Histone: Difference between revisions
No edit summary |
m (Robot: Automated text replacement (-{{WikiDoc Cardiology Network Infobox}} +, -<references /> +{{reflist|2}}, -{{reflist}} +{{reflist|2}})) |
||
| (2 intermediate revisions by 2 users not shown) | |||
| Line 29: | Line 29: | ||
| OPM protein = | | OPM protein = | ||
}} | }} | ||
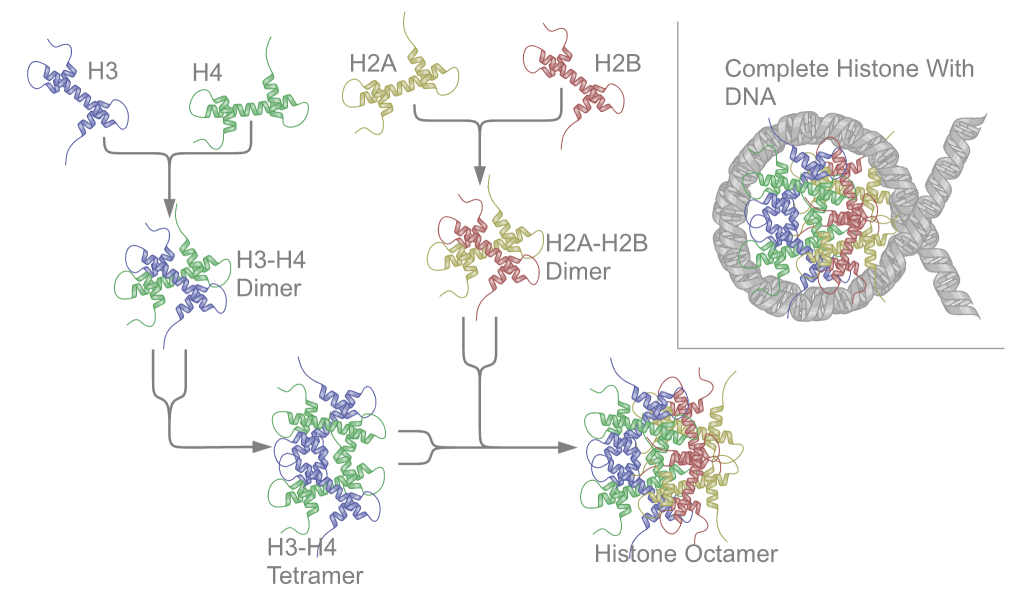
[[Image:Nucleosome structure.png|thumb| | [[Image:Nucleosome structure.png|thumb|right|300px|Schematic representation of the assembly of the core histones into the nucleosome.]] | ||
{{SI}} | {{SI}} | ||
{{CMG}} | {{CMG}} | ||
In [[biology]], '''histones''' are the chief [[protein]] components of [[chromatin]]. They act as spools around which [[DNA]] winds, and they play a role in [[gene regulation]]. Without histones, the unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA, but wound on the histones it has about 90 millimeters of chromatin, which, when duplicated and condensed during [[mitosis]], result in about 120 micrometers of [[chromosome]]s.<ref>{{cite journal |author=Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W |title=Histone H2A variants H2AX and H2AZ |journal=Curr Opin Genet Dev. |volume=12 |issue=2 |pages=162–9 |year=2002 |month=Apr |pmid=11893489 |url=http://linkinghub.elsevier.com/retrieve/pii/S0959437X02002824 |doi=10.1016/S0959-437X(02)00282-4}}</ref> | In [[biology]], '''histones''' are the chief [[protein]] components of [[chromatin]]. They act as spools around which [[DNA]] winds, and they play a role in [[gene regulation]]. Without histones, the unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA, but wound on the histones it has about 90 millimeters of chromatin, which, when duplicated and condensed during [[mitosis]], result in about 120 micrometers of [[chromosome]]s.<ref>{{cite journal |author=Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W |title=Histone H2A variants H2AX and H2AZ |journal=Curr Opin Genet Dev. |volume=12 |issue=2 |pages=162–9 |year=2002 |month=Apr |pmid=11893489 |url=http://linkinghub.elsevier.com/retrieve/pii/S0959437X02002824 |doi=10.1016/S0959-437X(02)00282-4}}</ref> | ||
| Line 222: | Line 222: | ||
==References== | ==References== | ||
{{reflist}} | {{reflist|2}} | ||
{{Chromo}} | {{Chromo}} | ||
[[Category:Epigenetics]] | [[Category:Epigenetics]] | ||
Latest revision as of 18:17, 4 September 2012
| Core histone H2A/H2B/H3/H4 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
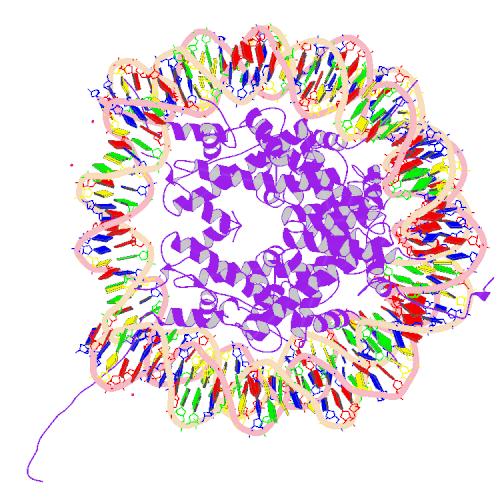 | |||||||||
| Identifiers | |||||||||
| Symbol | Histone | ||||||||
| Pfam | PF00125 | ||||||||
| InterPro | IPR007125 | ||||||||
| SCOP | 1hio | ||||||||
| SUPERFAMILY | 1hio | ||||||||
| |||||||||
| linker histone H1 and H5 family | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| Symbol | Linker_histone | ||||||||
| Pfam | PF00538 | ||||||||
| InterPro | IPR005818 | ||||||||
| SCOP | 1hst | ||||||||
| SUPERFAMILY | 1hst | ||||||||
| |||||||||
|
WikiDoc Resources for Histone |
|
Articles |
|---|
|
Most recent articles on Histone |
|
Media |
|
Evidence Based Medicine |
|
Clinical Trials |
|
Ongoing Trials on Histone at Clinical Trials.gov Clinical Trials on Histone at Google
|
|
Guidelines / Policies / Govt |
|
US National Guidelines Clearinghouse on Histone
|
|
Books |
|
News |
|
Commentary |
|
Definitions |
|
Patient Resources / Community |
|
Directions to Hospitals Treating Histone Risk calculators and risk factors for Histone
|
|
Healthcare Provider Resources |
|
Causes & Risk Factors for Histone |
|
Continuing Medical Education (CME) |
|
International |
|
|
|
Business |
|
Experimental / Informatics |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]
In biology, histones are the chief protein components of chromatin. They act as spools around which DNA winds, and they play a role in gene regulation. Without histones, the unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA, but wound on the histones it has about 90 millimeters of chromatin, which, when duplicated and condensed during mitosis, result in about 120 micrometers of chromosomes.[1]
Classes
There are a total of six classes of histones (H1, H2A, H2B, H3, H4, and H5) organized into two super classes as follows:
- core histones – H2A, H2B, H3 and H4
- linker histones – H1 and H5
Two each of the core histones assemble to form one octameric nucleosome core particle by wrapping 146 base pairs of DNA around the protein spool in 1.65 left-handed super-helical turn.[2] The linker histone H1 binds the nucleosome and the entry and exit sites of the DNA, thus locking the DNA into place[3] and allowing the formation of higher order structure. The most basic such formation is the 10 nm fiber or beads on a string conformation. This involves the wrapping of DNA around nucleosomes with approximately 50 base pairs of DNA spaced between each nucleosome (also referred to as linker DNA). The assembled histones and DNA is called chromatin. Higher order structures include the 30 nm fiber (forming an irregular zigzag) and 100 nm fiber, these being the structures found in normal cells. During mitosis and meiosis, the condensed chromosomes are assembled through interactions between nucleosomes and other regulatory proteins.
The following is a list of human histone proteins:
| Super family | Family | Subfamily | Members |
|---|---|---|---|
| Linker | |||
| H1 | |||
| H1F | H1F0, H1FNT, H1FOO, H1FX | ||
| H1H1 | HIST1H1A, HIST1H1B, HIST1H1C, HIST1H1D, HIST1H1E, HIST1H1T | ||
| Core | |||
| H2A | |||
| H2AF | H2AFB1, H2AFB2, H2AFB3, H2AFJ, H2AFV, H2AFX, H2AFY, H2AFY2, H2AFZ | ||
| H2A1 | HIST1H2AA, HIST1H2AB, HIST1H2AC, HIST1H2AD, HIST1H2AE, HIST1H2AG, HIST1H2AI, HIST1H2AJ, HIST1H2AK, HIST1H2AL, HIST1H2AM | ||
| H2A2 | HIST2H2AA3, HIST2H2AC | ||
| H2B | |||
| H2BF | H2BFM, H2BFO, H2BFS, H2BFWT | ||
| H2B1 | HIST1H2BA, HIST1H2BB, HIST1H2BC, HIST1H2BD, HIST1H2BE, HIST1H2BF, HIST1H2BG, HIST1H2BH, HIST1H2BI, HIST1H2BJ, HIST1H2BK, HIST1H2BL, HIST1H2BM, HIST1H2BN, HIST1H2BO | ||
| H2B2 | HIST2H2BE | ||
| H3 | |||
| H3A1 | HIST1H3A, HIST1H3B, HIST1H3C, HIST1H3D, HIST1H3E, HIST1H3F, HIST1H3G, HIST1H3H, HIST1H3I, HIST1H3J | ||
| H3A2 | HIST2H3C | ||
| H3A3 | HIST3H3 | ||
| H4 | |||
| H41 | HIST1H4A, HIST1H4B, HIST1H4C, HIST1H4D, HIST1H4E, HIST1H4F, HIST1H4G, HIST1H4H, HIST1H4I, HIST1H4J, HIST1H4K, HIST1H4L | ||
| H44 | HIST4H4 |
Structure
The nucleosome core is formed of two H2A-H2B dimers and a H3-H4 tetramer, forming two nearly symmetrical halves by tertiary structure (C2 symmetry; one macromolecule is the mirror image of the other)[2]. The H2A-H2B dimers and H3-H4 tetramer also show pseudodyad symmetry. The 4 'core' histones (H2A, H2B, H3 and H4) are relatively similar in structure and are highly conserved through evolution, all featuring a 'helix turn helix turn helix' motif (which allows the easy dimerisation). They also share the feature of long 'tails' on one end of the amino acid structure - this being the location of post-transcriptional modification (see below).
In all, histones make five types of interactions with DNA:
- Helix-dipoles from alpha-helices in H2B, H3, and H4 cause a net positive charge to accumulate at the point of interaction with negatively charged phosphate groups on DNA.
- Hydrogen bonds between the DNA backbone and the amide group on the main chain of histone proteins.
- Nonpolar interactions between the histone and deoxyribose sugars on DNA.
- Salt links and hydrogen bonds between side chains of basic amino acids (especially lysine and arginine) and phosphate oxygens on DNA.
- Non-specific minor groove insertions of the H3 and H2B N-terminal tails into two minor grooves each on the DNA molecule.
The highly basic nature of histones, aside from facilitating DNA-histone interactions, contributes to the water solubility of histones.[citation needed]
Histones are subject to posttranslational modification by enzymes primarily on their N-terminal tails, but also in their globular domains[citation needed]. Such modifications include methylation, citrullination, acetylation, phosphorylation, Sumoylation, ubiquitination, and ADP-ribosylation. This affects their function of gene regulation (see functions).
In general, genes that are active have less bound histone, while inactive genes are highly associated with histones during interphase[citation needed]. It also appears that the structure of histones has been evolutionarily conserved, as any deleterious mutations would be severely maladaptive.
Functions
Compacting DNA strands
Histones act as spools around which DNA winds. This enables the compaction necessary to fit the large genomes of eukaryotes inside cell nuclei: the compacted molecule is 30,000 times shorter than an unpacked molecule.
Histone modifications in chromatin regulation
Histones undergo posttranslational modifications which alter their interaction with DNA and nuclear proteins. The H3 and H4 histones have long tails protruding from the nucleosome which can be covalently modified at several places. Modifications of the tail include methylation, acetylation, phosphorylation, ubiquitination, sumoylation, citrullination, and ADP-ribosylation. The core of the histones (H2A and H3) can also be modified. Combinations of modifications are thought to constitute a code, the so-called "histone code."[4][5] Histone modifications act in diverse biological processes such as gene regulation, DNA repair and chromosome condensation (mitosis).[citation needed]
The common nomenclature of histone modifications is as follows:
- The name of the histone (e.g H3)
- The single letter amino acid abbreviation (e.g. K for Lysine) and the amino acid position in the protein
- The type of modification (Me: methyl, P: phosphate, Ac: acetyl, Ub: ubiquitin)
So H3K4me1 denotes the monomethylation of the 4th residue (a lysine) from the start (i.e., the N-terminal) of the H3 protein.
For a detailed example of histone modifications in transcription regulation see RNA polymerase control by chromatin structure and table.
Influence on gene expression in mammalian cells:
| Type of modification | ||||||
|---|---|---|---|---|---|---|
| [6][7][8][9] | H3K4 | H3K9 | H3K27 | H3K79 | H4K20 | H2BK5 |
| monomethylation | activation[7] | activation[6] | activation[6] | activation[6][8] | activation[6] | activation[6] |
| dimethylation | activation[8] | |||||
| trimethylations | activation[9] | repression[6] | repression[6] | repression[6]activation[8] | ||
| H3K9 | H3K14 | |||||
| acetylation | activation[9] | activation[9] |
History
Histones were discovered in 1884 by Albrecht Kossel. The word "histone" dates from the late 19th century and is from the German "Histon", of uncertain origin: perhaps from Greek histanai or from histos. Until the early 1990s, histones were dismissed as merely packing material for nuclear DNA. During the early 1990s, the regulatory functions of histones were discovered.[10]
Conservation across species
Histones are found in the nuclei of eukaryotic cells, and in certain Archaea, namely Euryarchaea, but not in bacteria. Archaeal histones may well resemble the evolutionary precursors to eukaryotic histones. Histone proteins are among the most highly conserved proteins in eukaryotes, emphasizing their important role in the biology of the nucleus.[citation needed]
Core histones are highly conserved proteins, that is, there are very few differences among the amino acid sequences of the histone proteins of different species. Linker histone usually has more than one form within a species and is also less conserved than the core histones.[citation needed]
There are some variant forms in some of the major classes. They share amino acid sequence homology and core structural similarity to a specific class of major histones but also have their own feature that is distinct from the major histones. These minor histones usually carry out specific functions of the chromatin metabolism. For example, histone H3-like CenpA is a histone only associated with centromere region of the chromosome. Histone H2A variant H2A.Z is associated with the promoters of actively transcribed genes and also involved in the formation of the heterochromatin. Another H2A variant H2A.X binds to the DNA with double strand breaks and marks the region undergoing DNA repair. Histone H3.3 is associated with the body of actively transcribed genes.[citation needed]
See also
- Nucleosome
- Chromatin
- Histone-Modifying Enzymes
- Histone deacetylase
- PRMT4 pathway
- Gene silencing
- Genetics
- Histone methyltransferase
- Histone acetyltransferase
External links
- Chromatin, Histones & Cathepsin; PMAP The Proteolysis Map-animation
- Nextbio
References
- ↑ Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W (2002). "Histone H2A variants H2AX and H2AZ". Curr Opin Genet Dev. 12 (2): 162–9. doi:10.1016/S0959-437X(02)00282-4. PMID 11893489. Unknown parameter
|month=ignored (help) - ↑ 2.0 2.1 Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ (1997). "Crystal structure of the nucleosome core particle at 2.8 A resolution". Nature. 389 (6648): 251–60. doi:10.1038/38444. PMID 9305837. PDB: 1AOI
- ↑ Farkas, Daniel (1996). DNA simplified: the hitchhiker's guide to DNA. Washington, D.C: AACC Press. ISBN 0-915274-84-1.
- ↑ Strahl BD, Allis CD (Jan 2000). "The language of covalent histone modifications". Nature. 403 (6765): 41–5. doi:10.1038/47412. PMID 10638745.
- ↑ Jenuwein T, Allis CD (Aug 2001). "Translating the histone code". Science. 293 (5532): 1074–80. doi:10.1126/science.1063127. PMID 11498575.
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 6.6 6.7 6.8 Barski A, Cuddapah S, Cui K; et al. (2007). "High-resolution profiling of histone methylations in the human genome". Cell. 129 (4): 823–37. doi:10.1016/j.cell.2007.05.009. PMID 17512414.
- ↑ 7.0 7.1 Benevolenskaya EV (2007). "Histone H3K4 demethylases are essential in development and differentiation". Biochem. Cell Biol. 85 (4): 435–43. doi:10.1139/o07-057. PMID 17713579.
- ↑ 8.0 8.1 8.2 8.3 Steger DJ, Lefterova MI, Ying L; et al. (2008). "DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells". Mol. Cell. Biol. 28 (8): 2825–39. doi:10.1128/MCB.02076-07. PMID 18285465.
- ↑ 9.0 9.1 9.2 9.3 Koch CM, Andrews RM, Flicek P; et al. (2007). "The landscape of histone modifications across 1% of the human genome in five human cell lines". Genome Res. 17 (6): 691–707. doi:10.1101/gr.5704207. PMID 17567990.
- ↑ Hulton CS, Seirafi A, Hinton JC, Sidebotham JM, Waddell L, Pavitt GD, Owen-Hughes T, Spassky A, Buc H, Higgins CF (1990). "Histone-like protein H1 (H-NS), DNA supercoiling, and gene expression in bacteria". Cell. 63 (3): 631–42. doi:10.1016/0092-8674(90)90458-Q. PMID 2171779. Unknown parameter
|month=ignored (help)
ar:هيستون ca:Histona cs:Histon da:Histon de:Histon fa:هیستون ko:히스톤 it:Istone he:היסטון lt:Histonas hu:Hiszton nl:Histon (eiwit) oc:Istòna sl:Histon sr:Хистони fi:Histoni sv:Histon uk:Гістони