Multiple myeloma pathophysiology: Difference between revisions
Hannan Javed (talk | contribs) |
Hannan Javed (talk | contribs) |
||
| Line 10: | Line 10: | ||
===Normal physiology and development of plasma cells=== | ===Normal physiology and development of plasma cells=== | ||
* [[Plasma cells]] are terminally differentiated [[B-cells]] which function to produce [[immunoglobulins]]. [[plasma cells]] arise from [[B lymphocytes]] through a series of events. These events take place in [[bone marrow]] and secondary [[Lymphoid organs|lymphoid tissues]]. | * [[Plasma cells]] are terminally differentiated [[B-cells]] which function to produce [[immunoglobulins]]. [[plasma cells]] arise from [[B lymphocytes]] through a series of events. These events take place in [[bone marrow]] and secondary [[Lymphoid organs|lymphoid tissues]]. | ||
* These events include [[immunoglobulin]] [[Heavy-chain immunoglobulin|heavy-chain]] (IgH) VDJ [[gene]] rearrangement, migration into [[Lymphoid tissue|lymphoid tissues]], [[somatic hypermutation]] in the [[Immunoglobulin heavy chain|IgH]] and [[Immunoglobulin light chains|light-chain]] [[Gene|genes]], [[antigen]] selection, switch in Ig class from [[IgM]] to [[IgG]], [[IgA]], [[IgD]], or [[IgE]], migration back to [[bone marrow]] and terminal [[differentiation]] into [[Plasma cell|plasma cells]].<ref name="pmid23176722" /> <ref name="pmid18070707" /><ref name="pmid25723853" /> | * These events include [[immunoglobulin]] [[Heavy-chain immunoglobulin|heavy-chain]] (IgH) VDJ [[gene]] rearrangement, migration into [[Lymphoid tissue|lymphoid tissues]], [[somatic hypermutation]] (SMH) in the [[Immunoglobulin heavy chain|IgH]] and [[Immunoglobulin light chains|light-chain]] [[Gene|genes]], [[antigen]] selection, switch in Ig class from [[IgM]] to [[IgG]], [[IgA]], [[IgD]], or [[IgE]], migration back to [[bone marrow]] and terminal [[differentiation]] into [[Plasma cell|plasma cells]].<ref name="pmid23176722" /> <ref name="pmid18070707" /><ref name="pmid25723853" /> | ||
'''Stem cells → Pre-B cells → Immature B-cells → Mature B-cells (naïve) → Activated B-cells → Memory B-cells and Plasmablasts → Plasma cells''' | '''Stem cells → Pre-B cells → Immature B-cells → Mature B-cells (naïve) → Activated B-cells → Memory B-cells and Plasmablasts → Plasma cells''' | ||
* During these events, [[B cells]] express variety of surface markers which may be used to denote their [[developmental]] stage such as [[CD19]], [[CD20]], [[CD27]], [[CD38]], [[CD10]], [[CD138]]. These markers and [[Immunoglobulin heavy chain|IgH]] chain [[gene]] sequences are important to define the nature of [[Multiple myeloma|myeloma]] cells.<ref name="pmid23176722" /><ref name="pmid18070707" /><ref name="pmid25723853" /> | * During these events, [[B cells]] express variety of surface markers which may be used to denote their [[developmental]] stage such as [[CD19]], [[CD20]], [[CD27]], [[CD38]], [[CD10]], [[CD138]]. These markers and [[Immunoglobulin heavy chain|IgH]] chain [[gene]] sequences are important to define the nature of [[Multiple myeloma|myeloma]] cells.<ref name="pmid23176722" /><ref name="pmid18070707" /><ref name="pmid25723853" /> | ||
| Line 21: | Line 21: | ||
=== Normal physiology and development of Immunoglobulins === | === Normal physiology and development of Immunoglobulins === | ||
'''Immunoglobulins''' (also known as '''Antibodies''') are [[protein]]s that are found in [[blood]] or other [[bodily fluid]]s of [[vertebrate]]s, and are used by the [[immune system]] to identify and neutralize foreign objects, such as [[bacterium|bacteria]] and [[virus]]es. They are made of a few basic structural units called ''chains''; each antibody has two large [[heavy chains]] '''H''' and two small [[light chain]]s '''L'''. There are several different types of [[antibody]] [[heavy chains]], and several different kinds of [[antibodies]], which are grouped into different ''[[isotype (immunology)|isotypes]]'' based on which heavy chain they possess. Five different antibody isotypes are known in mammals, which perform different roles, and help direct the appropriate immune response for each different type of foreign object they encounter.<ref name="Market">Eleonora Market, F. Nina Papavasiliou (2003) [http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371/journal.pbio.0000016 ''V(D)J Recombination and the Evolution of the Adaptive Immune System''] [[PLoS Biology]]1(1): e16.</ref><ref name="pmid8450761">{{cite journal |author=Litman GW, Rast JP, Shamblott MJ, ''et al'' |title=Phylogenetic diversification of immunoglobulin genes and the antibody repertoire |journal=Mol. Biol. Evol. |volume=10 |issue=1 |pages=60–72 |year=1993 |pmid=8450761 |doi=}}</ref> | * '''Immunoglobulins''' (also known as '''Antibodies''') are [[protein]]s that are found in [[blood]] or other [[bodily fluid]]s of [[vertebrate]]s, and are used by the [[immune system]] to identify and neutralize foreign objects, such as [[bacterium|bacteria]] and [[virus]]es. | ||
* They are made of a few basic structural units called ''chains''; each antibody has two large [[heavy chains]] '''H''' and two small [[light chain]]s '''L'''. There are several different types of [[antibody]] [[heavy chains]], and several different kinds of [[antibodies]], which are grouped into different ''[[isotype (immunology)|isotypes]]'' based on which heavy chain they possess.<ref name="Market" /><ref name="pmid8450761" /> | |||
* Five different antibody isotypes are known in mammals, which perform different roles, and help direct the appropriate immune response for each different type of foreign object they encounter.<ref name="Market">Eleonora Market, F. Nina Papavasiliou (2003) [http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371/journal.pbio.0000016 ''V(D)J Recombination and the Evolution of the Adaptive Immune System''] [[PLoS Biology]]1(1): e16.</ref><ref name="pmid8450761">{{cite journal |author=Litman GW, Rast JP, Shamblott MJ, ''et al'' |title=Phylogenetic diversification of immunoglobulin genes and the antibody repertoire |journal=Mol. Biol. Evol. |volume=10 |issue=1 |pages=60–72 |year=1993 |pmid=8450761 |doi=}}</ref> | |||
* [[Class switch recombination]] (CSR), a region specific deletion recombination process, generates different [[immunoglobulin]] (Ig) isotypes by replacing one switch region with another. This process leads to enhanced functionality of [[immunoglobulins]].<ref name="pmid246881083">{{cite journal |vauthors=Boyle EM, Davies FE, Leleu X, Morgan GJ |title=Understanding the multiple biological aspects leading to myeloma |journal=Haematologica |volume=99 |issue=4 |pages=605–12 |date=April 2014 |pmid=24688108 |pmc=3971069 |doi=10.3324/haematol.2013.097907 |url=}}</ref> | |||
* [[Class switch recombination]] (CSR), just like [[somatic hypermutation]] (SMH), requires the [[expression]] of activation-induced deaminase (AID) and both are dependent on creation of double-stranded [[DNA]] breaks (DSB) in the [[Immunoglobulin|Ig]] [[loci]].<ref name="pmid246881084">{{cite journal |vauthors=Boyle EM, Davies FE, Leleu X, Morgan GJ |title=Understanding the multiple biological aspects leading to myeloma |journal=Haematologica |volume=99 |issue=4 |pages=605–12 |date=April 2014 |pmid=24688108 |pmc=3971069 |doi=10.3324/haematol.2013.097907 |url=}}</ref> | |||
For more information on [[immunoglobulins]], [[Immunoglobulin|click here]]. | For more information on [[immunoglobulins]], [[Immunoglobulin|click here]]. | ||
Revision as of 15:33, 24 October 2018
|
Multiple myeloma Microchapters |
|
Diagnosis |
|---|
|
Treatment |
|
Case Studies |
|
Multiple myeloma pathophysiology On the Web |
|
American Roentgen Ray Society Images of Multiple myeloma pathophysiology |
|
Risk calculators and risk factors for Multiple myeloma pathophysiology |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1] Associate Editor(s)-in-Chief: Haytham Allaham, M.D. [2] Shyam Patel [3]
Overview
Multiple myeloma arises from post-germinal center plasma cells that are normally involved in production of human immunoglobulins.[1][2][3] Both sporadic events and familial contributions contribute to the pathogenesis of multiple myeloma. Renal involvement by multiple myeloma is catergorized into three entities: light chain cast nephropathy, monoclonal immunoglobulin deposition disease, and amyloidosis. Osseous involvement by multiple myeloma is based on cytokine and cellular interactions that lead to bone breakdown. On microscopic histopathological analysis, abundant eosinophilic cytoplasm, eccentrically placed nucleus, and Russell bodies are characteristic findings of multiple myeloma.[4]
Pathophysiology
Normal physiology and development of plasma cells
- Plasma cells are terminally differentiated B-cells which function to produce immunoglobulins. plasma cells arise from B lymphocytes through a series of events. These events take place in bone marrow and secondary lymphoid tissues.
- These events include immunoglobulin heavy-chain (IgH) VDJ gene rearrangement, migration into lymphoid tissues, somatic hypermutation (SMH) in the IgH and light-chain genes, antigen selection, switch in Ig class from IgM to IgG, IgA, IgD, or IgE, migration back to bone marrow and terminal differentiation into plasma cells.[5] [6][7]
Stem cells → Pre-B cells → Immature B-cells → Mature B-cells (naïve) → Activated B-cells → Memory B-cells and Plasmablasts → Plasma cells
- During these events, B cells express variety of surface markers which may be used to denote their developmental stage such as CD19, CD20, CD27, CD38, CD10, CD138. These markers and IgH chain gene sequences are important to define the nature of myeloma cells.[5][6][7]
- Plasma cells secrete antibodies which function in humoral immunity.[5][6] [7]
- Plasma cells are typically polyclonal and can respond to a variety of antigens, which helps combat infections. Under normal circumstances, there is no monoclonality amongst the plasma cell population in a person. These plasma cells are functionally intact in their ability to contribute to humoral immunity.[5][6][7]
- Transcription factors such as interferon regulatory factor 4 (IRF4), BCL6, B-lymphocyte-induced maturation protein 1 (BLIMP1, also known as PRDM1), paired box gene 5 (PAX5) and X box binding protein 1 (XBP1) play an important role in differentiation and survival of plasma cells.[8][9][10]
Interferon regulatory factor 4 (IRF4) → Down-regulation of BCL6 → Up-regulation of B-lymphocyte-induced maturation protein 1 (BLIMP1) → Down-regulation of paired box gene 5 (PAX5) and Up-regulation of X box binding protein 1 (XBP1).[11][12][13]
For more information on plasma cells, click here.
Normal physiology and development of Immunoglobulins
- Immunoglobulins (also known as Antibodies) are proteins that are found in blood or other bodily fluids of vertebrates, and are used by the immune system to identify and neutralize foreign objects, such as bacteria and viruses.
- They are made of a few basic structural units called chains; each antibody has two large heavy chains H and two small light chains L. There are several different types of antibody heavy chains, and several different kinds of antibodies, which are grouped into different isotypes based on which heavy chain they possess.[14][15]
- Five different antibody isotypes are known in mammals, which perform different roles, and help direct the appropriate immune response for each different type of foreign object they encounter.[14][15]
- Class switch recombination (CSR), a region specific deletion recombination process, generates different immunoglobulin (Ig) isotypes by replacing one switch region with another. This process leads to enhanced functionality of immunoglobulins.[16]
- Class switch recombination (CSR), just like somatic hypermutation (SMH), requires the expression of activation-induced deaminase (AID) and both are dependent on creation of double-stranded DNA breaks (DSB) in the Ig loci.[17]
For more information on immunoglobulins, click here.
Pathogenesis
The pathogenesis of multiple myeloma is complex and probably is a result of multiple and multi-step oncogenic events such as hyperdiploidy, deregulation of cyclin D1 and interaction of myeloma cells with marrow environment. A brief description of events thought to play a role in pathogenesis of multiple myeloma is given here.
Environmental and hereditary factors
- The Key environmental and hereditary factors thought to confer a greater risk of developing multiple myeloma or play a part in pathogenesis are summarized in the table below.[8][18][19][20][21][22][23][24][25][26][27]
| Environmental and hereditary risk factors |
|---|
|
| * Likely influenced by environmental and behavioral confounding factors. |
Chromosomal aberrations
- Two major sets of chromosomal abnormalities seen in multiple myeloma are translocations and numerical aberrations, either trisomy or monosomy.
| Chromosomal aberrations in multiple myeloma (MM) | ||
|---|---|---|
| Chromosomal Abnormality | Chromosome(s)/Protein(s) affected | Consequence |
| Trisomies | Odd-numbered chromosomes with the
exception of chromosomes 1, 13, and 21 |
|
| t(11;14)(q13;q32)
t(6;14q)(p21;32) t(12;14)(p13;q32) |
Cyclin D1
Cyclin D3 Cyclin D2 |
Over-expression; cell cycle dysregulation |
| t(4;14)(p16;q32) | FGFR3 or MMSET | Over-expression and activation; MM cell proliferation/apoptosis prevention MMSET
probably linked to crucial transforming event |
| t(14;16)(q32;q23)
t(14;20)(q32;q11) t(8;14)(q24;q32) |
c-MAF | Over-expression; involvement in IL-4 regulation |
| del 17p13 | p53 | Cell-cycle dysregulation/apoptosis |
| Monosomy 14 | Chromosome 14 | |
| Chromosome 13 deletion and monosomy | Chromosome 13 | |
| Gain(1q21) | Chromosome 1 | |
Pathophysiology of renal involvement
Abnormal antibody fragments are produced in multiple myeloma and are deposited in various organs, such as the kidneys. There are three major forms of renal damage in patients with multiple myeloma.
- Cast nephropathy: End-organ damage to the kidneys is typically due to light chain cast nephropathy. The pathophysiology of this type of renal involvement is based on light chain deposition in the renal tubules, which results in obstruction. Free light chains are readily filtered at the glomerulus and are reabsorbed in the proximal tubule of the nephron. This reabsorption occurs via the megalin-cubulin transport system.[28] In patients with multiple myeloma, there is excess production of free light chains, and the ability of the nephron to resorb light chains in the proximal tubule cannot meet the demands of the freely filtered light chains. This results in excess secretion of free light chains in the urine (known as Bence-Jones protein). Eosinophilic proteinaceous casts and crystalline structures can be seen. Cast formation occurs in the tubules due to excess abundance of free light chains that interact with Tamm-Horsfall proteins in the thick ascending loop of Henle.[28] Tubular obstruction ensues, triggering local inflammation which results in interstitial nephritis and fibrosis.[28] The onset of cast nephropathy can be very quick, requiring prompt treatment. Risk factors for development of cast nephropathy include monoclonal immunoglobulin secretion of >10 g/day, sepsis, and volume depletion.[29] Patients can also develop Fanconi syndrome, resulting in dysfunctional reabsorption ability by the proximal tubule, and type II renal tubular acidosis.
- Monoclonal immunoglobulin deposition disease (MIDD): In this subtype of renal involvement by multiple myeloma, the initial pathophysiological process is filtration of monoclonal immunoglobulins and subsequent deposition of immunoglobulins along the tubular or glomerular basement membrane.[29] Deposits of immunoglobulin can have a similar appearance as Kimmelstein-Wilson lesions (seen in diabetes). The immunoglobulins can appear fibroblast-like.
- Light chain amyloidosis: The pathophysiology of renal involvement by light chain amyloidosis begins with beta-pleated sheet formation in the tubules or glomeruli. Beta-pleated sheets form as a result of electrostatic interactions between heparan sulfate proteoglycan and amyloid proteins. Amyloid fibrils usually consist of immunoglobulin light chains (usually lambda light chain) and deposit in the basement membrane. The size of the fibrils vary from 7 to 10 nanometers. A diagnosis of this type of renal involvement is made by the visualization of apple green birefringence upon Congo red staining of the renal specimen.[29] It is frequently associated with nephrotic range proteinuria, in which greater than 3 grams of protein is excreted daily.
Pathophysiology of osseous involvement
The pathophysiology of bony involvement of multiple myeloma involves cytokines and cellular interactions.
- Cytokines: Multiple myeloma is one of the most common malignancies that creates lytic bony lesions. Other cancers that can create lytic bony lesions include renal cell carcinoma, lung cancer, breast cancer, thyroid cancer, and lymphoma. Lytic destruction has a distinct pathophysiology compared to the blastic destruction that is seen in prostate cancer. The pathophysiology of bony disease in multiple myeloma begins with stimulation of osteoclast production and suppression of osteoblast production. The steady state of bone metabolism is shifted in the direction of bone resorption. The molecular mechanism that governs osteoclast activation in multiple myeloma involves nuclear factor kappa B (NFkB), interleukin-3 (IL-3), interleukin-6 (IL-6), tumor necrosis factor-alpha (TNF-a), and CXCL12, also known as stromal cell-derived factor 1 (SDF1).[30]
- Cellular interactions: At the cellular level, bone breakdown is governed by interactions between multiple myeloma cells, osteoclasts, osteoblasts, mesenchymal stem cells, T lymphocytes and dendritic cells. Multiple myeloma cells chemoattract hematopoietic cells such as dendritic cells, which produce osteoclastic factors that contribute to bone breakdown. Dendritic cells can also transdifferentiate into osteoclasts.[30] Multiple myeloma cells suppress osteoblast production by inhibiting expression of the transcription factor RUNX2, which is critical for osteoblast activity. The immune microenvironment thus plays a major role in the formation of lytic lesions in patients with multiple myeloma. Malignant plasma cells infiltrate hematopoietic sites such as the red bone marrow where they interfere with the production of normal blood cells.[1][2][3][31] The distribution of multiple myeloma mirrors that of red bone marrow in older individuals, and thus multiple myeloma is mostly encountered in the axial skeleton and proximal appendicular skeleton such as:[1]
- Vertebrae (most common)
- Ribs
- Skull
- Shoulder girdle
- Pelvis
- Long bones
- Extra skeletal structures (extraosseous myeloma) (rare)
Gross Pathology
-
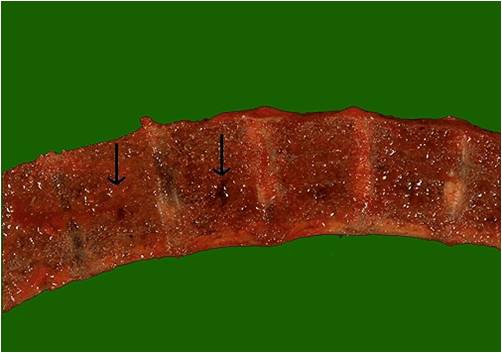
Vertebrae in multiple myeloma
(Image courtesy of Melih Aktan M.D.) -
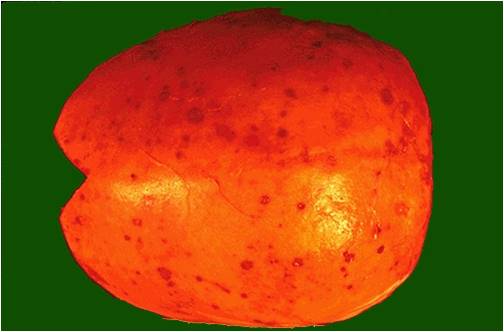
Calvarium in multiple myeloma.
(Image courtesy of Melih Aktan M.D.)
Microscopic Pathology
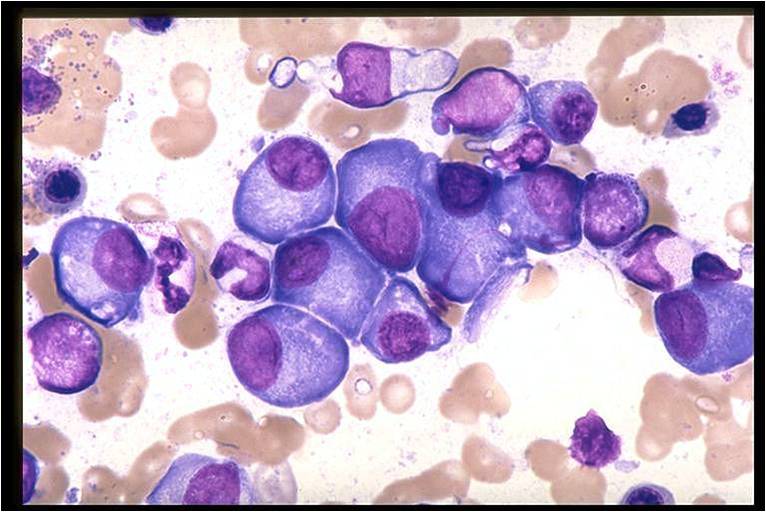
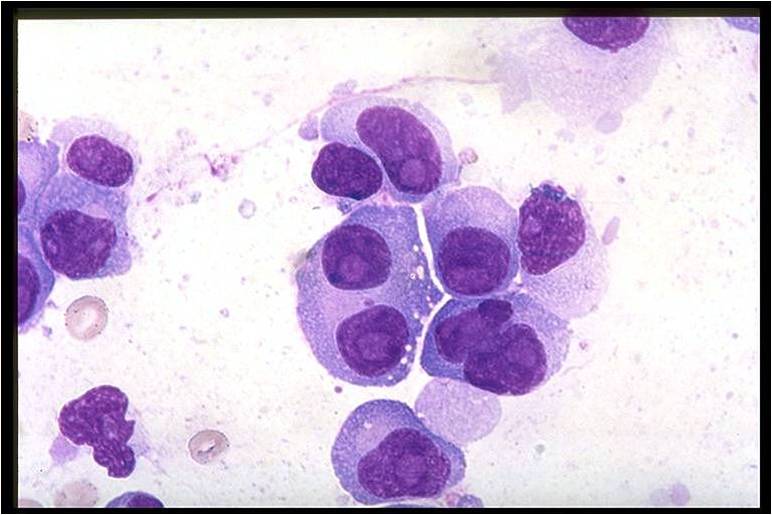
On microscopic histopathological analysis, multiple myeloma is characterized by the following:[4]
- Abundant eosinophilic cytoplasm
- Eccentrically placed nucleus
- Clock face morphology of the nucleus due to chromatin clumps around the edges
- Russell bodies which are eosinophilic, large (10-15 micrometres), homogenous immunoglobulin-containing inclusions
- Dutcher bodies which are PAS stain +ve intranuclear crystalline rods
- Shown below is a series of microscopic images seen in multiple myeloma:
-
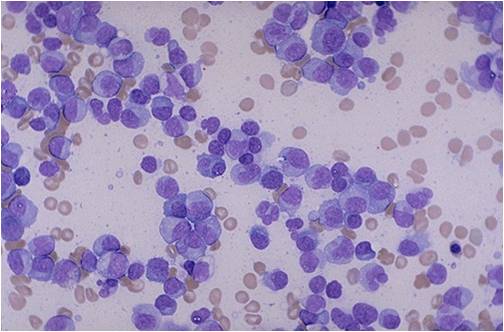
Multiple Myeloma slide showing plasma cells with large nucleus and scant cytoplasm [32]
-
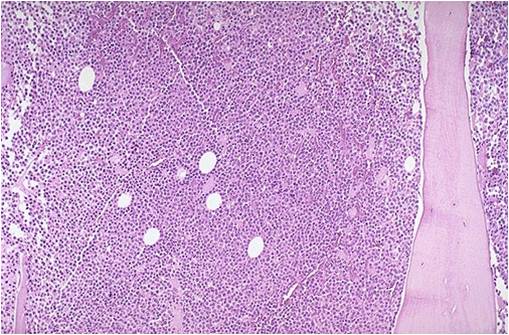
Bone marrow aspiration in multiple myeloma.
(Image courtesy of Melih Aktan M.D.) -
Bone marrow biopsy in multiple myeloma.
(Image courtesy of Melih Aktan M.D.) -
Bone marrow in multiple myeloma.
(Image courtesy of Melih Aktan M.D.) -
Bone marrow in multiple myeloma.
(Image courtesy of Melih Aktan M.D.) -
Multiple myeloma slide with intermediate magnification[4]
-
Multiple myeloma slide with high magnification[4]
-
Multiple myeloma slide with russell bodies[4]
References
- ↑ 1.0 1.1 1.2 Multiple myeloma. Radiopaedia (2015)http://radiopaedia.org/articles/multiple-myeloma-1 Accessed on September, 20th 2015
- ↑ 2.0 2.1 Multiple myeloma. Wikipedia (2015)https://en.wikipedia.org/wiki/Multiple_myeloma#Pathophysiology Accessed on September, 20th 2015
- ↑ 3.0 3.1 Multiple myeloma. Medlineplus (2015)https://www.nlm.nih.gov/medlineplus/multiplemyeloma.html Accessed on September, 20th 2015
- ↑ 4.0 4.1 4.2 4.3 4.4 Multiple myeloma. Librepathology (2015)http://www.wikidoc.org/index.php?title=Multiple_myeloma_pathophysiology&action=edit§ion=1 Accessed on September, 20th 2015
- ↑ 5.0 5.1 5.2 5.3 Borrello I (November 2012). "Can we change the disease biology of multiple myeloma?". Leuk. Res. 36 Suppl 1: S3–12. doi:10.1016/S0145-2126(12)70003-6. PMC 3698609. PMID 23176722.
- ↑ 6.0 6.1 6.2 6.3 Chng WJ, Glebov O, Bergsagel PL, Kuehl WM (December 2007). "Genetic events in the pathogenesis of multiple myeloma". Best Pract Res Clin Haematol. 20 (4): 571–96. doi:10.1016/j.beha.2007.08.004. PMC 2198931. PMID 18070707.
- ↑ 7.0 7.1 7.2 7.3 Hengeveld PJ, Kersten MJ (February 2015). "B-cell activating factor in the pathophysiology of multiple myeloma: a target for therapy?". Blood Cancer J. 5: e282. doi:10.1038/bcj.2015.3. PMC 4349256. PMID 25723853.
- ↑ 8.0 8.1 Boyle EM, Davies FE, Leleu X, Morgan GJ (April 2014). "Understanding the multiple biological aspects leading to myeloma". Haematologica. 99 (4): 605–12. doi:10.3324/haematol.2013.097907. PMC 3971069. PMID 24688108.
- ↑ Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH (July 2001). "Plasma cell differentiation requires the transcription factor XBP-1". Nature. 412 (6844): 300–7. doi:10.1038/35085509. PMID 11460154.
- ↑ Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD (October 2011). "The genetic network controlling plasma cell differentiation". Semin. Immunol. 23 (5): 341–9. doi:10.1016/j.smim.2011.08.010. PMID 21924923.
- ↑ Boyle EM, Davies FE, Leleu X, Morgan GJ (April 2014). "Understanding the multiple biological aspects leading to myeloma". Haematologica. 99 (4): 605–12. doi:10.3324/haematol.2013.097907. PMC 3971069. PMID 24688108.
- ↑ Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH (July 2001). "Plasma cell differentiation requires the transcription factor XBP-1". Nature. 412 (6844): 300–7. doi:10.1038/35085509. PMID 11460154.
- ↑ Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD (October 2011). "The genetic network controlling plasma cell differentiation". Semin. Immunol. 23 (5): 341–9. doi:10.1016/j.smim.2011.08.010. PMID 21924923.
- ↑ 14.0 14.1 Eleonora Market, F. Nina Papavasiliou (2003) V(D)J Recombination and the Evolution of the Adaptive Immune System PLoS Biology1(1): e16.
- ↑ 15.0 15.1 Litman GW, Rast JP, Shamblott MJ; et al. (1993). "Phylogenetic diversification of immunoglobulin genes and the antibody repertoire". Mol. Biol. Evol. 10 (1): 60–72. PMID 8450761.
- ↑ Boyle EM, Davies FE, Leleu X, Morgan GJ (April 2014). "Understanding the multiple biological aspects leading to myeloma". Haematologica. 99 (4): 605–12. doi:10.3324/haematol.2013.097907. PMC 3971069. PMID 24688108.
- ↑ Boyle EM, Davies FE, Leleu X, Morgan GJ (April 2014). "Understanding the multiple biological aspects leading to myeloma". Haematologica. 99 (4): 605–12. doi:10.3324/haematol.2013.097907. PMC 3971069. PMID 24688108.
- ↑ Morgan GJ, Davies FE, Linet M (July 2002). "Myeloma aetiology and epidemiology". Biomed. Pharmacother. 56 (5): 223–34. PMID 12199621.
- ↑ Wadhera RK, Rajkumar SV (October 2010). "Prevalence of monoclonal gammopathy of undetermined significance: a systematic review". Mayo Clin. Proc. 85 (10): 933–42. doi:10.4065/mcp.2010.0337. PMC 2947966. PMID 20713974.
- ↑ Landgren O, Kyle RA, Pfeiffer RM, Katzmann JA, Caporaso NE, Hayes RB, Dispenzieri A, Kumar S, Clark RJ, Baris D, Hoover R, Rajkumar SV (May 2009). "Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study". Blood. 113 (22): 5412–7. doi:10.1182/blood-2008-12-194241. PMC 2689042. PMID 19179464.
- ↑ Wang SS, Voutsinas J, Chang ET, Clarke CA, Lu Y, Ma H, West D, Lacey JV, Bernstein L (July 2013). "Anthropometric, behavioral, and female reproductive factors and risk of multiple myeloma: a pooled analysis". Cancer Causes Control. 24 (7): 1279–89. doi:10.1007/s10552-013-0206-0. PMC 3684420. PMID 23568533.
- ↑ Grulich AE, van Leeuwen MT, Falster MO, Vajdic CM (July 2007). "Incidence of cancers in people with HIV/AIDS compared with immunosuppressed transplant recipients: a meta-analysis". Lancet. 370 (9581): 59–67. doi:10.1016/S0140-6736(07)61050-2. PMID 17617273.
- ↑ Kachuri L, Demers PA, Blair A, Spinelli JJ, Pahwa M, McLaughlin JR, Pahwa P, Dosman JA, Harris SA (October 2013). "Multiple pesticide exposures and the risk of multiple myeloma in Canadian men". Int. J. Cancer. 133 (8): 1846–58. doi:10.1002/ijc.28191. PMID 23564249.
- ↑ Singh J, Dudley AW, Kulig KA (December 1990). "Increased incidence of monoclonal gammopathy of undetermined significance in blacks and its age-related differences with whites on the basis of a study of 397 men and one woman in a hospital setting". J. Lab. Clin. Med. 116 (6): 785–9. PMID 2246554.
- ↑ Kristinsson SY, Björkholm M, Goldin LR, Blimark C, Mellqvist UH, Wahlin A, Turesson I, Landgren O (November 2009). "Patterns of hematologic malignancies and solid tumors among 37,838 first-degree relatives of 13,896 patients with multiple myeloma in Sweden". Int. J. Cancer. 125 (9): 2147–50. doi:10.1002/ijc.24514. PMC 2737604. PMID 19582882.
- ↑ Broderick P, Chubb D, Johnson DC, Weinhold N, Försti A, Lloyd A, Olver B, Ma Y, Dobbins SE, Walker BA, Davies FE, Gregory WA, Childs JA, Ross FM, Jackson GH, Neben K, Jauch A, Hoffmann P, Mühleisen TW, Nöthen MM, Moebus S, Tomlinson IP, Goldschmidt H, Hemminki K, Morgan GJ, Houlston RS (November 2011). "Common variation at 3p22.1 and 7p15.3 influences multiple myeloma risk". Nat. Genet. 44 (1): 58–61. doi:10.1038/ng.993. PMC 5108406. PMID 22120009.
- ↑ Chubb D, Weinhold N, Broderick P, Chen B, Johnson DC, Försti A, Vijayakrishnan J, Migliorini G, Dobbins SE, Holroyd A, Hose D, Walker BA, Davies FE, Gregory WA, Jackson GH, Irving JA, Pratt G, Fegan C, Fenton JA, Neben K, Hoffmann P, Nöthen MM, Mühleisen TW, Eisele L, Ross FM, Straka C, Einsele H, Langer C, Dörner E, Allan JM, Jauch A, Morgan GJ, Hemminki K, Houlston RS, Goldschmidt H (October 2013). "Common variation at 3q26.2, 6p21.33, 17p11.2 and 22q13.1 influences multiple myeloma risk". Nat. Genet. 45 (10): 1221–1225. doi:10.1038/ng.2733. PMC 5053356. PMID 23955597.
- ↑ 28.0 28.1 28.2 Finkel KW, Cohen EP, Shirali A, Abudayyeh A, American Society of Nephrology Onco-Nephrology Forum (2016). "Paraprotein-Related Kidney Disease: Evaluation and Treatment of Myeloma Cast Nephropathy". Clin J Am Soc Nephrol. 11 (12): 2273–2279. doi:10.2215/CJN.01640216. PMC 5142056. PMID 27526708.
- ↑ 29.0 29.1 29.2 Heher EC, Rennke HG, Laubach JP, Richardson PG (2013). "Kidney disease and multiple myeloma". Clin J Am Soc Nephrol. 8 (11): 2007–17. doi:10.2215/CJN.12231212. PMC 3817918. PMID 23868898.
- ↑ 30.0 30.1 Yaccoby S (2010). "Advances in the understanding of myeloma bone disease and tumour growth". Br J Haematol. 149 (3): 311–21. doi:10.1111/j.1365-2141.2010.08141.x. PMC 2864366. PMID 20230410.
- ↑ What is multiple myeloma. Canadian Cancer Society (2015) http://www.cancer.ca/en/cancer-information/cancer-type/multiple-myeloma/multiple-myeloma/?region=mb Accessed on September, 20th 2015
- ↑ http://picasaweb.google.com/mcmumbi/USMLEIIImages
![Multiple Myeloma slide showing plasma cells with large nucleus and scant cytoplasm [32]](/images/a/a5/Multiple_Myeloma.jpg)
![Multiple myeloma slide with intermediate magnification[4]](/images/4/40/Multiple_myeloma_intermed_mag.jpg)
![Multiple myeloma slide with high magnification[4]](/images/d/d5/Multiple_myeloma.jpg)
![Multiple myeloma slide with russell bodies[4]](/images/a/ad/Russell_bodies.jpg)