centric heterochromatin is usually formed on alpha satellite DNA in humans; however, there have been cases where centric heterochromatin and centromeres have formed on originally euchromatin domains lacking alpha satellite DNA; this usually happens as a result of a chromosome breakage event and the formed centromere is called a neocentromere.[21]
Centric heterochromatin domains are flanked by pericentric heterochromatin.[21]
Acetyl groups
Acetylation (or in IUPAC nomenclature ethanoylation) describes a reaction that introduces an acetyl functional group into a chemical compound. (Deacetylation is the removal of the acetyl group.)
In histone acetylation and deacetylation, histone proteins are acetylated and deacetylated on lysine residues in the N-terminal tail as part of gene regulation. Typically, these reactions are catalyzed by enzymes with histone acetyltransferase (HAT) or histone deacetylase (HDAC) activity, although HATs and HDACs can modify the acetylation status of non-histone proteins as well.[22]
There are "nearly 50,000 acetylated sites [punctate sites of modified histones] in the human genome that correlate with active transcription start sites and CpG islands and tend to cluster within gene-rich loci."[1]
"[L]ysine acetylation almost always correlates with chromatin accessibility and transcriptional activity".[1]
Acyl groups
Adenyl groups
Adenylylation,[23][24] more commonly known as AMPylation, is a process in which an adenosine monophosphate (AMP) molecule is covalently attached to the amino acid side chain of a protein.[25] This covalent addition of AMP to a hydroxyl side chain of the protein is a posttranslational modification.[26]
Amidal groups
Aminal groups
Carbamyl groups
Carboxyl groups
Citrulinyl groups
Desmosinal groups
Desmosine in urine, plasma or sputum samples can be a marker for elastin breakdown due to high elastase activity related to certain diseases.[27][28]
Detyrosinal groups
Diphthal groups
Disulfidyl groups
Flavinal groups
Formyl groups
Glutamyl groups
Glycyl groups
Glycosyl groups
Hydroxyl groups
Imidazolinonal groups
The property of photoconversion in Kaede is contributed by the tripeptide, His62-Tyr63-Gly64, that acts as a green chromophore that can be converted to red.[29] Once Kaede is synthesized, a chromophore, 4-(p-hydroxybenzylidene)-5-imidazolinone, derived from the tripeptide mediates green fluorescence in Kaede. When exposed to UV, Kaede protein undergoes un conventional cleavage between the amide nitrogen and the α carbon (Cα) at His62 via a formal β-elimination reaction. Followed by the formation of a double bond between His62-Cα and –Cβ, the π-conjugation is extended to the imidazole ring of His62. A new chromophore, 2-[(1E)-2-(5-imidazolyl)ethenyl]-4-(p-hydroxybenzylidene)-5-imidazolinone, is formed with the red-emitting property.
GFP has a beta barrel structure consisting of eleven β-strands with a pleated sheet arrangement, with an alpha helix containing the covalently bonded chromophore 4-(p-hydroxybenzylidene)imidazolidin-5-one (HBI) running through the center.[30][31][32] Five shorter alpha helices form caps on the ends of the structure. The beta barrel structure is a nearly perfect cylinder, 42Å long and 24Å in diameter (some studies have reported a diameter of 30Å[33]),[31] creating what is referred to as a "β-can" formation, which is unique to the GFP-like family.[32] HBI, the spontaneously modified form of the tripeptide Ser65–Tyr66–Gly67, is nonfluorescent in the absence of the properly folded GFP scaffold and exists mainly in the un-ionized phenol form in wtGFP.[34] Inward-facing sidechains of the barrel induce specific cyclization reactions in Ser65–Tyr66–Gly67 that induce ionization of HBI to the phenolate form and chromophore formation. This process of post-translational modification is referred to as maturation.[35] The hydrogen-bonding network and electron-stacking interactions with these sidechains influence the color, intensity and photostability of GFP and its numerous derivatives.[36] The tightly packed nature of the barrel excludes solvent molecules, protecting the chromophore fluorescence from quenching by water. In addition to the auto-cyclization of the Ser65-Tyr66-Gly67, a 1,2-dehydrogenation reaction occurs at the Tyr66 residue.[33] Besides the three residues that form the chromophore, residues such as Gln94, Arg96, His148, Thr203, and Glu222 all act as stabilizers. The residues of Gln94, Arg96, and His148 are able to stabilize by delocalizing the chromophore charge. Arg96 is the most important stabilizing residue due to the fact that it prompts the necessary structural realignments that are necessary from the HBI ring to occur. Any mutation to the Arg96 residue would result in a decrease in the development rate of the chromophore because proper electrostatic and steric interactions would be lost. Tyr66 is the recipient of hydrogen bonds and does not ionize in order to produce favorable electrostatics.[37]
Iminal groups
Mannosyl groups
Methyl groups
Methylation is "the addition of a methyl group replacing a hydrogen atom.
DNA methylation in vertebrates typically occurs at CpG sites (cytosine-phosphate-guanine sites, that is, where a cytosine is directly followed by a guanine in the DNA sequence). This methylation results in the conversion of the cytosine to 5-methylcytosine. The formation of Me-CpG is catalyzed by the enzyme DNA methyltransferase. Human DNA has about 80%-90% of CpG sites methylated, but there are certain areas, known as CpG islands, that are GC-rich (made up of about 65% CG residues), wherein none are methylated. These are associated with the promoters of 56% of mammalian genes, including all ubiquitously expressed genes. One to two percent of the human genome are CpG clusters, and there is an inverse relationship between CpG methylation and transcriptional activity.
"Non-CpG methylation (CNG and CNN) ... has been observed at a low frequency in the early mouse embryo"[1]
Protein methylation typically takes place on arginine or lysine amino acid residues in the protein sequence.[38] Arginine can be methylated once (monomethylated arginine) or twice, with either both methyl groups on one terminal nitrogen (asymmetric dimethylated arginine) or one on both nitrogens (symmetric dimethylated arginine) by peptidylarginine methyltransferases (PRMTs). Lysine can be methylated once, twice or three times by lysine methyltransferases. Protein methylation has been most-studied in the histones. The transfer of methyl groups from S-adenosyl methionine to histones is catalyzed by enzymes known as histone methyltransferases. Histones that are methylated on certain residues can act epigenetically to repress or activate gene expression.[39][40]
Myristoyl groups
Palmitoyl groups
Phosphoryl groups
Phosphorylation is the addition of a phosphate (PO43-) group to a protein or other organic molecule.
Kinases phosphorylate proteins and phosphatases dephosphorylate proteins.
Reversible phosphorylation of proteins is an important regulatory mechanism that occurs in both prokaryotic and eukaryotic organisms.[41][42][43][44]
Phosphoryl groups attach to histones at serine and threonine sites.[1]
Porphyl groups
Prenyl groups
Ribosyl groups
ADP-ribosylation is the addition of one or more ADP-ribose moieties to a protein.[45][46] It is a reversible post-translational modification that is involved in many cellular processes, including cell signaling, DNA repair, gene regulation and apoptosis.[47][48]
Succinimidal groups
Sulfal groups
Sulfiliminal groups
Sulfilimine bonds stabilize collagen IV strands found in the extracellular matrix[49] and arose at least 500 mya.[50] These bonds covalently connect hydroxylysine and methionine residues of adjacent polypeptide strands to form a larger collagen trimer.
Sumoyl groups
Topaquinyl groups
Tryptophanyl groups
Tryptophan tryptophylquinone (TTQ)[51] is an enzyme cofactor, generated by posttranslational modification of amino acids within the protein. Methylamine dehydrogenase (MADH), an amine dehydrogenase, requires TTQ for its catalytic function.[52]
Tyrosylquinonal groups
Ubiquityl groups
"The core histones that make up the nucleosome are subject to ... modifications, including ubiquitination [that occurs] primarily at specific positions within the amino-terminal histone tails."[1]
Hypotheses
- The epigenome around A1BG is opened as if for any gene rather than a specific promoter, enhancer, or other transcription related factor.
Acknowledgements
The content on this page was first contributed by: Henry A. Hoff.
See also
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Bradley E. Bernstein, Alexander Meissner, Eric S. Lander (February 23, 2007). "The Mammalian Epigenome" (PDF). Cell. 128 (4): 669–81. doi:10.1016/j.cell.2007.01.033. Retrieved 19 December 2011.
- ↑ Conley, A.B., King Jordan, I. (2012). Endogenous Retroviruses and the Epigenome. In: Witzany, G. (ed). Viruses: Essential Agents of Life, Springer, Dordrecht, pp. 309-323.
- ↑ Darwin 1794–1796, Vol I, section XXXIX
- ↑ CommonFund (March 5, 2018). A Scientific Illustration of How Epigenetic Mechanisms Can Affect Health. Bethesda, Maryland USA: National Institutes of Health. Retrieved 24 December 2018.
- ↑ genome. San Francisco, California: Wikimedia Foundation, Inc. 16 October 2012. Retrieved 30 October 2012.
- ↑ Ridley, M. (2006). Genome. New York, NY: Harper Perennial. ISBN 0-06-019497-9
- ↑ Human Genome.
- ↑ Harp JM, Hanson BL, Timm DE, Bunick GJ (2000-04-06). X-ray structure of the nucleosome core particle at 2.5 A resolution. RCSB Protein Data Bank (PDB). doi:10.2210/pdb1eqz/pdb. Retrieved 8 October 2012.
- ↑ Harp JM, Hanson BL, Timm DE, Bunick GJ (December 2000). "Asymmetries in the nucleosome core particle at 2.5 A resolution". Acta Crystallographica Section D. 56 (Pt 12): 1513–34. doi:10.1107/S0907444900011847. PMID 11092917. PDB ID: 1EQZ.
- ↑ Reece, Jane; Campbell, Neil (2006). Biology. San Francisco: Benjamin Cummings. ISBN 0-8053-6624-5.
- ↑ Alberts, Bruce (2002). Molecular biology of the cell (4th ed.). New York: Garland Science. p. 207. ISBN 0-8153-4072-9.
- ↑ Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ (September 1997). "Crystal structure of the nucleosome core particle at 2.8 A resolution". Nature. 389 (6648): 251–60. doi:10.1038/38444. PMID 9305837.
- ↑ Monday, Tanmoy (July 2010). "Characterization of the RNA content of chromatin". Genome Res. 20 (7): 899–907. doi:10.1101/gr.103473.109. PMC 2892091. PMID 20404130.
- ↑ Chromatin Network Home Page. Retrieved 2008-11-18.
- ↑ Dame, R.T. (May 2005). "The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin". Molecular Microbiology. 56 (4): 858–870. doi:10.1111/j.1365-2958.2005.04598.x. PMID 15853876.
- ↑ Portoso M, Cavalli G (2008). The Role of RNAi and Noncoding RNAs in Polycomb Mediated Control of Gene Expression and Genomic Programming, In: RNA and the Regulation of Gene Expression: A Hidden Layer of Complexity. Caister Academic Press. ISBN 978-1-904455-25-7.
- ↑ euchromatin. San Francisco, California: Wikimedia Foundation, Inc. 2 January 2012. Retrieved 30 October 2012.
- ↑ Lohe, A.R.; et al. (August 1, 1993). "Mapping Simple Repeated DNA Sequences in Heterochromatin of Drosophila Melanogaster". Genetics. 134 (4): 1149–74. ISSN 0016-6731. PMC 1205583. PMID 8375654.
- ↑ Lu, B.Y.; et al. (June 1, 2000). "Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila". Genetics. 155 (2): 699–708. ISSN 0016-6731. PMC 1461102. PMID 10835392.
- ↑ C-Banding. Retrieved 2015-12-02.
- ↑ 21.0 21.1 21.2 Molecular Biology of the Cell. pp. 229–231. ISBN 978-0-8153-4105-5.
- ↑ Sadoul K, Boyault C, Pabion M, Khochbin S (February 2008). "Regulation of protein turnover by acetyltransferases and deacetylases". Biochimie. 90 (2): 306–12. doi:10.1016/j.biochi.2007.06.009. PMID 17681659.
- ↑ Han KK, Martinage A (1992). "Post-translational chemical modification(s) of proteins". Int. J. Biochem. 24 (1): 19–28. PMID 1582530.
- ↑ Garrett, R.H., and C.M. Grisham. Biochemistry. 3rd ed. Belmont, CA: Thomas, 2007. 815-20
- ↑ Itzen, Aymelt, Wulf Blankenfeldt, and Roger S. Goody. "Adenylylation: renaissance of a forgotten post-translational modification." Trends in Biochemical Sciences 36.4 (2011): 221-228. Print.
- ↑ Woolery, Andrew. "AMPylation: something old is new again." Frontiers in Microbiology 1 (2010): 1-18. Print.
- ↑ https://web.archive.org/web/20140224040311/http://www.millipore.com/catalogue/item/263275-5mg. Retrieved 2014-02-19. Missing or empty
|title=(help) - ↑ Ma, S; Turino, G. M.; Lin, Y. Y. (2011). "Quantitation of desmosine and isodesmosine in urine, plasma, and sputum by LC-MS/MS as biomarkers for elastin degradation". Journal of Chromatography B. 879 (21): 1893–8. doi:10.1016/j.jchromb.2011.05.011. PMID 21621489.
- ↑ Mizuno, H.; Mal, T. K.; Tong, K. I.; Ando, R.; Furuta, T.; Ikura, M.; Miyawaki, A. (2003). "Photo-induced peptide cleavage in the green-to-red conversion of a fluorescent protein". Molecular Cell. 12 (4): 1051–1058. doi:10.1016/s1097-2765(03)00393-9. PMID 14580354.
- ↑ Tsien RY (1998). "The green fluorescent protein" (PDF). Annual Review of Biochemistry. 67: 509–44. doi:10.1146/annurev.biochem.67.1.509. PMID 9759496.
- ↑ 31.0 31.1 Ormö M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ (Sep 1996). "Crystal structure of the Aequorea victoria green fluorescent protein". Science. 273 (5280): 1392–5. Bibcode:1996Sci...273.1392O. doi:10.1126/science.273.5280.1392. PMID 8703075. Unknown parameter
|s2cid=ignored (help) - ↑ 32.0 32.1 Yang F, Moss LG, Phillips GN (Oct 1996). "The molecular structure of green fluorescent protein" (PDF). Nature Biotechnology. 14 (10): 1246–51. doi:10.1038/nbt1096-1246. hdl:1911/19233. PMID 9631087. Unknown parameter
|s2cid=ignored (help) - ↑ 33.0 33.1 Brejc, K.; Sixma, T. K.; Kitts, P. A.; Kain, S. R.; Tsien, R. Y.; Ormö, M.; Remington, S. J. Structural basis for dual excitation and photoisomerization of the Aequorea victoria green fluorescent protein. Proc. Natl. Acad. Sci. U. S. A.. 1997, 94 (6), 2306-2311.
- ↑ Bokman SH, Ward WW (1982). "Reversible denaturation of Aequorea green-fluorescent protein: physical separation and characterization of the renatured protein". Biochemistry. 21 (19): 4535–4540. doi:10.1021/bi00262a003. PMID 6128025.
- ↑ Pouwels LJ, Zhang L, Chan NH, Dorrestein PC, Wachter RM (Sep 2008). "Kinetic isotope effect studies on the de novo rate of chromophore formation in fast- and slow-maturing GFP variants". Biochemistry. 47 (38): 10111–22. doi:10.1021/bi8007164. PMC 2643082. PMID 18759496.
- ↑ Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA (Jul 2010). "Fluorescent proteins and their applications in imaging living cells and tissues". Physiological Reviews. 90 (3): 1103–63. doi:10.1152/physrev.00038.2009. PMID 20664080. Unknown parameter
|s2cid=ignored (help) - ↑ Stepaneko, O. V.; Verkhusha, V. V.; Shavlovsky, M. M.; Kuznetsova, I. M.; Uversky, V. N.; Turoverov, K. K. Understanding the role of Arg96 in structure and stability of green fluorescent protein. Proteins: Struct., Funct., Bioinf. 1999, 73 (3), 539-551.
- ↑ Christopher Walsh (2006). "Chapter 5 - Protein Methylation". Posttranslational modification of proteins: expanding nature's inventory (PDF). Roberts and Co. Publishers. ISBN 0-9747077-3-2.
- ↑ Grewal SI, Rice JC (2004). "Regulation of heterochromatin by histone methylation and small RNAs". Curr. Opin. Cell Biol. 16 (3): 230–8. doi:10.1016/j.ceb.2004.04.002. PMID 15145346.
- ↑ Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI (2001). "Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly". Science. 292 (5514): 110–3. Bibcode:2001Sci...292..110N. doi:10.1126/science.1060118. PMID 11283354.
- ↑ Cozzone AJ (1988). "Protein phosphorylation in prokaryotes". Annu. Rev. Microbiol. 42: 97–125. doi:10.1146/annurev.mi.42.100188.000525. PMID 2849375.
- ↑ Stock JB, Ninfa AJ, Stock AM (December 1989). "Protein phosphorylation and regulation of adaptive responses in bacteria". Microbiol. Rev. 53 (4): 450–90. PMC 372749. PMID 2556636.
- ↑ Chang C, Stewart RC (July 1998). "The Two-Component System . Regulation of Diverse Signaling Pathways in Prokaryotes and Eukaryotes". Plant Physiol. 117 (3): 723–31. doi:10.1104/PPSOE.117.3.723. PMC 1539182. PMID 9662515.
- ↑ Barford D, Das AK, Egloff MP (1998). "The structure and mechanism of protein phosphatases: insights into catalysis and regulation". Annu Rev Biophys Biomol Struct. 27: 133–64. doi:10.1146/annurev.biophys.27.1.133. PMID 9646865.
- ↑ Belenky P, Bogan KL, Brenner C (2007). "NAD+ metabolism in health and disease" (PDF). Trends Biochem. Sci. 32 (1): 12–9. doi:10.1016/j.tibs.2006.11.006. PMID 17161604.
- ↑ Ziegler M (2000). "New functions of a long-known molecule. Emerging roles of NAD in cellular signaling". Eur. J. Biochem. 267 (6): 1550–64. doi:10.1046/j.1432-1327.2000.01187.x. PMID 10712584.
- ↑ Berger F, Ramírez-Hernández MH, Ziegler M (2004). "The new life of a centenarian: signalling functions of NAD(P)". Trends Biochem. Sci. 29 (3): 111–8. doi:10.1016/j.tibs.2004.01.007. PMID 15003268.
- ↑ Corda D, Di Girolamo M (2003). "NEW EMBO MEMBER'S REVIEW: Functional aspects of protein mono-ADP-ribosylation". EMBO J. 22 (9): 1953–8. doi:10.1093/emboj/cdg209. PMC 156081. PMID 12727863.
- ↑ Vanacore R, Ham AL, Voehler M, Sanders CR, Conrads TP, Veenstra TD, Sharpless KB, Dawson PE, Hudson BG (September 4, 2009). "A sulfilimine bond identified in collagen IV". Science. 325 (5945): 1230–1234. Bibcode:2009Sci...325.1230V. doi:10.1126/science.1176811. PMC 2876822. PMID 19729652.
- ↑ A unique covalent bond in basement membrane is a primordial innovation for tissue evolution PNAS
- ↑ Victor L. Davidson and Aimin Liu (2012 Jan 28). "Tryptophan tryptophylquinone biosynthesis: A radical approach to posttranslational modification". National Center for Biotechnology Information, U.S. National Library of Medicine. doi:10.1016/j.bbapap.2012.01.008. Check date values in:
|date=(help) - ↑ Davidson VL, Liu A: Uncovering novel biochemistry in the mechanism of tryptophan tryptophylquinone cofactor biosynthesis Curr. Op. Chem. Biol. 2009, 13: 469-474
External links
- CS1 maint: Multiple names: authors list
- CS1 maint: Uses authors parameter
- CS1 maint: Explicit use of et al.
- Pages with citations lacking titles
- Pages with citations having bare URLs
- Pages with citations using unsupported parameters
- CS1 errors: dates
- Pages with broken file links
- Resources last modified in January 2021