WBR0935
Jump to navigation
Jump to search
| Author | PageAuthor::William J Gibson (Reviewed by Serge Korjian) |
|---|---|
| Exam Type | ExamType::USMLE Step 1 |
| Main Category | MainCategory::Biochemistry |
| Sub Category | SubCategory::Hematology, SubCategory::General Principles |
| Prompt | [[Prompt::Lenalidomide is a highly effective chemotherapeutic agent against multiple myeloma. A scientist is investigating the anti-neoplatic mechanism of lenalidomide. She finds that addition of lenalidomide to multiple myeloma cells decreases the protein abundance of the plasma cell essential transcription factors IKZF1 and IKZF3. She finds that lenalidomide binds to the CRBN-CRL4 complex, which then induces a post-translational modification of IKZF1 and IKZF3. Which of the following enzymatic activities does CRBN-CRL4 most likely possess?]] |
| Answer A | AnswerA::Glycosyltransferase |
| Answer A Explanation | [[AnswerAExp::Glycosylation refers to the addition of a carbohydrate to a protein. The glycosylation of proteins does not target them for degradation. Glycosylation is often necessary for the proper function or folding of the modified protein. One example of a heavily glycosylated protein family is the heparin sulfate proteoglycans.]] |
| Answer B | AnswerB::Farnesyltransferase |
| Answer B Explanation | AnswerBExp::Farnesyl groups can be added to proteins for their proper localization to cell membranes. Farnesylation does not target proteins for degradation. |
| Answer C | AnswerC::Gamma-carboxylase |
| Answer C Explanation | AnswerCExp::Vitamin K is gamma-carboxylated for its normal function. |
| Answer D | AnswerD::Hydroxylase |
| Answer D Explanation | AnswerDExp::Hydroxylation refers to the addition of an –OH group to a compound. Hydroxylation is an important step in the synthesis of steroid compounds. Proline hydroxylation is accomplished with Vitamin C as a cofactor in the synthesis of collagen. |
| Answer E | AnswerE::Ubiquitin ligase |
| Answer E Explanation | AnswerEExp::Proteins can be targeted for degradation by the addition of ubiquitin molecules to lysine residues. |
| Right Answer | RightAnswer::E |
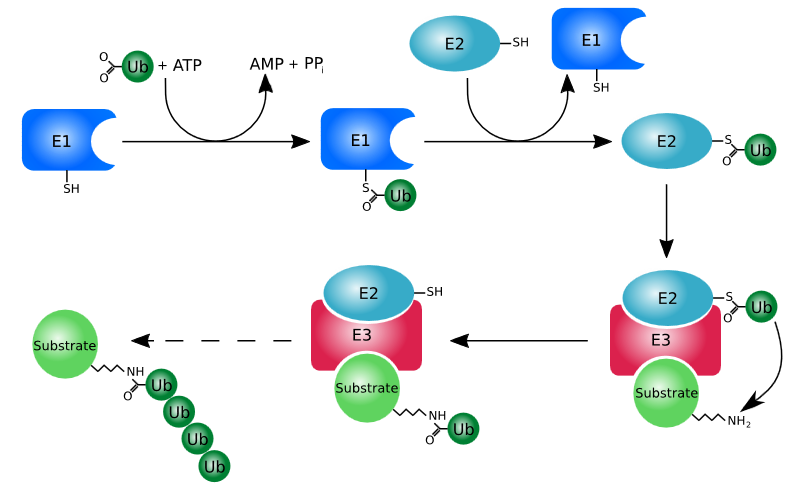
| Explanation | [[Explanation::Upon addition of lenalidomide, two transcription factors are post-translationally modified and their abundance is shown to decrease i.e. they are most likely degraded. The post-translational modification that induces degradation of proteins is polyubiquitination. Ubiquitination is a process whereby ubiquitin is attached to a substrate protein leading to several downstream effects. Polyubiquitination, or the addition of multiple ubiquitin molecules in a chain to a certain protein, targets the protein for degradation by the proteasome, an enzymatic complex that breaks down unneeded proteins via proteolysis. It is important to note that not every addition of a ubiquitin molecule to a protein tags the latter for degradation, this process only occurs with polyubiquitination. Ubiquitination requires three main enzymes ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3). The process is demonstrated below.
|
| Approved | Approved::Yes |
| Keyword | WBRKeyword::Ubiquitin, WBRKeyword::Post-translational modification, WBRKeyword::Protein, WBRKeyword::Translation, WBRKeyword::Molecular biology, WBRKeyword::Cell |
| Linked Question | Linked:: |
| Order in Linked Questions | LinkedOrder:: |