MALT lymphoma pathophysiology
For more information about H.pylori infection click here.
|
MALT lymphoma Microchapters |
|
Diagnosis |
|---|
|
Treatment |
|
Case Studies |
|
MALT lymphoma pathophysiology On the Web |
|
American Roentgen Ray Society Images of MALT lymphoma pathophysiology |
|
Risk calculators and risk factors for MALT lymphoma pathophysiology |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]; Associate Editor(s)-in-Chief: Sujit Routray, M.D. [2]
Overview
MALT lymphoma is a form of lymphoma involving the mucosa-associated lymphoid tissue (MALT), frequently of the stomach, but virtually any mucosal site can be afflicted. It is a cancer originating from B cells in the marginal zone of the MALT. The evolution of gastric MALT lymphoma is a multistage process starting with the infection of H. pylori resulting in the recruitment of B- and T-cells and other inflammatory cells to the gastric mucosa.[1] Genes involved in the pathogenesis of MALT lymphoma include FOXP1 and BCL6. Chromosomal translocations are also involved in the pathogenesis of MALT lymphoma, which include t(1;14)(p22;q32), t(11;18)(q21;q21), t(14;18)(q32;q21), and t(3;14)(p14.1;q32). Gastric MALT lymphoma is frequently associated with chronic inflammation as a result of the presence of Helicobacter pylori (72-98%).[2] Chronic immune stimulation is also suspected in the pathogenesis of non-gastric MALT lymphoma, and hence often have a history of autoimmune disorders, such as Hashimoto's thyroiditis, Sjögren's syndrome, Celiac disease, and relapsing polychondritis.[3] On microscopic histopathological analysis, MALT lymphoma is characterized by the presence of dense diffuse lymphoid infiltrate of marginal‐zone cells in lamina propria with prominent lymphoepithelial lesions and consisting of small atypical cells with monocytoid features.[4][5] A characteristic feature of MALT lymphoma is the presence of neoplastic cells within epithelial structures with associated destruction of the glandular architecture to form lymphoepithelial lesions.[6] The neoplastic cells of MALT lymphoma may be positive for B-cell associated antigens (CD19, CD20, CD22, CD79a) that co-express BCL-2, and are negative for CD5, CD10, CD43, and cyclin D1.[3]
Pathogenesis
- MALT lymphoma (MALToma) is a form of lymphoma involving the mucosa-associated lymphoid tissue (MALT), frequently of the stomach, but virtually any mucosal site can be afflicted. It is a cancer originating from B cells in the marginal zone of the MALT.
- The evolution of gastric MALT lymphoma is a multistage process starting with the infection of H. pylori resulting in the recruitment of B- and T-cells and other inflammatory cells to the gastric mucosa.[1]
- The infiltrated B-cells are stimulated by the H. pylori-specific T-cells and undergo malignant transformation due to the acquisition of genetic abnormalities. One example is the association between the H. pylori infection and gastric MALT lymphoma, in which H. pylori stimulates tumor cell growth when coincubated with helper T-cells.
- Epithelial cells are activated by chronic infectious stimuli, expressing high levels of HLA-DR and costimulatory molecules, including CD80, on their surface. These cells may be able to present antigens along with HLA molecules to T-cells. CD40 ligand molecules expressed on the activated T-cells can react with the CD40 molecule on B-cells, upregulating B-cell expression of CD80. This surface protein can react with the CD28 molecule on CD4 T-cells, strongly activating the latter.
- Activated CD4 T-cells can stimulate B-cells through CD40L-CD40 interaction, in conjunction with the action of various cytokines and chemokines. This interaction among epithelial cells, T-cells and B-cells may allow these cells to survive cooperatively in lymphoepithelial lesions and not to undergo apoptosis. Lymphoepithelial lesions are thought to be the origin of lymphomas.
- The transition from polyclonal to a monoclonal lesion is facilitated by chronic stimulation with exogenous or autoantigens, thereby increasing the frequency of their transformation. MALT lymphoma with H. pylori-dependent alterations like trisomies 3, 12, or 18 can progress and become H. pylori-independent.
- Eventually, MALT lymphoma may transform into high-grade tumors. Complete inactivation of the tumor suppressor gene p53, homologous deletion of the P16 gene, and chromosomal translocation of cMYC and BCL6 are associated with the transformation of MALT lymphoma. MALT lymphomas, devoid of t(11;18)(q21;q21) with an amplification at 3q27, are prone to high-grade transformation. On the other hand, MALT lymphomas with t(11;18)(q21;q21) are H. pylori-independent but rarely transform to aggressive lymphoma.[1]
Genetics
- Development of MALT lymphoma is the result of multiple genetic mutations.
- Genes involved in the pathogenesis of MALT lymphoma are tabulated below:[1]

1) Translocations
- There are four main recurrent chromosomal translocations associated with the pathogenesis of MALT lymphomas: t(1;14)(p22;q32), t(11;18)(q21;q21), t(14;18)(q32;q21), and t(3;14)(p14.1;q32).
- The frequency of genetic aberrations is also dependent on the primary site of disease.
- Translocation t(11;18)(q21;q21) was mainly found in pulmonary and gastric lymphoma, whereas t(14;18)(q32;q21) was most detected in ocular adnexal, orbit, skin, and salivary gland MALT lymphoma.[1]
- The t(1;14)(p22;q32) translocation occurs in 1-2% of MALT lymphomas and has been reported in the stomach, lung, and skin.The entire coding sequence of the BCL10 gene on chromosome 1 is relocated to the immunoglobulin heavy chain (IgH) enhancer region on chromosome 14 resulting in the nuclear overexpression of the BCL10 protein.
- The t(1;14)(p22;q32) translocation has exclusively been reported in MALT lymphoma, and these cases typically display additional genomic alterations. Patients with advanced stage MALT lymphoma exhibit this translocation and do not respond to Helicobacter pylori (H. pylori) eradication.
- The t(14;18)(q32;q21) translocation occurring in 15-20% of MALT lymphomas brings the MALT1 gene under the transcriptional control of the IgH enhancer region on chromosome 14. This translocation occurs more frequently in nongastrointestinal MALT lymphomas. In contrast to t(11;18)(q21;q21), the t(14;18)(q32;q21) is frequently associated with other cytogenetic abnormalities. t(14;18)(q32;q21) positive cases also show an overexpression of the BCL10 protein but display cytoplasmatic localization in contrast to t(1;14)(p22;q32) and t(11;18)(q21;q21) positive MALT lymphomas.
- The t(11;18)(q21;q21) translocation is the most common translocation, occurring in 15–40% of all MALT lymphomas. This translocation is restricted to MALT lymphomas and has not been found in nodal or splenic marginal zone lymphomas (MZL). In most of these translocation-positive cases, it is the sole chromosomal aberration and only in exceptional cases has it been detected in de novo diffuse large B-cell lymphoma arising at mucosal sites. The t(11;18)(q21;q21) has been found in MALT lymphomas at a number of different anatomic sites, including lung, stomach, intestine, and, less commonly, skin, orbit, and salivary gland. It has also been associated with cases that do not respond to H. pylori eradication and is rarely seen in transformed MALT lymphomas. The t(11;18)(q21;q21) translocation represents the fusion of the apoptosis inhibitor 2—named BIRC2 (API2)—gene on chromosome 11 and the MALT lymphoma associated translocation 1 (MALT1) gene on chromosome 18. Breakpoints observed in this translocation are clustered in the region of intron 7 and exon 8 of the BIRC2 gene and introns 4, 6, 7, and 8 of the MALT1 gene. High frequencies of deletions and duplications in both genes are also found, which implies that multiple double-strand DNA breaks (DSBs) must have occurred during the translocation process appearing as a result from illegitimate nonhomologous end joining after double stranded breaks. The resulting fusion transcript always comprises the N-terminal BIRC2 with three intact baculovirus inhibitor of apoptosis repeat (BIR) domains and the C-terminal MALT1 region containing an intact caspase-like domain. t(11;18)(q21;q21) cases show a nuclear overexpression of the BCL10 protein.
- The t(3;14)(p14.1;q32) translocation has been most recently described and establishes the juxtaposition of the transcription factor FOXP1 next to the enhancer region of the IgH chain genes. Overexpression of FOX1P analysed by chromatin immunoprecipitation in lymphoma cells demonstrates that FOX1P acts as transcriptional repressor of multiple proapoptotic genes repressing caspase-dependent apoptosis.
- The occurrence of the recurrent translocations t(1;14)(p22;q32), t(14;18)(q32;q21), and t(11;18)(q21;q21) in MALT lymphoma, constitutively activating the NF-κB pathway by the association of BCL10 and MALT1 in malignant lymphocytes, defines this pathway as an oncogenic event. Physiologically, BCL10 binds to the Ig-like domain of MALT1, and this binding induces the MALT1 oligomerization. The BCL10-MALT1 complex promotes the ubiquitylation of IκB kinase-γ and NF-κB is released to translocate into the nucleus and to transactivate genes, such as those encoding factors for cytokines and growth factors for cellular activation, proliferation, and survival. In MALT lymphoma with t(1;14)(p22;q32), BCL10 is believed to form oligomers through its CARD domain without the need for upstream signaling and thus triggers the MALT1 oligomerization and aberrant NF-κB activation. In lymphoma cases with t(14;18)(q32;q21), MALT1 is overexpressed. MALT1 does not possess a structural domain mediating self-oligomerization and it does not activate NF-κB in vitro. It seems likely that MALT1 interacts with and stabilizes BCL10, causing its accumulation in the cytoplasm of t(14;18)(q32;q21) positive tumor cells resulting in oligomerization of MALT1 and activation of NF-κB. In t(11;18)(q21;q21) positive MALT lymphomas, the BIR domain of the BIRC2-MALT1 mediates self-oligomerization, which in turn leads to NF-κB activation.
- However, two different transgenic mice—overexpressing either of the two translocations, BCL10 or BIRC2-MALT1, seen frequently in MALT lymphomas—develop splenic marginal zone hyperplasia, but not lymphoma. However, Sagaert et al. reported lymphoma development when BIRC2-MALT1 mice were exposed to antigen stimulation. Altogether, these data indicate that in MALT lymphoma chromosome translocations alone are not sufficient for full malignant transformation. Cooperation with a chronic infectious process seems to be necessary for lymphomagenesis. Recently, a novel molecular mechanism of the BIRC2-MALT1 fusion protein has been identified. Nie et al. demonstrated that the tumor suppressor gene LIMA1 binds BIRC2 and is proteolytically cleaved by MALT1 through its paracaspase activity. This cleavage generates a LIM domain—only (LMO)—containing fragment with oncogenic properties in vitro and in vivo.[1]
2) Numeric Chromosomal Aberration: Trisomies and Deletions
- Other cytogenetic alterations include trisomies 3, 12, and/or 18, which are present as a sole abnormality in 22% of the cases, but they are often associated with one of the four main translocations described above.
- Taji et al. detected trisomy 3 as the most common aberration in gastrointestinal MALT lymphomas with a frequency of up to 35%. Partial trisomies of chromosomes 3 and 18 also have been observed, as published by Krugmann et al. In contrast, Ott et al. reported an incidence of only 20% trisomy 3 in low-grade MALT lymphoma and an even lower rate in high-grade ones. The genetic mechanism by which trisomy 3 may contribute to lymphomagenesis has not yet been experimentally addressed. However, an increased gene dosage effect resulting from higher copy numbers of genes relevant to lymphoma development is likely to explain the biological consequences underlying chromosomal trisomies. Several promising candidate genes are located on chromosome 3 and have been implicated in lymphomagenesis, such as the protooncogene BCL6 and the transcription factor FOXP1. One of our previous studies describes CCR4—a chemokine receptor genomically located on chromosome 3 (3p24)—highly expressed in trisomy 3 + MALT lymphoma whereas transcripts for this chemokine receptor were missing in trisomy 3− MALT lymphomas.
- Apart from the typical chromosomal translocations, TNFAIP3 (A20) has been identified as frequently deleted in ocular adnexal MALT lymphoma as detected by array comparative genomic hybridization. As an important player in the NF-κB pathway by various mechanisms, TNFAIP3 acts as a tumor suppressor gene in various lymphoma subtypes. In ocular adnexal MALT lymphoma, complete TNFAIP3 inactivation is associated with poor lymphoma-free survival. TNFAIP3 deletion occurred in MALT lymphoma of the ocular adnexa (19%), salivary gland (8%), thyroid (11%), and liver (0.5%), but not, or at almost undetectable frequencies, in the lung, stomach, and skin. However, TNFAIP3 inactivation alone is not sufficient for malignant transformation but nevertheless may represent a promising future therapeutic target.
3) Somatic Mutations
- The number of studies investigating somatic mutations in MALT lymphoma is low and a whole genome sequencing approach has not yet been done. Our group reported somatic missense mutations in PIM1 and cMyc in 46% and 30% of MALT lymphomas (gastric and extragastric origin) and in 30% and 41% of transformed MALT lymphomas and 72% of primary cutaneous marginal zone B cell lymphomas (PCMZL), considered as integral part of MALT lymphomas. Du et al. [55] detected missense and frameshift mutations in p53 in 20.8% of MALT lymphoma and 30% of transformed MALT lymphoma (both mainly of gastric origin). Mutation analysis of NF-κB signal pathway-related genes—TNFAIP3, Card11, CD79B, and Myd88, known to be frequently mutated in aggressive lymphomas, demonstrated missense or frameshift mutations in 6% of MALT lymphoma cases in the Myd88 locus and in 28.6% of ocular adnexal MALT lymphomas mutations in the TNFAIP3 locus.
- Liu et al. [62] reported that Card11 and CD79B were not affected in their cohort of ocular adnexal MALT lymphomas.
- These genetic lesions are not restricted to MALT lymphoma. Rinaldi et al. performed a comprehensive analysis of genomic DNA copy number changes in more than 200 samples of MZL and demonstrated a distinct distribution of lesions in different subtypes (MALT lymphoma, nodal MZL, and splenic MZL). Whereas 3q and 18q gains were common in all three subtypes, del (6q23)(TNFAIP3) could be used for differentiation between MALT lymphoma and splenic MZL.
Associated Conditions
- Gastric MALT lymphoma is frequently associated with chronic inflammation as a result of the presence of Helicobacter pylori (72-98%).[2]
- Chronic immune stimulation is also suspected in the pathogenesis of non-gastric MALT lymphoma, and hence often have a history of autoimmune disorders, such as:[3]
- MALT lymphoma may be associated with infectious agents, which include:[5]
- Ocular adnexal MALT lymphoma and Chlamydia psittaci
- Salivary gland MALT lymphoma and hepatitis C virus
- Small intestinal MALT lymphoma and Campylobacter jejuni
- Cutaneous MALT lymphoma and Borrelia afzelii infection
Gross Pathology
MALT lymphoma starts in the tissues or organs outside of the lymph nodes (extranodal). MALT lymphoma develops in mucosa-associated lymphoid tissue, in the mucosa, or tissue that lines body organs or body cavities including:[7][3]
- Gastrointestinal tract (30-40%)
- The stomach is the most common location for MALT lymphoma, but they can also occur in the small bowel and colon.
- Lungs
- Eyes, including the orbit
- Skin
- Salivary glands
- Thyroid gland
- Breasts
| MALT lymphoma pathophysiology | |
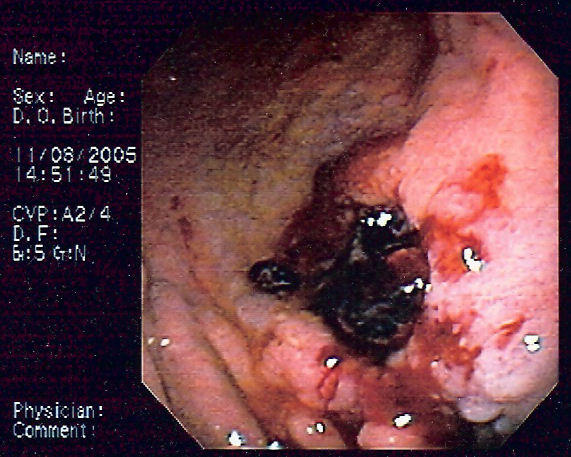 | |
|---|---|
| Endoscopic image of gastric MALT lymphoma taken in body of stomach in patient who presented with upper GI hemorrhage. Appearance is similar to gastric ulcer with adherent clot. |
Microscopic pathology
- On microscopic histopathological analysis, MALT lymphoma is characterized by the presence of dense diffuse lymphoid infiltrate of marginal‐zone cells in lamina propria with prominent lymphoepithelial lesions and consisting of small atypical cells with monocytoid features.[8][5]
- A characteristic feature of MALT lymphoma is the presence of neoplastic cells within epithelial structures with associated destruction of the glandular architecture to form lymphoepithelial lesions.[9]
- The morphology of the neoplastic cells is variable with small mature lymphocytes, cells resembling centrocytes (centrocyte-like cells), or marginal zone/monocytoid B cells.
- Plasmacytoid or plasmacytic differentiation is frequent.
- Lymphoid follicles are ubiquitous to MALT lymphoma but may be indistinct as they are often overrun or colonized by the neoplastic cells. Large transformed B cells are present scattered among the small cell population. If these large cells are present in clusters or sheets, a diagnosis of associated large B-cell lymphoma should be considered.
Immunohistochemistry
- The neoplastic cells of MALT lymphoma may be positive for B-cell associated antigens (CD19, CD20, CD22, CD79a) that co-express BCL-2, and are negative for CD5, CD10, CD43, and cyclin D1.[3]
- The tumor cells may also express complement receptors (CD21 and CD35).
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Troppan, Katharina; Wenzl, Kerstin; Neumeister, Peter; Deutsch, Alexander (2015). "Molecular Pathogenesis of MALT Lymphoma". Gastroenterology Research and Practice. 2015: 1–10. doi:10.1155/2015/102656. ISSN 1687-6121.
- ↑ 2.0 2.1 Parsonnet J, Hansen S, Rodriguez L, Gelb A, Warnke R, Jellum E, Orentreich N, Vogelman J, Friedman G (1994). "Helicobacter pylori infection and gastric lymphoma". N Engl J Med. 330 (18): 1267–71. PMID 8145781.
- ↑ 3.0 3.1 3.2 3.3 3.4 Kinkade, Zoe; Esan, Olukemi A.; Rosado, Flavia G.; Craig, Michael; Vos, Jeffrey A. (2015). "Ileal mucosa-associated lymphoid tissue lymphoma presenting with small bowel obstruction: a case report". Diagnostic Pathology. 10 (1). doi:10.1186/s13000-015-0353-6. ISSN 1746-1596.
- ↑ Taal, B G; Boot, H; van Heerde, P; de Jong, D; Hart, A A; Burgers, J M (1 October 1996). "Primary non-Hodgkin lymphoma of the stomach: endoscopic pattern and prognosis in low versus high grade malignancy in relation to the MALT concept". Gut. 39 (4): 556–561. doi:10.1136/gut.39.4.556.
- ↑ 5.0 5.1 5.2 Bacon CM, Du MQ, Dogan A (2007). "Mucosa-associated lymphoid tissue (MALT) lymphoma: a practical guide for pathologists". J Clin Pathol. 60 (4): 361–72. doi:10.1136/jcp.2005.031146. PMC 2001121. PMID 16950858.
- ↑ Janusz, edited by Jankowski, (2012). Handbook of Gastrointestinal Cancer (2 ed.). Chicester: John Wiley and Sons Ltd. pp. 243–244. ISBN 978-0-470-65624-2.
- ↑ Extranodal marginal zone of mucosa-associated lymphoid tissue (MALT lymphoma). Canadian Cancer Society 2016. http://www.cancer.ca/en/cancer-information/cancer-type/non-hodgkin-lymphoma/non-hodgkin-lymphoma/types-of-nhl/malt-lymphoma/?region=on. Accessed on January 28, 2016
- ↑ Taal, B G; Boot, H; van Heerde, P; de Jong, D; Hart, A A; Burgers, J M (1 October 1996). "Primary non-Hodgkin lymphoma of the stomach: endoscopic pattern and prognosis in low versus high grade malignancy in relation to the MALT concept". Gut. 39 (4): 556–561. doi:10.1136/gut.39.4.556.
- ↑ Janusz, edited by Jankowski, (2012). Handbook of Gastrointestinal Cancer (2 ed.). Chicester: John Wiley and Sons Ltd. pp. 243–244. ISBN 978-0-470-65624-2.