Acute myeloid leukemia pathophysiology: Difference between revisions
No edit summary |
|||
| Line 21: | Line 21: | ||
=== Normal Hematopoeisis === | === Normal Hematopoeisis === | ||
* Hematopoeisis is tightly regulated under physiological conditions via a number of lineage-specific growth | * [[Hematopoeisis]] is tightly regulated under physiological conditions via a number of lineage-specific growth factors and lineage-specific growth signalling pathways. | ||
* The differentiation of myeloid stem cells into mature myelocytes is controlled by lineage-specific transcription factors that regulate the expression of lineage-specific genes. | * The differentiation of [[Myeloid cells|myeloid stem cells]] into mature myelocytes is controlled by lineage-specific [[transcription factors]] that regulate the expression of lineage-specific [[genes]]. | ||
* Normal hematopoeisis is dependent upon specific growth factor receptors. The two key growth factor receptors involved are as following: | * Normal hematopoeisis is dependent upon specific [[Growth factor receptor|growth factor receptors]]. The two key [[Growth factor receptor|growth factor receptors]] involved are as following: | ||
** Growth factor receptors with intrinsic tyrosine kinase activity (expressed on CD34+ hematopoietic progenitor cells, examples include, receptors for the platelet-derived growth factors (PDGFs) PDGFR A and B, the receptor for M-CSF, | ** [[Growth factor receptor|Growth factor receptors]] with intrinsic [[tyrosine kinase]] activity (expressed on [[CD34|CD34+]] [[hematopoietic]] progenitor [[cells]], examples include, receptors for the [[Platelet-derived growth factor|platelet-derived growth factors]] ([[Platelet-derived growth factor|PDGFs]]) PDGFR A and B, the receptor for [[Macrophage colony-stimulating factor|macrophage colony stimulating factor]] ([[Macrophage colony-stimulating factor|M-CSF]]), FMS, and the receptors for KL (SCF) and FL, Kit and Flt3)<ref name="pmid14977821">{{cite journal |vauthors=Müller-Tidow C, Schwäble J, Steffen B, Tidow N, Brandt B, Becker K, Schulze-Bahr E, Halfter H, Vogt U, Metzger R, Schneider PM, Büchner T, Brandts C, Berdel WE, Serve H |title=High-throughput analysis of genome-wide receptor tyrosine kinase expression in human cancers identifies potential novel drug targets |journal=Clin. Cancer Res. |volume=10 |issue=4 |pages=1241–9 |date=February 2004 |pmid=14977821 |doi= |url=}}</ref> | ||
** Growth factor receptors without intrinsic tyrosine kinase activity and depend upon intracellular kinases of the Src and JAK families (examples include, the JAK-STAT pathway)<ref name="pmid11248555">{{cite journal |vauthors=Ihle JN |title=The Stat family in cytokine signaling |journal=Curr. Opin. Cell Biol. |volume=13 |issue=2 |pages=211–7 |date=April 2001 |pmid=11248555 |doi= |url=}}</ref> | ** Growth factor receptors without intrinsic [[tyrosine kinase]] activity and depend upon [[intracellular]] [[kinases]] of the Src and [[Janus kinase|Janus Kinase]] ([[JAK]] ) families (examples include, the [[JAK-STAT pathway]])<ref name="pmid11248555">{{cite journal |vauthors=Ihle JN |title=The Stat family in cytokine signaling |journal=Curr. Opin. Cell Biol. |volume=13 |issue=2 |pages=211–7 |date=April 2001 |pmid=11248555 |doi= |url=}}</ref> | ||
'''Role Altered Signal Transduction and Autonomous Proliferaion (Protein Tyrosine Kinase Activation)''' | '''Role Altered Signal Transduction and Autonomous Proliferaion (Protein Tyrosine Kinase Activation)''' | ||
* Activation of tyrosine kinase receptors is followed by signal transduction via intracellular signal cascades leading to alteration of transcription within the cell nucleus.<ref name="pmid11057895">{{cite journal |vauthors=Schlessinger J |title=Cell signaling by receptor tyrosine kinases |journal=Cell |volume=103 |issue=2 |pages=211–25 |date=October 2000 |pmid=11057895 |doi= |url=}}</ref> | * Activation of [[tyrosine kinase]] receptors is followed by [[signal transduction]] via [[intracellular]] signal cascades leading to alteration of [[transcription]] within the [[cell nucleus]].<ref name="pmid11057895">{{cite journal |vauthors=Schlessinger J |title=Cell signaling by receptor tyrosine kinases |journal=Cell |volume=103 |issue=2 |pages=211–25 |date=October 2000 |pmid=11057895 |doi= |url=}}</ref> | ||
* An important pathway that leads to cellular proliferation is the Ras-MAP Kinase pathway, where Ras is activated by binding of GTP. Ras-bound GTP in turn triggers a cascade of events that finally lead to activation of serine/threonine kinases. Consequently, there is an activation of MAP kinases, which phosphorylate important transcriptional regulators of cell cycle.<ref name="pmid8316835">{{cite journal |vauthors=Skolnik EY, Batzer A, Li N, Lee CH, Lowenstein E, Mohammadi M, Margolis B, Schlessinger J |title=The function of GRB2 in linking the insulin receptor to Ras signaling pathways |journal=Science |volume=260 |issue=5116 |pages=1953–5 |date=June 1993 |pmid=8316835 |doi= |url=}}</ref><ref name="pmid8479541">{{cite journal |vauthors=Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J |title=Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling |journal=Nature |volume=363 |issue=6424 |pages=85–8 |date=May 1993 |pmid=8479541 |doi=10.1038/363085a0 |url=}}</ref><ref name="pmid1465135">{{cite journal |vauthors=Rozakis-Adcock M, McGlade J, Mbamalu G, Pelicci G, Daly R, Li W, Batzer A, Thomas S, Brugge J, Pelicci PG, Schlessinger J, Pawson T |title=Association of the Shc and Grb2/Sem5 SH2-containing proteins is implicated in activation of the Ras pathway by tyrosine kinases |journal=Nature |volume=360 |issue=6405 |pages=689–92 |date=December 1992 |pmid=1465135 |doi=10.1038/360689a0 |url=}}</ref><ref name="pmid8562934">{{cite journal |vauthors=Carow CE, Levenstein M, Kaufmann SH, Chen J, Amin S, Rockwell P, Witte L, Borowitz MJ, Civin CI, Small D |title=Expression of the hematopoietic growth factor receptor FLT3 (STK-1/Flk2) in human leukemias |journal=Blood |volume=87 |issue=3 |pages=1089–96 |date=February 1996 |pmid=8562934 |doi= |url=}}</ref> | * An important pathway that leads to cellular proliferation is the [[Ras (protein)|Ras-MAP Kinase pathway]], where [[Ras gene|Ras]] is activated by binding of [[guanosine triphosphate]] ([[Guanosine triphosphate|GTP]]). [[Ras]]-bound [[Guanosine|GTP]] in turn triggers a cascade of events that finally lead to activation of [[serine]]/[[threonine]] [[kinases]]. Consequently, there is an activation of [[MAP kinases]], which [[phosphorylate]] important [[Transcriptional regulation|transcriptional]] regulators of [[cell cycle]].<ref name="pmid8316835">{{cite journal |vauthors=Skolnik EY, Batzer A, Li N, Lee CH, Lowenstein E, Mohammadi M, Margolis B, Schlessinger J |title=The function of GRB2 in linking the insulin receptor to Ras signaling pathways |journal=Science |volume=260 |issue=5116 |pages=1953–5 |date=June 1993 |pmid=8316835 |doi= |url=}}</ref><ref name="pmid8479541">{{cite journal |vauthors=Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J |title=Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling |journal=Nature |volume=363 |issue=6424 |pages=85–8 |date=May 1993 |pmid=8479541 |doi=10.1038/363085a0 |url=}}</ref><ref name="pmid1465135">{{cite journal |vauthors=Rozakis-Adcock M, McGlade J, Mbamalu G, Pelicci G, Daly R, Li W, Batzer A, Thomas S, Brugge J, Pelicci PG, Schlessinger J, Pawson T |title=Association of the Shc and Grb2/Sem5 SH2-containing proteins is implicated in activation of the Ras pathway by tyrosine kinases |journal=Nature |volume=360 |issue=6405 |pages=689–92 |date=December 1992 |pmid=1465135 |doi=10.1038/360689a0 |url=}}</ref><ref name="pmid8562934">{{cite journal |vauthors=Carow CE, Levenstein M, Kaufmann SH, Chen J, Amin S, Rockwell P, Witte L, Borowitz MJ, Civin CI, Small D |title=Expression of the hematopoietic growth factor receptor FLT3 (STK-1/Flk2) in human leukemias |journal=Blood |volume=87 |issue=3 |pages=1089–96 |date=February 1996 |pmid=8562934 |doi= |url=}}</ref> | ||
* As a consequence of these, there is autonomous increased proliferation of cells. | * As a consequence of these, there is autonomous increased proliferation of cells. | ||
'''Role of Altered Gene Expression and Differentiation Blockade''' | '''Role of Altered Gene Expression and Differentiation Blockade''' | ||
* Altered gene expression leads to autonomous | * Altered [[gene expression]] leads to autonomous [[Cell proliferation|cellular proliferation]] with defect in regulatory pathways involved in [[Cell proliferation|cellular proliferation]]. | ||
* Chromosomal translocations and point mutations | * [[Chromosomal translocation|Chromosomal translocations]] and [[point mutations]] play pivotal role in generating a differentiation blockade on [[myeloid cells]]. | ||
* This results in a disruption in transcription factors | * This results in a disruption in [[transcription factors]]. | ||
* Transcription factors affected by chromosomal rearrangement (translocations) include: | * [[Transcription factors]] affected by [[Chromosomal aberration|chromosomal rearrangement]] (translocations) include: | ||
** Core binding factor complex [t(8;21)-Runx1-MTG8 fusion; Inversion of chromosome 16-yielding the CBFβ-MYH11 fusion; t(3,21)-generating the RUNX1-EVI1 fusion]<ref name="pmid12773394">{{cite journal |vauthors=Follows GA, Tagoh H, Lefevre P, Hodge D, Morgan GJ, Bonifer C |title=Epigenetic consequences of AML1-ETO action at the human c-FMS locus |journal=EMBO J. |volume=22 |issue=11 |pages=2798–809 |date=June 2003 |pmid=12773394 |pmc=156747 |doi=10.1093/emboj/cdg250 |url=}}</ref><ref name="pmid12732181">{{cite journal |vauthors=Hiebert SW, Reed-Inderbitzin EF, Amann J, Irvin B, Durst K, Linggi B |title=The t(8;21) fusion protein contacts co-repressors and histone deacetylases to repress the transcription of the p14ARF tumor suppressor |journal=Blood Cells Mol. Dis. |volume=30 |issue=2 |pages=177–83 |date=2003 |pmid=12732181 |doi= |url=}}</ref><ref name="pmid11283671">{{cite journal |vauthors=Pabst T, Mueller BU, Harakawa N, Schoch C, Haferlach T, Behre G, Hiddemann W, Zhang DE, Tenen DG |title=AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia |journal=Nat. Med. |volume=7 |issue=4 |pages=444–51 |date=April 2001 |pmid=11283671 |doi=10.1038/86515 |url=}}</ref> | ** Core binding factor complex [t(8;21)-Runx1-MTG8 fusion; Inversion of chromosome 16-yielding the CBFβ-MYH11 fusion; t(3,21)-generating the RUNX1-EVI1 fusion]<ref name="pmid12773394">{{cite journal |vauthors=Follows GA, Tagoh H, Lefevre P, Hodge D, Morgan GJ, Bonifer C |title=Epigenetic consequences of AML1-ETO action at the human c-FMS locus |journal=EMBO J. |volume=22 |issue=11 |pages=2798–809 |date=June 2003 |pmid=12773394 |pmc=156747 |doi=10.1093/emboj/cdg250 |url=}}</ref><ref name="pmid12732181">{{cite journal |vauthors=Hiebert SW, Reed-Inderbitzin EF, Amann J, Irvin B, Durst K, Linggi B |title=The t(8;21) fusion protein contacts co-repressors and histone deacetylases to repress the transcription of the p14ARF tumor suppressor |journal=Blood Cells Mol. Dis. |volume=30 |issue=2 |pages=177–83 |date=2003 |pmid=12732181 |doi= |url=}}</ref><ref name="pmid11283671">{{cite journal |vauthors=Pabst T, Mueller BU, Harakawa N, Schoch C, Haferlach T, Behre G, Hiddemann W, Zhang DE, Tenen DG |title=AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia |journal=Nat. Med. |volume=7 |issue=4 |pages=444–51 |date=April 2001 |pmid=11283671 |doi=10.1038/86515 |url=}}</ref> | ||
** Retinoic acid receptor (RAR) [t(15;17), which generates the PML-RARα fusion]<ref name="pmid9486655">{{cite journal |vauthors=Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara FF, Zamir I, Seiser C, Grignani F, Lazar MA, Minucci S, Pelicci PG |title=Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia |journal=Nature |volume=391 |issue=6669 |pages=815–8 |date=February 1998 |pmid=9486655 |doi=10.1038/35901 |url=}}</ref><ref name="pmid9462740">{{cite journal |vauthors=He LZ, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A, Pandolfi PP |title=Distinct interactions of PML-RARalpha and PLZF-RARalpha with co-repressors determine differential responses to RA in APL |journal=Nat. Genet. |volume=18 |issue=2 |pages=126–35 |date=February 1998 |pmid=9462740 |doi=10.1038/ng0298-126 |url=}}</ref> | ** Retinoic acid receptor (RAR) [t(15;17), which generates the PML-RARα fusion]<ref name="pmid9486655">{{cite journal |vauthors=Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara FF, Zamir I, Seiser C, Grignani F, Lazar MA, Minucci S, Pelicci PG |title=Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia |journal=Nature |volume=391 |issue=6669 |pages=815–8 |date=February 1998 |pmid=9486655 |doi=10.1038/35901 |url=}}</ref><ref name="pmid9462740">{{cite journal |vauthors=He LZ, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A, Pandolfi PP |title=Distinct interactions of PML-RARalpha and PLZF-RARalpha with co-repressors determine differential responses to RA in APL |journal=Nat. Genet. |volume=18 |issue=2 |pages=126–35 |date=February 1998 |pmid=9462740 |doi=10.1038/ng0298-126 |url=}}</ref> | ||
Revision as of 16:37, 2 October 2018
| https://https://www.youtube.com/watch?v=itkRVTqfPsE%7C350}} |
|
Acute myeloid leukemia Microchapters |
|
Diagnosis |
|---|
|
Treatment |
|
Case Studies |
|
Acute myeloid leukemia pathophysiology On the Web |
|
American Roentgen Ray Society Images of Acute myeloid leukemia pathophysiology |
|
Risk calculators and risk factors for Acute myeloid leukemia pathophysiology |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]; Associate Editor(s)-in-Chief: Syed Hassan A. Kazmi BSc, MD [2] Raviteja Guddeti, M.B.B.S. [3] Carlos A Lopez, M.D. [4]
Overview
Acute myeloid leukemia arises from myeloblasts, which are hematologic white cells that are normally involved in hematopoiesis. Genetic translocations involved in the pathogenesis of acute myeloid leukemia include translocations between chromosome 8 and 21 t(8;21) and translocations between chromosome 15 and 17 t(15;17). Inversions in the chromosomal translocations in chromosome 16 inv(16) are also involved in the pathogenesis of acute myeloid leukemia.
Pathophysiology
- The malignant cell in acute myeloid leukemia is the myeloblast. In normal hematopoiesis, the myeloblast is an immature precursor of myeloid white blood cells; a normal myeloblast will gradually mature into a mature white blood cell. However, in acute myeloid leukemia a single myeloblast accumulates genetic changes which "freeze" the cell in its immature state and prevent differentiation.[1] Such a mutation alone does not cause leukemia; however, when such a "differentiation arrest" is combined with other mutations which disrupt genes controlling proliferation, the result is the uncontrolled growth of an immature clone of cells, leading to the clinical entity of acute myeloid leukemia.[2]
- Much of the diversity and heterogeneity of acute myeloid leukemia stems from the fact that leukemic transformation can occur at a number of different steps along the differentiation pathway.[3]
- Modern classification schemes for acute myeloid leukemia recognize that the characteristics and behavior of the leukemic cell (and the leukemia) may depend on the stage at which differentiation was halted.
Normal Hematopoeisis
- Hematopoeisis is tightly regulated under physiological conditions via a number of lineage-specific growth factors and lineage-specific growth signalling pathways.
- The differentiation of myeloid stem cells into mature myelocytes is controlled by lineage-specific transcription factors that regulate the expression of lineage-specific genes.
- Normal hematopoeisis is dependent upon specific growth factor receptors. The two key growth factor receptors involved are as following:
- Growth factor receptors with intrinsic tyrosine kinase activity (expressed on CD34+ hematopoietic progenitor cells, examples include, receptors for the platelet-derived growth factors (PDGFs) PDGFR A and B, the receptor for macrophage colony stimulating factor (M-CSF), FMS, and the receptors for KL (SCF) and FL, Kit and Flt3)[4]
- Growth factor receptors without intrinsic tyrosine kinase activity and depend upon intracellular kinases of the Src and Janus Kinase (JAK ) families (examples include, the JAK-STAT pathway)[5]
Role Altered Signal Transduction and Autonomous Proliferaion (Protein Tyrosine Kinase Activation)
- Activation of tyrosine kinase receptors is followed by signal transduction via intracellular signal cascades leading to alteration of transcription within the cell nucleus.[6]
- An important pathway that leads to cellular proliferation is the Ras-MAP Kinase pathway, where Ras is activated by binding of guanosine triphosphate (GTP). Ras-bound GTP in turn triggers a cascade of events that finally lead to activation of serine/threonine kinases. Consequently, there is an activation of MAP kinases, which phosphorylate important transcriptional regulators of cell cycle.[7][8][9][10]
- As a consequence of these, there is autonomous increased proliferation of cells.
Role of Altered Gene Expression and Differentiation Blockade
- Altered gene expression leads to autonomous cellular proliferation with defect in regulatory pathways involved in cellular proliferation.
- Chromosomal translocations and point mutations play pivotal role in generating a differentiation blockade on myeloid cells.
- This results in a disruption in transcription factors.
- Transcription factors affected by chromosomal rearrangement (translocations) include:
- Core binding factor complex [t(8;21)-Runx1-MTG8 fusion; Inversion of chromosome 16-yielding the CBFβ-MYH11 fusion; t(3,21)-generating the RUNX1-EVI1 fusion][11][12][13]
- Retinoic acid receptor (RAR) [t(15;17), which generates the PML-RARα fusion][14][15]
- MLL protein [Activator protein of Hox gene promoters, Hox gene promoters in turn promote self-renewal of immature myeloid cells][16]
- Hox proteins[17][18]
- Point mutations in myeloid transcription factors include:
Evasion of Apoptosis (Protein Tyrosine Kinase Activation)
- The increased expression of Bcl-2 pro-survival molecule plays a key role in evasion of programmed cell death in AML.[21]
- PI 3-kinase activate the AKT serine/threonine kinase, and this kinase in turn phosphorylates BAD and releases the BCL-2 anti-apoptotic molecule[22][23]
- The RUNX1-MTG8 fusion protein of AML represses the expression of p14ARF and promotes destabilization of p53 (a tumor supressor gene)[24][25][26]
Self-Renewal
- The myeloid cells in acute myeolid leukemia have an ability to self-renew without being committed to a specific cell lineage.[27]
- The self-renewing capacity of myeloid cells in AMLs is thought to be mediated via the following:
- Fusion of ALK tyrosine kinase with nucleophosmin protein (NPM)[28]
- Mutated FLT3-ITD[29][30]
- RUNX1-MTG8, PML-RARα , and PLZF-RARα fusions can all induce the expression of β-catenin and γ-catenin (plako-globin) proteins[31][32]
- The Wnt signalling pathway has also been shown to be involved in self-renewal of myeloid cells.[33]
Genetics
Specific cytogenetic abnormalities can be found in many patients with acute myeloid leukemia; the types of chromosomal abnormalities often have prognostic significance.[34] The chromosomal translocations encode abnormal fusion proteins, usually transcription factors whose altered properties may cause the "differentiation arrest."[35] For example, in acute promyelocytic leukemia, the t(15;17) translocation produces a PML-RARα fusion protein which binds to the retinoic acid receptor element in the promoters of several myeloid-specific genes and inhibits myeloid differentiation.[36]. Acute myeloid leukemia M2 subtype is characterized by a translocation of a part of chromosome 8 to chromosome 21, written as t(8;21). On both sides of the chromosome, now containing pieces from two chromosomes, the DNA codes for different proteins. These two proteins are now being created as one single large protein, with a different effect in the body as the two proteins originally coded by the two different chromosomes. The two different proteins that are fused together are: RUNX1 and ETO.
The clinical signs and symptoms of acute myeloid leukemia result from the fact that, as the leukemic clone of cells grows, it tends to displace or interfere with the development of normal blood cells in the bone marrow.[37] This leads to neutropenia, anemia, and thrombocytopenia. The symptoms of acute myeloid leukemia are in turn often due to the low numbers of these normal blood elements. In rare cases, patients can develop a chloroma, or solid tumor of leukemic cells outside the bone marrow, which can cause various symptoms depending on its location.
Microscopic Pathology
Description of pictures according the classification of Acute myeloid leukemia system.
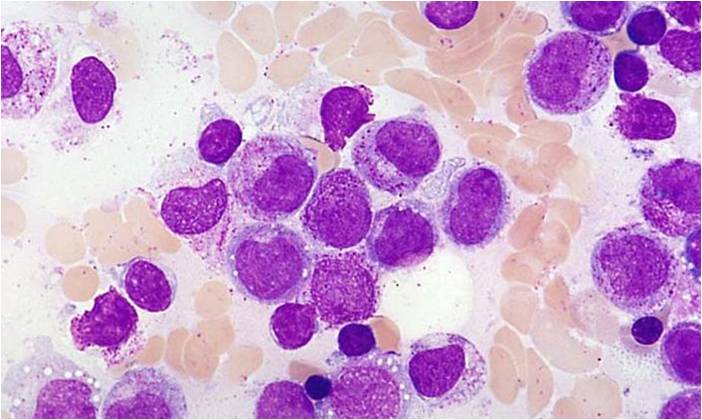
Acute myeloid leukemia M0 classification
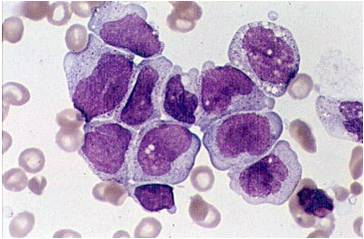
- Acute myeloid leukemia M0 with lack of obvious myeloid differentiation by routine histologic examination and presence of myeloperoxidase in <3% of blasts. Morphologically, blasts are small to large with no granules or Auer rods.
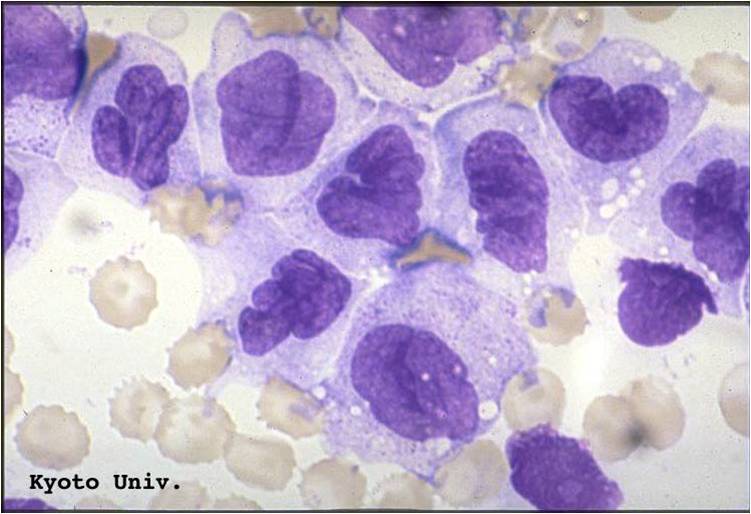
- Acute myeloid leukemia-M1 with presence of more than 90% myeloblasts in blood.
- Acute myeloid leukemia-M1 peroxidase.
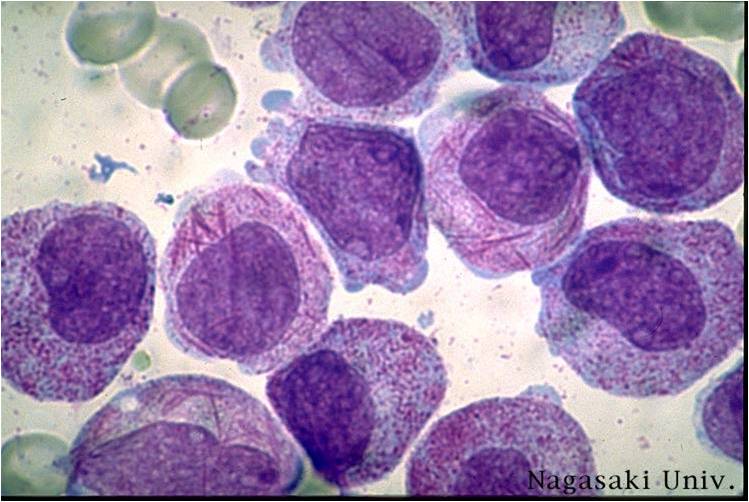
Acute myeloid leukemia M2 classification
- Acute myeloid leukemia-M2: Presence of granules can be noted.
- Acute myeloid leukemia-M2: Large myeloblasts with prominent nucleoli.
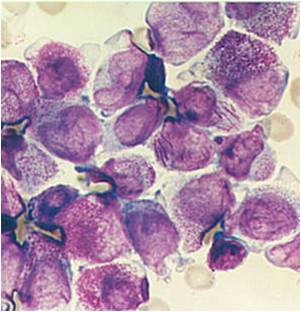
Acute myeloid leukemia M3 classification
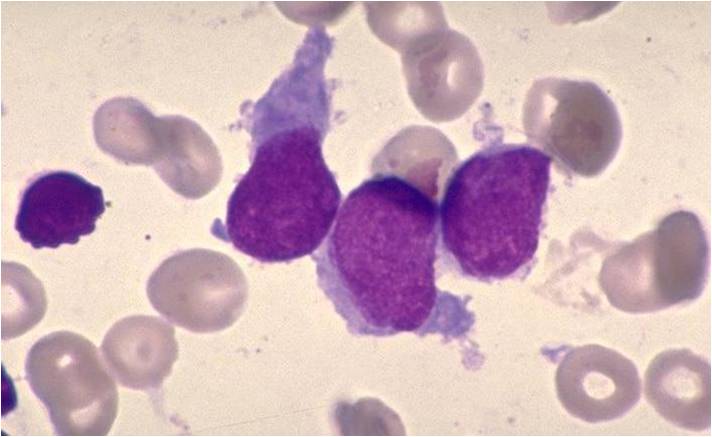
- Acute myeloid leukemia-M3: Also called promyelocytic leukemia. Hypergranular morphology with most cells containing abundant large granules.
- Acute myeloid leukemia-M3: Ruptured cells are releasing their granules free onto the slide. Presence of Auer rods can be noticed.
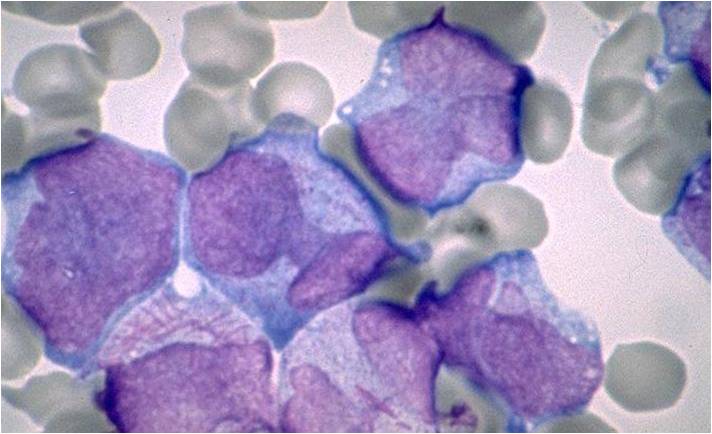
Acute myeloid leukemia M5a and M5b classification
- Acute myeloid leukemia-M5a: >80% monoblasts in the marrow.
- Acute myeloid leukemia-M5a: large monoblasts with fine nuclear chromatin and prominent nucleoli. Note the absence of Auer rods.
- Acute myeloid leukemia-M5b: <80% monoblasts in the marrow.
Acute myeloid leukemia M7 classification
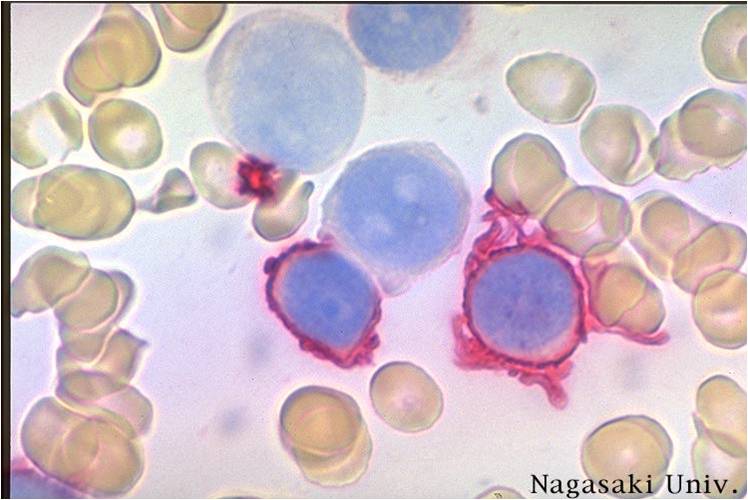
- Acute myeloid leukemia-M7: Irregular cytoplasmic border is often noted in some of the megakaryoblasts and occasionally projections resembling budding atypical platelets are present.
- Megakaryoblasts are usually medium-sized to large cells with a high nuclear-cytoplasmic ratio
- Nuclear chromatin is dense and homogeneous
- Variable basophilic cytoplasm which may be vacuolated
- An irregular cytoplasmic border is often noted in some of the megakaryoblasts and occasionally projections resembling budding atypical platelets are present
- Megakaryoblasts lack myeloperoxidase (MPO) activity and stain negatively with Sudan black B
- Clumps or granules in the cytoplasm
- PAS staining varies from negative to focal or granular positivity, to strongly positive staining
- More precise identification is by immunophenotyping or with electron microscopy (EM)
- Immunophenotyping using MoAb to megakaryocytic restricted antigen (CD41 and CD61) may be diagnostic
(Images shown below are courtesy of Melih Aktan MD., Istanbul Medical Faculty - Turkey, and Kyoto University - Japan)
-
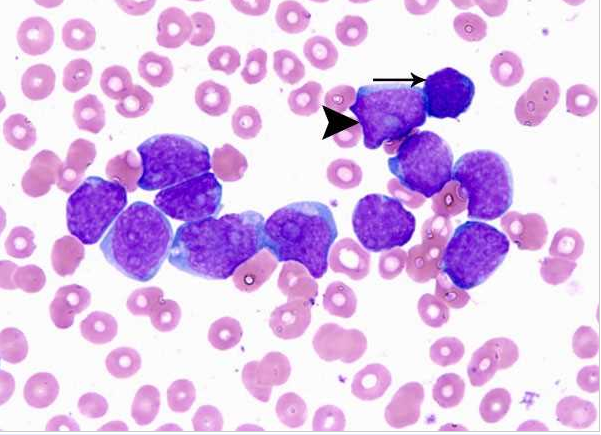
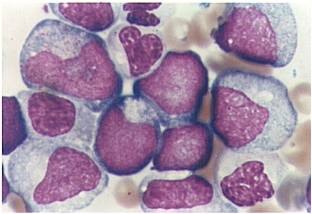
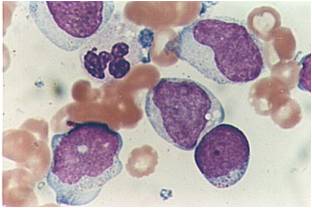
Acute myeloid leukemia-M0 - lack of obvious myeloid differentiation by routine histologic examination and presence of myeloperoxidase in <3% of blasts. Morphologically, blasts are small to large with no granules or Auer rods.
-
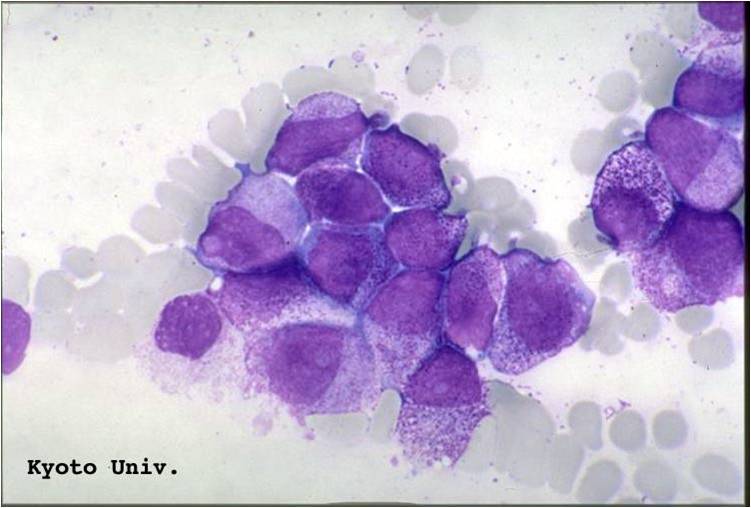
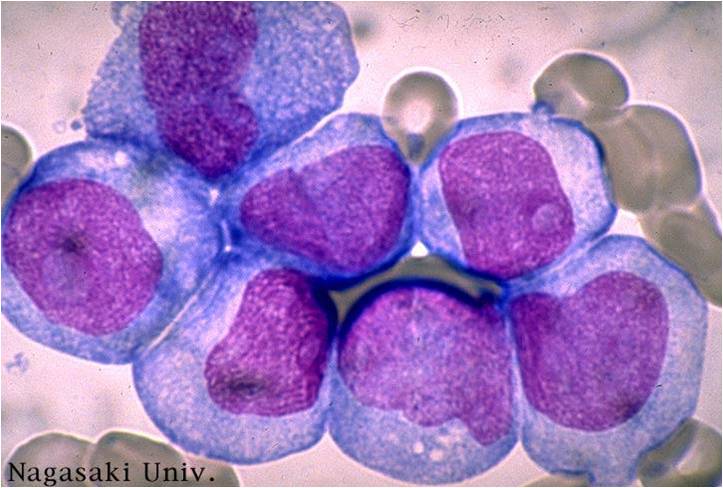
Acute myeloid leukemia-M1 - presence of more than 90% myeloblasts in blood.
-
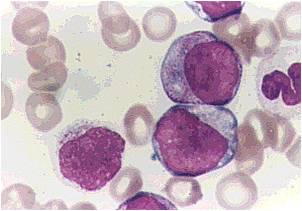
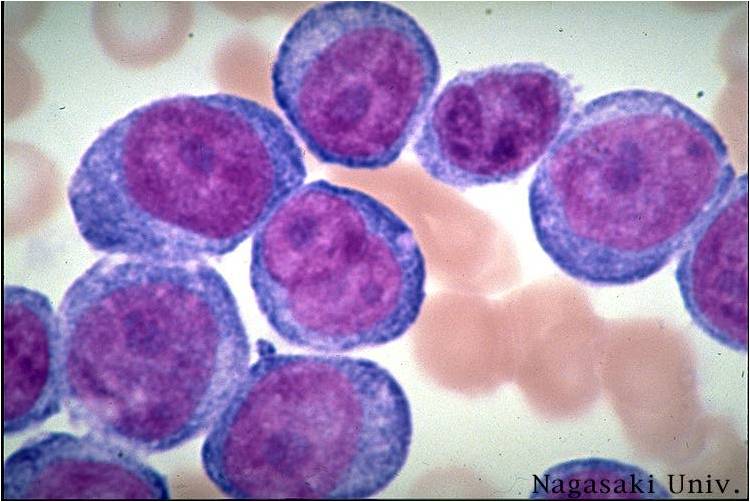
Acute myeloid leukemia-M1 peroxidase.
-
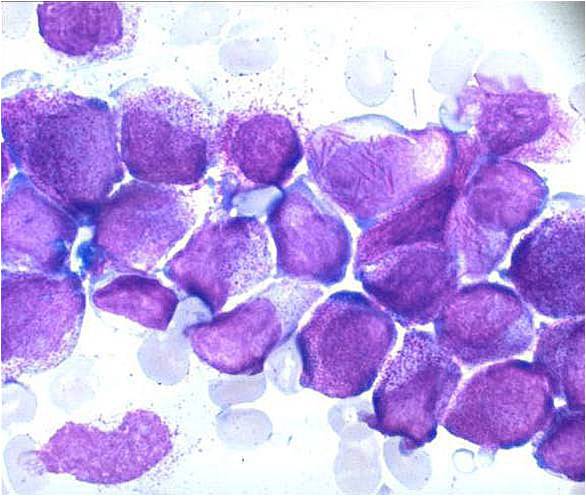
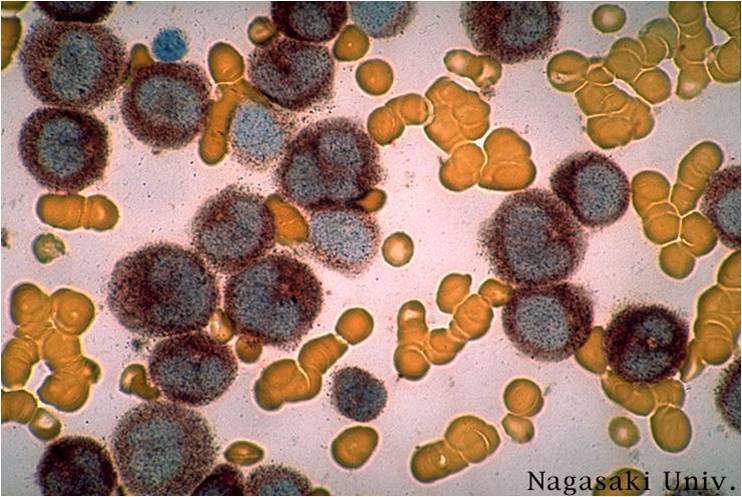
Acute myeloid leukemia-M1 - more cytoplasm than M0, but still no granules.
-
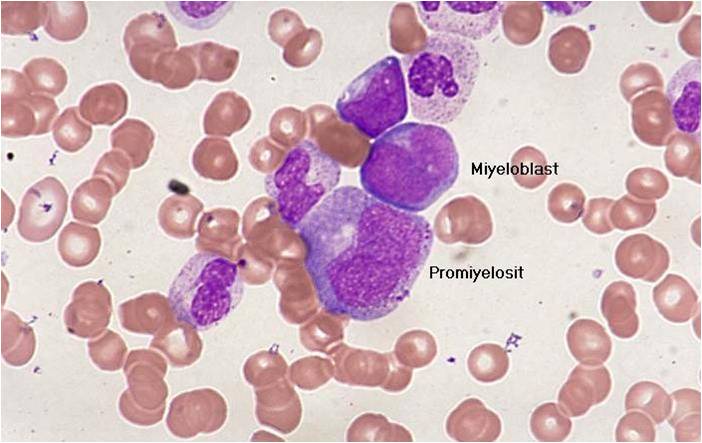
Acute myeloid leukemia-M1.
-
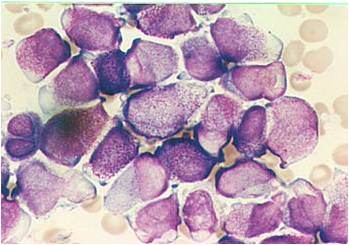
Acute myeloid leukemia-M1.
-
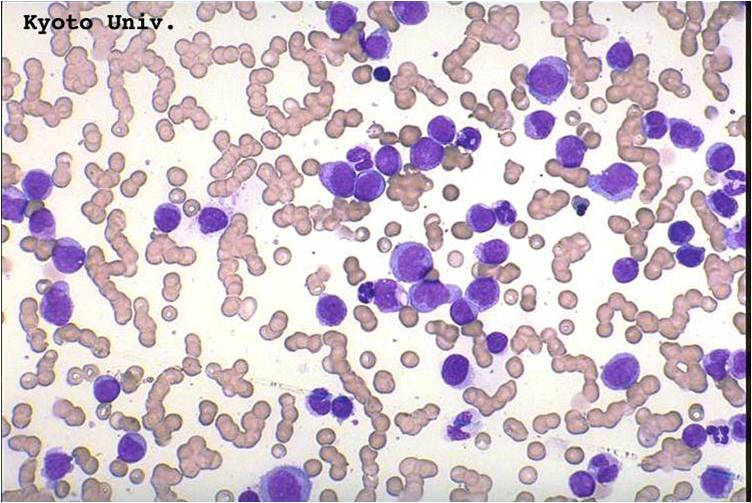
AML-M2 - presence of granules can be noted.
-
AML-M2 - large myeloblasts with prominent nucleoli.
-
AML-M2 - maturing myeloid elements including band and segmented neutrophils in the background.
-
AML-M2 - presence of a few maturing myeloid elements.
-
AML-M3 - also called promyelocytic leukemia. Hypergranular morphology with most cells containing abundant large granules.
-
AML-M3 - ruptured cells are releasing their granules free onto the slide. Presence of Auer rods can be noticed.
-
AML-M3 - presence of abundant fine granules.
-
AML-M3 - polarity of cytoplasmic granulation. In many cells the granules tend to polarize toward one portion of the cytoplasm and the nucleus on the opposite side.
-
AML-M3 Auer bodies.
-
AML-M3 Auer bodies.
-
AML-M3 variation.
-
AML-M4 - large monoblasts with abundant cytoplasm and moderately to intensely basophilic. The monoblasts will have lacy nuclear chromatin and one or more predominant nucleoli.
-
AML-M5a - >80% monoblasts in the marrow.
-
AML-M5a - large monoblasts with fine nuclear chromatin and prominent nucleoli. Note the absence of Auer rods.
-
AML-M5a (alpha naphtyle acetat).
-
AML-M5b - <80% monoblasts in the marrow.
-
AML-M5b - moderate amounts of cytoplasm and large nuclei with fine chromatin but without prominent nucleoli. Note presence of nuclear folds and creases, which are a characteristic feature of promonocytes.
-
AML-M7 - irregular cytoplasmic border is often noted in some of the megakaryoblasts and occasionally projections resembling budding atypical platelets are present.
-
AML-M7 - megakaryoblasts are usually medium sized to large cells with a high nuclear-cytoplasmic ratio. Nuclear chromatin is dense and homogeneous.
References
- ↑ Fialkow PJ: Clonal origin of human tumors. Biochim Biophys Acta 1976;458:283–321. PMID 1067873
- ↑ Fialkow PJ, Janssen JW, Bartram CR: Clonal remissions in acute nonlymphocytic leukemia: Evidence for a multistep pathogenesis of the malignancy. Blood 1991;77:1415–1517. PMID 2009365
- ↑ Bonnet D, Dick JE: Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997;3:730–737. PMID 9212098
- ↑ Müller-Tidow C, Schwäble J, Steffen B, Tidow N, Brandt B, Becker K, Schulze-Bahr E, Halfter H, Vogt U, Metzger R, Schneider PM, Büchner T, Brandts C, Berdel WE, Serve H (February 2004). "High-throughput analysis of genome-wide receptor tyrosine kinase expression in human cancers identifies potential novel drug targets". Clin. Cancer Res. 10 (4): 1241–9. PMID 14977821.
- ↑ Ihle JN (April 2001). "The Stat family in cytokine signaling". Curr. Opin. Cell Biol. 13 (2): 211–7. PMID 11248555.
- ↑ Schlessinger J (October 2000). "Cell signaling by receptor tyrosine kinases". Cell. 103 (2): 211–25. PMID 11057895.
- ↑ Skolnik EY, Batzer A, Li N, Lee CH, Lowenstein E, Mohammadi M, Margolis B, Schlessinger J (June 1993). "The function of GRB2 in linking the insulin receptor to Ras signaling pathways". Science. 260 (5116): 1953–5. PMID 8316835.
- ↑ Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J (May 1993). "Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling". Nature. 363 (6424): 85–8. doi:10.1038/363085a0. PMID 8479541.
- ↑ Rozakis-Adcock M, McGlade J, Mbamalu G, Pelicci G, Daly R, Li W, Batzer A, Thomas S, Brugge J, Pelicci PG, Schlessinger J, Pawson T (December 1992). "Association of the Shc and Grb2/Sem5 SH2-containing proteins is implicated in activation of the Ras pathway by tyrosine kinases". Nature. 360 (6405): 689–92. doi:10.1038/360689a0. PMID 1465135.
- ↑ Carow CE, Levenstein M, Kaufmann SH, Chen J, Amin S, Rockwell P, Witte L, Borowitz MJ, Civin CI, Small D (February 1996). "Expression of the hematopoietic growth factor receptor FLT3 (STK-1/Flk2) in human leukemias". Blood. 87 (3): 1089–96. PMID 8562934.
- ↑ Follows GA, Tagoh H, Lefevre P, Hodge D, Morgan GJ, Bonifer C (June 2003). "Epigenetic consequences of AML1-ETO action at the human c-FMS locus". EMBO J. 22 (11): 2798–809. doi:10.1093/emboj/cdg250. PMC 156747. PMID 12773394.
- ↑ Hiebert SW, Reed-Inderbitzin EF, Amann J, Irvin B, Durst K, Linggi B (2003). "The t(8;21) fusion protein contacts co-repressors and histone deacetylases to repress the transcription of the p14ARF tumor suppressor". Blood Cells Mol. Dis. 30 (2): 177–83. PMID 12732181.
- ↑ 13.0 13.1 Pabst T, Mueller BU, Harakawa N, Schoch C, Haferlach T, Behre G, Hiddemann W, Zhang DE, Tenen DG (April 2001). "AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia". Nat. Med. 7 (4): 444–51. doi:10.1038/86515. PMID 11283671.
- ↑ Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara FF, Zamir I, Seiser C, Grignani F, Lazar MA, Minucci S, Pelicci PG (February 1998). "Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia". Nature. 391 (6669): 815–8. doi:10.1038/35901. PMID 9486655.
- ↑ He LZ, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A, Pandolfi PP (February 1998). "Distinct interactions of PML-RARalpha and PLZF-RARalpha with co-repressors determine differential responses to RA in APL". Nat. Genet. 18 (2): 126–35. doi:10.1038/ng0298-126. PMID 9462740.
- ↑ Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL (November 2002). "MLL targets SET domain methyltransferase activity to Hox gene promoters". Mol. Cell. 10 (5): 1107–17. PMID 12453418.
- ↑ Thorsteinsdottir U, Sauvageau G, Humphries RK (December 1997). "Hox homeobox genes as regulators of normal and leukemic hematopoiesis". Hematol. Oncol. Clin. North Am. 11 (6): 1221–37. PMID 9443054.
- ↑ Nakamura T, Largaespada DA, Shaughnessy JD, Jenkins NA, Copeland NG (February 1996). "Cooperative activation of Hoxa and Pbx1-related genes in murine myeloid leukaemias". Nat. Genet. 12 (2): 149–53. doi:10.1038/ng0296-149. PMID 8563752.
- ↑ Tiesmeier J, Czwalinna A, Müller-Tidow C, Krauter J, Serve H, Heil G, Ganser A, Verbeek W (November 2003). "Evidence for allelic evolution of C/EBPalpha mutations in acute myeloid leukaemia". Br. J. Haematol. 123 (3): 413–9. PMID 14616999.
- ↑ Mueller BU, Pabst T, Osato M, Asou N, Johansen LM, Minden MD, Behre G, Hiddemann W, Ito Y, Tenen DG (August 2002). "Heterozygous PU.1 mutations are associated with acute myeloid leukemia". Blood. 100 (3): 998–1007. PMID 12130514.
- ↑ Lisovsky M, Estrov Z, Zhang X, Consoli U, Sanchez-Williams G, Snell V, Munker R, Goodacre A, Savchenko V, Andreeff M (November 1996). "Flt3 ligand stimulates proliferation and inhibits apoptosis of acute myeloid leukemia cells: regulation of Bcl-2 and Bax". Blood. 88 (10): 3987–97. PMID 8916965.
- ↑ Franke TF, Hornik CP, Segev L, Shostak GA, Sugimoto C (December 2003). "PI3K/Akt and apoptosis: size matters". Oncogene. 22 (56): 8983–98. doi:10.1038/sj.onc.1207115. PMID 14663477.
- ↑ Nosaka T, Kawashima T, Misawa K, Ikuta K, Mui AL, Kitamura T (September 1999). "STAT5 as a molecular regulator of proliferation, differentiation and apoptosis in hematopoietic cells". EMBO J. 18 (17): 4754–65. doi:10.1093/emboj/18.17.4754. PMC 1171548. PMID 10469654.
- ↑ Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW (October 1998). "Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling". Genes Dev. 12 (19): 3008–19. PMC 317198. PMID 9765203.
- ↑ Pomerantz J, Schreiber-Agus N, Liégeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee HW, Cordon-Cardo C, DePinho RA (March 1998). "The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53". Cell. 92 (6): 713–23. PMID 9529248.
- ↑ Zhang Y, Xiong Y, Yarbrough WG (March 1998). "ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways". Cell. 92 (6): 725–34. PMID 9529249.
- ↑ Buchholz F, Refaeli Y, Trumpp A, Bishop JM (August 2000). "Inducible chromosomal translocation of AML1 and ETO genes through Cre/loxP-mediated recombination in the mouse". EMBO Rep. 1 (2): 133–9. doi:10.1038/sj.embor.embor610. PMC 1084259. PMID 11265752.
- ↑ Redner RL, Rush EA, Faas S, Rudert WA, Corey SJ (February 1996). "The t(5;17) variant of acute promyelocytic leukemia expresses a nucleophosmin-retinoic acid receptor fusion". Blood. 87 (3): 882–6. PMID 8562957.
- ↑ Kiyoi H, Towatari M, Yokota S, Hamaguchi M, Ohno R, Saito H, Naoe T (September 1998). "Internal tandem duplication of the FLT3 gene is a novel modality of elongation mutation which causes constitutive activation of the product". Leukemia. 12 (9): 1333–7. PMID 9737679.
- ↑ Kelly LM, Kutok JL, Williams IR, Boulton CL, Amaral SM, Curley DP, Ley TJ, Gilliland DG (June 2002). "PML/RARalpha and FLT3-ITD induce an APL-like disease in a mouse model". Proc. Natl. Acad. Sci. U.S.A. 99 (12): 8283–8. doi:10.1073/pnas.122233699. PMC 123059. PMID 12060771.
- ↑ Muller-Tidow, C.; Steffen, B.; Cauvet, T.; Tickenbrock, L.; Ji, P.; Diederichs, S.; Sargin, B.; Kohler, G.; Stelljes, M.; Puccetti, E.; Ruthardt, M.; deVos, S.; Hiebert, S. W.; Koeffler, H. P.; Berdel, W. E.; Serve, H. (2004). "Translocation Products in Acute Myeloid Leukemia Activate the Wnt Signaling Pathway in Hematopoietic Cells". Molecular and Cellular Biology. 24 (7): 2890–2904. doi:10.1128/MCB.24.7.2890-2904.2004. ISSN 0270-7306.
- ↑ . doi:10.1101/gad.14.11.1319. Missing or empty
|title=(help) - ↑ Austin TW, Solar GP, Ziegler FC, Liem L, Matthews W (May 1997). "A role for the Wnt gene family in hematopoiesis: expansion of multilineage progenitor cells". Blood. 89 (10): 3624–35. PMID 9160667.
- ↑ Abeloff, Martin et al. (2004), pp. 2831–32.
- ↑ Greer, John P., et al. Wintrobe's Clinical Hematology, 11th ed. Philadelphia: Lippincott, Williams, and Wilkins, 2004. p. 2045–2062
- ↑ Melnick A, Licht JD. Deconstructing a disease: RARα its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood 1999;93:3167–3215. PMID 10233871
- ↑ Abeloff, Martin et al. (2004), p. 2828.