Polio causes: Difference between revisions
Joao Silva (talk | contribs) |
Joao Silva (talk | contribs) |
||
| Line 20: | Line 20: | ||
==Structure== | ==Structure== | ||
The genome of poliovirus consists of a single positive-sense RNA molecule, of approximately 7740 nucleotides. At the 5' end of the RNA molecule are coded 88 nucleotides that interact to form a ''clover leaf structure'', which is involved in the replication process.<ref name="pmid15885840">{{cite journal| author=Mueller S, Wimmer E, Cello J| title=Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event. | journal=Virus Res | year= 2005 | volume= 111 | issue= 2 | pages= 175-93 | pmid=15885840 | doi=10.1016/j.virusres.2005.04.008 | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=15885840 }} </ref> | |||
<!-- | <!-- | ||
The 3′ NTR contains two stem loops that may interact to form a tertiary pseudoknot structure (Pilipenko et al., 1992, p. 2557; Jacobson et al., 1993, p. 2558). The very 3′ end of the viral RNA is com- prised of a genetically encoded stretch of poly(A) varying around 60 adenylate residues in length (Yogo and Wimmer, 1972). Unlike most eukaryotic mRNAs the poliovirus ge- nomic RNA does not carry the m7G cap structure essential for translational initiation of normal eukaryotic mRNA. In- stead a small viral protein, VPg, is covalently linked to the 5′ terminus (Lee et al., 1977). Contrary to canonical eukaryotic translational initiation, cap-independent translation of the po- liovirus genome is initiated by binding of the host transla- tional machinery to a highly structured RNA cis-acting ele- ment within the 5′ non-translated region of the genome. This element has been termed internal ribosomal entry site (IRES) for its propensity to initiate translation regardless of its loca- tion within an RNA molecule (Jang et al., 1989, 1988; Molla et al., 1992). First discovered in encephalomyocarditis virus and poliovirus (Jang et al., 1988; Pelletier and Sonenberg, 1988), later in other picornaviruses (Bandyopadhyay et al., 1992; Borman and Jackson, 1992), IRES-dependent transla- tion is now established as a novel mechanism of eukaryotic cap-independent protein synthesis. More and more IRES elements have been discovered not only in viral genomes, but in the 5′ untranslated region of many cellular mRNAs (reviewed in Hellen and Sarnow, 2001) | |||
--> | --> | ||
Revision as of 18:46, 28 August 2014
|
Polio Microchapters |
|
Causes |
|---|
|
Diagnosis |
|
Treatment |
|
Case Studies |
|
Polio causes On the Web |
|
American Roentgen Ray Society Images of Polio causes |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]
Overview
Polio is a highly infectious disease caused by Poliovirus that invades the nervous system. Poliovirus are small (27–30 nm), nonenveloped viruses with capsids enclosing a single-stranded, positive-sense RNA genome about 7,500 nucleotides long. Person-to-person spread of poliovirus via the fecal-oral route is the most important route of transmission, although the oral-oral route may account for some cases.
Taxonomy
Biology
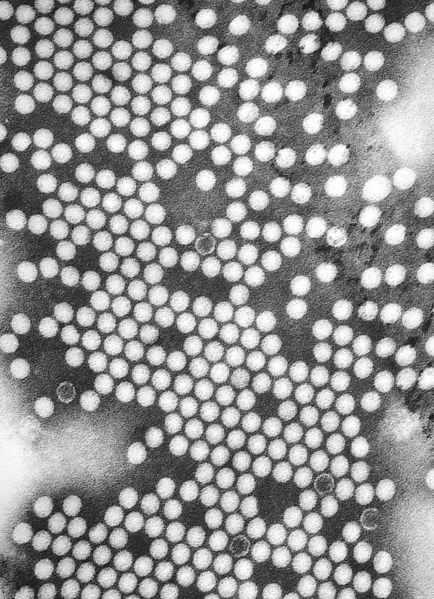 |
Poliovirus is a member of the genus enterovirus, family Picornaviridae. Enteroviruses are small, nonenveloped, positive stranded RNA viruses. Other members of the family include: Rhinovirus, Hepatovirus, Cardiovirus and Apthovirus. Poliovirus is a transient inhabitant of the gastrointestinal tract, stable at an acid pH.[2][3] Enteroviruses in general do not cause disease, or are responsible for mild symptoms. Disease syndromes resulting from viral spread to other secondary regions are rare. Despite rare, these syndromes may lead to severe disease complications, seldom with fatal outcomes.
There are three poliovirus serotype (P1, P2, and P3) that replicate efficiently in the gastrointestinal tract. There is minimal heterotypic immunity between the three serotypes. That is, immunity to one serotype does not produce significant immunity to the other serotypes. The poliovirus is rapidly inactivated by heat, formaldehyde, chlorine, and ultraviolet light.[2]
The characteristics of poliovirus make it a good model for viral study, specifically: high viral titers, stable capsid and ease of purification, along with a low bio-safety requirement.[3]
Structure
The genome of poliovirus consists of a single positive-sense RNA molecule, of approximately 7740 nucleotides. At the 5' end of the RNA molecule are coded 88 nucleotides that interact to form a clover leaf structure, which is involved in the replication process.[3]
Tropism
Natural Reservoir
Humans are the only known reservoir of poliovirus, which is transmitted most frequently by persons with inapparent infections. There is no asymptomatic carrier state except in immune deficient persons.
References
- ↑ "http://phil.cdc.gov/phil/details.asp". External link in
|title=(help) - ↑ 2.0 2.1 "Polyomavirus" (PDF).
- ↑ 3.0 3.1 3.2 Mueller S, Wimmer E, Cello J (2005). "Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event". Virus Res. 111 (2): 175–93. doi:10.1016/j.virusres.2005.04.008. PMID 15885840.