Hereditary epithelial ovarian tumors
Jump to navigation
Jump to search
|
Epithelial ovarian tumors Microchapters |
|
Differentiating Epithelial Ovarian Tumors from other Diseases |
|---|
|
Diagnosis |
|
Treatment |
|
Case Studies |
|
Hereditary epithelial ovarian tumors On the Web |
|
American Roentgen Ray Society Images of Hereditary epithelial ovarian tumors |
|
Risk calculators and risk factors for Hereditary epithelial ovarian tumors |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]Associate Editor(s)-in-Chief: Hannan Javed, M.D.[2]
Hereditary epithelial ovarian carcinoma: an overview of hereditary syndromes and the genes mutations
Hereditary breast and ovarian cancer (HBOC)
- Hereditary breast and ovarian cancer (HBOC) is an autosomal dominant disorder caused by mutations in BRCA1 and BRCA2 genes that are responsible for DNA repair in homologous recombination pathway.[1][2]
- Individuals with this disorder are at risk of developing breast (lifetime risk is 30-80%) and ovarian cancer (lifetime risk is 30-50%), along with other malignancies such as pancreatic, stomach, laryngeal, fallopian tube and prostate cancer.[1][2]
- The reason for increased susceptibility to ovarian and epithelial cancer is not fully understood but but may be explained by repression of the transcription of hormone-mediated signalling factors or production of reactive oxygen species during menstrual cycle mediating DNA damage.[2][3][4]
| Malignancies associated with BRCA mutations (Hereditary breast and ovarian cancer syndrome)[5] |
|---|
|
Lynch Syndrome
- Lynch syndrome (LS), also known as hereditary nonpolyposis colon cancer (HNPCC), is characterized by germline mutations in DNA mismatch repair genes MLH1, MSH2, MSH6, MLH3, and PMS2.[1][6][7]
- A simplified version of repair mechanism by mismatch repair genes products is described below:[7][8]
MutS homologs (MSHs) recognize the DNA mismatch → MutS homologs (MSHs) recruit MutL homologs (MLHs) → excision of mismatched DNA → DNA polymerase re-synthesizes DNA.
- Accounted for 10-15% of all ovarian cancers, this syndrome is caused by inherited mutation in one allele and then loss of second allele (secondary hit).[1][9]
- The most common malignancies in Lynch syndrome are colorectal carcinoma and gynecological cancers, endometrial carcinoma being the most common among gynecological malignancies followed by ovarian carcinoma.[9]
- Other malignancies that have been observed in lynch syndrome are gastric cancer, small bowel malignancies, hepatobiliary epithelial carcinoma, uroepithelial epithelial carcinoma and brain tumors.[9][10]
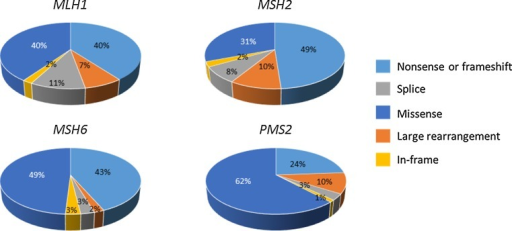
Li-Fraumeni Syndrome
- Li-Fraumeni Syndrome is an autosomal dominant disorder caused by germline mutation in TP53, the most mutated gene in human cancers. The most common of the mutations are missense mutations.[12][13]
- TP53 encodes for a transcription factor that responds to various cell signals and is a major regulator of the cell cycle. It is involved in variety of cellular functions such as cellular proliferation and cell cycle, apoptosis, and stability & integrity of the genome.[14][13]
- Mutations in TP53 resulting defective or decreased p53 are not only implicated in pathogenesis but also impact prognosis, causing worse survival rate among the individuals with the mutations.[14][15]
- These mutations are most commonly observed in epithelial ovarian cancer (47%), colorectal carcinoma (43%), head/neck cancer (42%), and esophageal cancer (41%). Breast cancer, sarcoma and brain, and adrenocortical carcinoma account for majority of the tumors encountered in Li-Fraumeni syndrome.[14][16]
Site-Specific Ovarian Cancer
- A term used to describe families in which there are several relatives with epithelial ovarian cancer but no other co-existent malignancies that are associated with other hereditary syndromes associated with epithelial ovarian cancer.[17]
- A hypothesis is that it is caused by gen/genes that are yet to be identified. Site-specific ovarian cancer appears to be transmitted in autosomal-dominant fashion in some families but some studies have suggested the risk to be as low as 5%.[17][18]
Cowden syndrome
- An autosomal-dominant syndrome , caused by mutations in PTEN gene, has been associated with a variety of neoplastic/non-neoplastic lesions and clinical manifestations throughout the body including:[17][19][20]
- Epithelial ovarian cancer
- Hamartomatous lesions of skin and organs
- Macrocephaly
- Breast cancer
- Thyroid cancer
- Endometrial cancer
RAD51
- RAD51 is a recombinase that binds with eight BRC repeats of BRCA2. This allows RAD51 to be recruited to double stranded DNA breaks, an essential step in homologous recombination double stranded DNA repair.[2][21][22][12]
- Some studies have suggested risk for developing ovarian cancer in RAD51 mutations is as high as six-fold. There is also an increased risk for developing breast cancer.[12][23][24]
PALB2
- Partner and localizer of BRCA2 (PALB2) physically connects BRCA1 and BRCA2 through N-terminal coiled-coil domain and the C terminus. This BRCA2 interacting protein plays an essential role in DNA repair.[2][25][26]
- The association of PALB2 with ovarian cancer has not be fully established but an increased risk for breast cancer, pancreatic cancer and ovarian cancer has been observed in some studies.[6][27][28]
CHEK2
- CHEK2 gene encodes for a protein called checkpoint kinase 2 (CHK2). It interacts with other regulators and tumor suppressors such as TP53 to play a role in tumor suppression through cell-cycle regulation and apoptosis.[29][30]
- There are conflicting results regarding association of CHEK2 with ovarian cancers. Some studies have suggested no association but the limitations were observed because of focus on only certain allelic mutations in CHEK2.[6][31]
Mre11 Complex
- Mre11 Complex is involved in DNA repair and comprises of meiotic recombination 11 (MRE11), RAD50 and Nijmegen breakage syndrome 1 (NBS1; also known as nibrin).[6][32]
- This complex plays an essential role in homologous recombination mediated DNA repair, non-homologous end-joining (NHEJ) and alternative non-homologous end-joining (A-NHEJ) pathways, all involved in double stranded DNA repair.[32][33]
- Some studies have suggested an increased susceptibility to ovarian and breast cancers in hereditary mutations in Mre11 complex.[6][34]
BARD1
- This gene encodes for a peptide that interacts with BRCA1 and forms a heterodiamer that plays a role in homologous recombination mediated repair of double stranded DNA breaks.[35][36]
- Mutations in BARD1 have been associated with breast and ovarian cancer.[6][37]
BRIP1
- BRCA1-interacting protein 1 (BRIP1) encodes for a helicase that interacts with BRCA1 in homologous recombination mediated repair of double stranded DNA breaks.[38][39]
- Mutation in BRIP1 gene association with familial ovarian cancer have been demonstrated in some studies. There also been proposed risk for breast cancer but it has yet to be established.[39][40]
References
- ↑ 1.0 1.1 1.2 1.3 Lynch HT, Casey MJ, Snyder CL, Bewtra C, Lynch JF, Butts M, Godwin AK (April 2009). "Hereditary ovarian carcinoma: heterogeneity, molecular genetics, pathology, and management". Mol Oncol. 3 (2): 97–137. doi:10.1016/j.molonc.2009.02.004. PMC 2778287. PMID 19383374.
- ↑ 2.0 2.1 2.2 2.3 2.4 Roy R, Chun J, Powell SN (December 2011). "BRCA1 and BRCA2: different roles in a common pathway of genome protection". Nat. Rev. Cancer. 12 (1): 68–78. doi:10.1038/nrc3181. PMC 4972490. PMID 22193408.
- ↑ Fan S, Wang J, Yuan R, Ma Y, Meng Q, Erdos MR, Pestell RG, Yuan F, Auborn KJ, Goldberg ID, Rosen EM (May 1999). "BRCA1 inhibition of estrogen receptor signaling in transfected cells". Science. 284 (5418): 1354–6. PMID 10334989.
- ↑ Hamada J, Nakata D, Nakae D, Kobayashi Y, Akai H, Konishi Y, Okada F, Shibata T, Hosokawa M, Moriuchi T (February 2001). "Increased oxidative DNA damage in mammary tumor cells by continuous epidermal growth factor stimulation". J. Natl. Cancer Inst. 93 (3): 214–9. PMID 11158190.
- ↑ Famorca-Tran J, Roux G (2015). "The Consequences of a BRCA Mutation in Women". J Adv Pract Oncol. 6 (3): 194–210. PMC 4625626. PMID 26557407.
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 Toss A, Tomasello C, Razzaboni E, Contu G, Grandi G, Cagnacci A, Schilder RJ, Cortesi L (2015). "Hereditary ovarian cancer: not only BRCA 1 and 2 genes". Biomed Res Int. 2015: 341723. doi:10.1155/2015/341723. PMC 4449870. PMID 26075229.
- ↑ 7.0 7.1 Martín-López JV, Fishel R (June 2013). "The mechanism of mismatch repair and the functional analysis of mismatch repair defects in Lynch syndrome". Fam. Cancer. 12 (2): 159–68. doi:10.1007/s10689-013-9635-x. PMC 4235668. PMID 23572416.
- ↑ Hsieh P, Yamane K (2008). "DNA mismatch repair: molecular mechanism, cancer, and ageing". Mech. Ageing Dev. 129 (7–8): 391–407. doi:10.1016/j.mad.2008.02.012. PMC 2574955. PMID 18406444.
- ↑ 9.0 9.1 9.2 Sehgal R, Sheahan K, O'Connell PR, Hanly AM, Martin ST, Winter DC (June 2014). "Lynch syndrome: an updated review". Genes (Basel). 5 (3): 497–507. doi:10.3390/genes5030497. PMC 4198913. PMID 24978665.
- ↑ Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Clendenning M, Sotamaa K, Prior T, Westman JA, Panescu J, Fix D, Lockman J, LaJeunesse J, Comeras I, de la Chapelle A (December 2008). "Feasibility of screening for Lynch syndrome among patients with colorectal cancer". J. Clin. Oncol. 26 (35): 5783–8. doi:10.1200/JCO.2008.17.5950. PMC 2645108. PMID 18809606.
- ↑ Peltomäki P (July 2016). "Update on Lynch syndrome genomics". Fam. Cancer. 15 (3): 385–93. doi:10.1007/s10689-016-9882-8. PMC 4901089. PMID 26873718.
- ↑ 12.0 12.1 12.2 Toss A, Tomasello C, Razzaboni E, Contu G, Grandi G, Cagnacci A, Schilder RJ, Cortesi L (2015). "Hereditary ovarian cancer: not only BRCA 1 and 2 genes". Biomed Res Int. 2015: 341723. doi:10.1155/2015/341723. PMC 4449870. PMID 26075229.
- ↑ 13.0 13.1 Miller M, Shirole N, Tian R, Pal D, Sordella R (2016). "The Evolution of TP53 Mutations: From Loss-of-Function to Separation-of-Function Mutants". J Cancer Biol Res. 4 (4). PMC 5298884. PMID 28191499.
- ↑ 14.0 14.1 14.2 Toss A, Tomasello C, Razzaboni E, Contu G, Grandi G, Cagnacci A, Schilder RJ, Cortesi L (2015). "Hereditary ovarian cancer: not only BRCA 1 and 2 genes". Biomed Res Int. 2015: 341723. doi:10.1155/2015/341723. PMC 4449870. PMID 26075229.
- ↑ Tabori U, Baskin B, Shago M, Alon N, Taylor MD, Ray PN, Bouffet E, Malkin D, Hawkins C (March 2010). "Universal poor survival in children with medulloblastoma harboring somatic TP53 mutations". J. Clin. Oncol. 28 (8): 1345–50. doi:10.1200/JCO.2009.23.5952. PMID 20142599.
- ↑ Levine AJ, Momand J, Finlay CA (June 1991). "The p53 tumour suppressor gene". Nature. 351 (6326): 453–6. doi:10.1038/351453a0. PMID 2046748.
- ↑ 17.0 17.1 17.2 Shulman LP, Dungan JS (2010). "Cancer genetics: risks and mechanisms of cancer in women with inherited susceptibility to epithelial ovarian cancer". Cancer Treat. Res. 156: 69–85. doi:10.1007/978-1-4419-6518-9_6. PMC 3086477. PMID 20811826.
- ↑ Stratton JF, Pharoah P, Smith SK, Easton D, Ponder BA (May 1998). "A systematic review and meta-analysis of family history and risk of ovarian cancer". Br J Obstet Gynaecol. 105 (5): 493–9. PMID 9637117.
- ↑ Lachlan KL, Lucassen AM, Bunyan D, Temple IK (September 2007). "Cowden syndrome and Bannayan Riley Ruvalcaba syndrome represent one condition with variable expression and age-related penetrance: results of a clinical study of PTEN mutation carriers". J. Med. Genet. 44 (9): 579–85. doi:10.1136/jmg.2007.049981. PMC 2597943. PMID 17526800.
- ↑ Kalin A, Merideth MA, Regier DS, Blumenthal GM, Dennis PA, Stratton P (February 2013). "Management of reproductive health in Cowden syndrome complicated by endometrial polyps and breast cancer". Obstet Gynecol. 121 (2 Pt 2 Suppl 1): 461–4. doi:http://10 1097/AOG.0b013e318270444f Check
|doi=value (help). PMC 3799979. PMID 23344409. - ↑ Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC (October 2010). "The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA". Nat. Struct. Mol. Biol. 17 (10): 1263–5. doi:10.1038/nsmb.1905. PMC 4041013. PMID 20729858.
- ↑ Carreira A, Hilario J, Amitani I, Baskin RJ, Shivji MK, Venkitaraman AR, Kowalczykowski SC (March 2009). "The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51". Cell. 136 (6): 1032–43. doi:10.1016/j.cell.2009.02.019. PMC 2669112. PMID 19303847.
- ↑ Loveday C, Turnbull C, Ramsay E, Hughes D, Ruark E, Frankum JR, Bowden G, Kalmyrzaev B, Warren-Perry M, Snape K, Adlard JW, Barwell J, Berg J, Brady AF, Brewer C, Brice G, Chapman C, Cook J, Davidson R, Donaldson A, Douglas F, Greenhalgh L, Henderson A, Izatt L, Kumar A, Lalloo F, Miedzybrodzka Z, Morrison PJ, Paterson J, Porteous M, Rogers MT, Shanley S, Walker L, Eccles D, Evans DG, Renwick A, Seal S, Lord CJ, Ashworth A, Reis-Filho JS, Antoniou AC, Rahman N (August 2011). "Germline mutations in RAD51D confer susceptibility to ovarian cancer". Nat. Genet. 43 (9): 879–882. doi:10.1038/ng.893. PMC 4845885. PMID 21822267.
- ↑ Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, Niederacher D, Freund M, Lichtner P, Hartmann L, Schaal H, Ramser J, Honisch E, Kubisch C, Wichmann HE, Kast K, Deissler H, Engel C, Müller-Myhsok B, Neveling K, Kiechle M, Mathew CG, Schindler D, Schmutzler RK, Hanenberg H (May 2010). "Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene". Nat. Genet. 42 (5): 410–4. doi:10.1038/ng.569. PMID 20400964.
- ↑ Sy SM, Huen MS, Chen J (April 2009). "PALB2 is an integral component of the BRCA complex required for homologous recombination repair". Proc. Natl. Acad. Sci. U.S.A. 106 (17): 7155–60. doi:10.1073/pnas.0811159106. PMC 2678481. PMID 19369211.
- ↑ Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM (June 2006). "Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2". Mol. Cell. 22 (6): 719–29. doi:10.1016/j.molcel.2006.05.022. PMID 16793542.
- ↑ Casadei S, Norquist BM, Walsh T, Stray S, Mandell JB, Lee MK, Stamatoyannopoulos JA, King MC (March 2011). "Contribution of inherited mutations in the BRCA2-interacting protein PALB2 to familial breast cancer". Cancer Res. 71 (6): 2222–9. doi:10.1158/0008-5472.CAN-10-3958. PMC 3059378. PMID 21285249.
- ↑ Poumpouridou N, Kroupis C (December 2011). "Hereditary breast cancer: beyond BRCA genetic analysis; PALB2 emerges". Clin. Chem. Lab. Med. 50 (3): 423–34. doi:10.1515/cclm-2011-0840. PMID 22505525.
- ↑ "CHEK2 gene - Genetics Home Reference - NIH".
- ↑ Cai Z, Chehab NH, Pavletich NP (September 2009). "Structure and activation mechanism of the CHK2 DNA damage checkpoint kinase". Mol. Cell. 35 (6): 818–29. doi:10.1016/j.molcel.2009.09.007. PMID 19782031.
- ↑ Meijers-Heijboer H, van den Ouweland A, Klijn J, Wasielewski M, de Snoo A, Oldenburg R, Hollestelle A, Houben M, Crepin E, van Veghel-Plandsoen M, Elstrodt F, van Duijn C, Bartels C, Meijers C, Schutte M, McGuffog L, Thompson D, Easton D, Sodha N, Seal S, Barfoot R, Mangion J, Chang-Claude J, Eccles D, Eeles R, Evans DG, Houlston R, Murday V, Narod S, Peretz T, Peto J, Phelan C, Zhang HX, Szabo C, Devilee P, Goldgar D, Futreal PA, Nathanson KL, Weber B, Rahman N, Stratton MR (May 2002). "Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations". Nat. Genet. 31 (1): 55–9. doi:10.1038/ng879. PMID 11967536.
- ↑ 32.0 32.1 Stracker TH, Petrini JH (February 2011). "The MRE11 complex: starting from the ends". Nat. Rev. Mol. Cell Biol. 12 (2): 90–103. doi:10.1038/nrm3047. PMC 3905242. PMID 21252998.
- ↑ Lamarche BJ, Orazio NI, Weitzman MD (September 2010). "The MRN complex in double-strand break repair and telomere maintenance". FEBS Lett. 584 (17): 3682–95. doi:10.1016/j.febslet.2010.07.029. PMC 2946096. PMID 20655309.
- ↑ Heikkinen K, Karppinen SM, Soini Y, Mäkinen M, Winqvist R (December 2003). "Mutation screening of Mre11 complex genes: indication of RAD50 involvement in breast and ovarian cancer susceptibility". J. Med. Genet. 40 (12): e131. PMC 1735331. PMID 14684699.
- ↑ Westermark UK, Reyngold M, Olshen AB, Baer R, Jasin M, Moynahan ME (November 2003). "BARD1 participates with BRCA1 in homology-directed repair of chromosome breaks". Mol. Cell. Biol. 23 (21): 7926–36. PMID 14560035.
- ↑ Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE (October 2001). "Structure of a BRCA1-BARD1 heterodimeric RING-RING complex". Nat. Struct. Biol. 8 (10): 833–7. doi:10.1038/nsb1001-833. PMID 11573085.
- ↑ Klonowska K, Ratajska M, Czubak K, Kuzniacka A, Brozek I, Koczkowska M, Sniadecki M, Debniak J, Wydra D, Balut M, Stukan M, Zmienko A, Nowakowska B, Irminger-Finger I, Limon J, Kozlowski P (May 2015). "Analysis of large mutations in BARD1 in patients with breast and/or ovarian cancer: the Polish population as an example". Sci Rep. 5: 10424. doi:10.1038/srep10424. PMID 25994375.
- ↑ [++++https://ghr.nlm.nih.gov/gene/BRIP1 "BRIP1 gene - Genetics Home Reference - NIH"] Check
|url=value (help). - ↑ 39.0 39.1 Ring KL, Garcia C, Thomas MH, Modesitt SC (November 2017). "Current and future role of genetic screening in gynecologic malignancies". Am. J. Obstet. Gynecol. 217 (5): 512–521. doi:10.1016/j.ajog.2017.04.011. PMID 28411145.
- ↑ Weber-Lassalle N, Hauke J, Ramser J, Richters L, Groß E, Blümcke B, Gehrig A, Kahlert AK, Müller CR, Hackmann K, Honisch E, Weber-Lassalle K, Niederacher D, Borde J, Thiele H, Ernst C, Altmüller J, Neidhardt G, Nürnberg P, Klaschik K, Schroeder C, Platzer K, Volk AE, Wang-Gohrke S, Just W, Auber B, Kubisch C, Schmidt G, Horvath J, Wappenschmidt B, Engel C, Arnold N, Dworniczak B, Rhiem K, Meindl A, Schmutzler RK, Hahnen E (January 2018). "BRIP1 loss-of-function mutations confer high risk for familial ovarian cancer, but not familial breast cancer". Breast Cancer Res. 20 (1): 7. doi:10.1186/s13058-018-0935-9. PMC 5784717. PMID 29368626.