Hepatitis B pathophysiology
|
Hepatitis B |
|
Diagnosis |
|
Treatment |
|
Case Studies |
|
Hepatitis B pathophysiology On the Web |
|
American Roentgen Ray Society Images of Hepatitis B pathophysiology |
|
Risk calculators and risk factors for Hepatitis B pathophysiology |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]
Overview
Pathogenesis
Intracellular HBV is non-cytopathic and causes little or no damage to the cell.[1]
Life Cycle
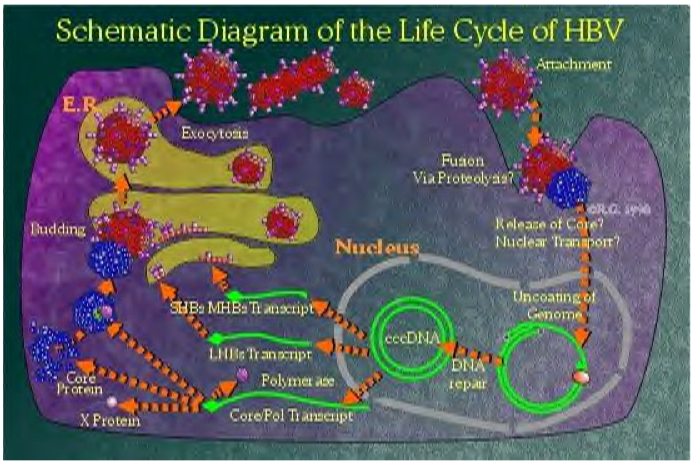 |
- The HBV virion binds to a receptor at the surface of the hepatocyte.[1]
- Severeal cellular receptors have been identified, including the transferrin receptor, the asialoglycoprotien receptor molecule, and human liver endonexin. However, the mechanism of HBsAg binding to a specific receptor to enter cells has not been established yet. Viral nucleocapsids enter the cell and reach the nucleus, where the viral genome is delivered.[1][3][4]
- In the nucleus, second-strand DNA synthesis is completed and the gaps in both strands are repaired to yield a covalently closed circular (ccc) supercoiled DNA molecule that serves as a template for transcription of four viral RNAs that are 3.5, 2.4, 2.1, and 0.7 kb long.[5][6]
HBV and Antibody Nomenclature
| Nomenclature | Full Name | Description |
|---|---|---|
| HBV | Hepatitis B Virus (complete infectious virion) | The 42 nm, double-shelled particle, that consists of a 7 nm thick outer shell and a 27 nm inner core. The core contains a small, circular, partially double-stranded DNA molecule and an endogenous DNA polymerase. This is the prototype agent for the family epadnaviridae. |
| HBsAg | ||
| HBcAg | ||
| HBeAg | ||
| Anti-HBs Anti-HBc Anti-HBe |
- These transcripts are polyadenylated and transported to the cytoplasm, where they are translated into the viral nucleocapsid and precore antigen (C, pre-C), polymerase (P), envelope L (large), M (medium), S (small)), and transcriptional transactivating proteins (X).[7][8]
- The envelope proteins insert themselves as integral membrane proteins into the lipid membrane of the endoplasmic reticulum (ER). The 3.5 kb species, spanning the entire genome and termed pregenomic RNA (pgRNA), is packaged together with HBV polymerase and a protein kinase into core particles where it serves as a template for reverse transcription of negative-strand DNA. The RNA to DNA conversion takes place inside the particles.
- The new, mature, viral nucleocapsids can then follow two different intracellular pathways, one of which leads to the formation and secretion of new virions, whereas the other leads to amplification of the viral genome inside the cell nucleus. In the virion assembly pathway, the nucleocapsids reach the ER, where they associate with the envelope proteins and bud into the lumen of the ER, from which they are secreted via the Golgi apparatus out of the cell.[1]
Associated Conditions
Hepatitis B is often associated with hepatocellular carcinoma. More than 85% of hepatocellular tumours examined harbor integrated HBV DNA, often multiple copies per cell. The viral DNA integrants are usually highly rearranged, with deletions, inversions, and sequence reiterations all commonly observed. Most of these rearrangements ablate viral gene expression, but the integrations alter the host DNA.[9][10][1]
Interestingly, tumours are clonal with respect to these integrants: every cell in the tumour contains an identical complement of HBV insertions. This implies that the integration event(s) preceded the clonal expansion of the cells. How integration is achieved is still not well understood. Since integration is not an obligatory step in the hepadnaviral replication cycle, and hepadnaviruses have no virus-encoded integration machinery, HBV DNA is probably assimilated into the nucleus by host mechanisms.
There is no similarity in the pattern of integration between different tumours, and variation is seen both in the integration site(s) and in the number of copies or partial copies of the viral genome.[1]
Transmission
Transmission results from exposure to infectious blood or body fluids containing blood. Possible forms of transmission include (but are not limited to) unprotected sexual contact, blood transfusions, re-use of contaminated needles and syringes, and vertical transmission from mother to child during childbirth. Without intervention, a mother who is positive for the hepatitis B surface antigen confers a 20% risk of passing the infection to her offspring at the time of birth. This risk is as high as 90% if the mother is also positive for the hepatitis B e antigen. HBV can also be transmitted between family members within households, possibly by contact of nonintact skin or mucous membrane with secretions or saliva containing HBV.[11]
Immunopathogenesis
During HBV infection, the host immune response causes both hepatocellular damage and viral clearance. While the innate immune response does not play a significant role in these processes, the adaptive immune response, particularly virus-specific cytotoxic T lymphocytes (CTLs), contributes to nearly all of the liver injury associated with HBV infection. By killing infected cells and by producing antiviral cytokines capable of purging HBV from viable hepatocytes, CTLs also eliminate the virus.[12]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 "Hepatitis B" (PDF).
- ↑ "http://www.who.int/en/". External link in
|title=(help) - ↑ Nathanson, Neal (1997). Viral pathogenesis. Philadelphia: Lippincott-Raven. ISBN 0781702976.
- ↑ Guidotti LG, Martinez V, Loh YT, Rogler CE, Chisari FV (1994). "Hepatitis B virus nucleocapsid particles do not cross the hepatocyte nuclear membrane in transgenic mice". J Virol. 68 (9): 5469–75. PMC 236947. PMID 8057429.
- ↑ Nathanson, Neal (1997). Viral pathogenesis. Philadelphia: Lippincott-Raven. ISBN 0781702976.
- ↑ Plotkin, Stanley (1999). Vaccines. Philadelphia: W.B. Saunders Co. ISBN 0721674437.
- ↑ Nathanson, Neal (1997). Viral pathogenesis. Philadelphia: Lippincott-Raven. ISBN 0781702976.
- ↑ Mandell, Gerald (2005). Mandell, Douglas, and Bennett's principles and practice of infectious diseases. New York: Elsevier/Churchill Livingstone. ISBN 0443066434.
- ↑ Fields, Bernard (2007). Fields virology. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins. ISBN 0781760607.
- ↑ Mandell, Gerald (2005). Mandell, Douglas, and Bennett's principles and practice of infectious diseases. New York: Elsevier/Churchill Livingstone. ISBN 0443066434.
- ↑ name="pmid791124">Petersen NJ, Barrett DH, Bond WW, Berquist KR, Favero MS, Bender TR, Maynard JE (1976). "Hepatitis B surface antigen in saliva, impetiginous lesions, and the environment in two remote Alaskan villages". Appl. Environ. Microbiol. 32 (4): 572–574. PMID 791124.
- ↑ {{cite journal | author=Iannacone M. et al | title=Pathogenetic and antiviral immune responses against hepatitis B virus | journal=Future Virology | year=2006 | pages=189-196 | volume=1 | issue=2