Salmonella: Difference between revisions
Joao Silva (talk | contribs) No edit summary |
Joao Silva (talk | contribs) |
||
| Line 27: | Line 27: | ||
|} | |} | ||
''Salmonella'' is a [[Gram-negative]] bacterium, facultatively anaerobic, non-spore-forming bacilli. It measures 2 to 3 by 0.4 to 0.6 μm. Salmonella reduce nitrates, produce acid on glucose fermentation and are non producers of cytochrome oxidase. | |||
<!-- | |||
17 All organisms except S. Gallinarum-Pullorum are motile as a result of peritrichous flagella, and most do not ferment lactose. However, approximately 1% of organisms are able to ferment lactose and therefore may not be detected if only MacConkey agar or other semiselective media are used to identify Salmonella based on colorimetric assay for fermentation of lactose. The differential metabolism of sugars can be used to distin- guish many Salmonella serotypes; serotype Typhi is the only organism that does not produce gas on sugar fermentation.17 | |||
Freshly passed stool is preferred for the isolation of Salmonella and should be plated directly onto agar plates. Low-selective media, such as MacConkey agar and deoxycholate agar, and intermediate-selective media, such as Salmonella-Shigella, xylose-lysine-deoxycholate, or Hektoen agar, are widely used to screen for both Salmonella and Shi- gella species. Selective chromogenic media, such as CHROMagar, are more specific than other selective media, reduce the need for confirma- tory testing and time to identification, and increasingly are used for the primary isolation and presumptive identification of Salmonella from clinical stool specimens.18 | |||
In addition to plating stool onto primary media, tetrathionate- and selenite-based enrichment broths are often used to facilitate the recov- ery of low numbers of organisms.18 Highly Salmonella-selective media, such as selenite with brilliant green, should be reserved for use in stool cultures of suspected carriers and under special circumstances, such as outbreaks. Bismuth sulfite agar, which contains an indicator of hydro- gen sulfite production and does not contain lactose, is preferred for the isolation of S. Typhi and can be used for the detection of the 1% of Salmonella strains (including most Salmonella serogroup C strains) that ferment lactose.19 After primary isolation, possible Salmonella iso- lates can be tested in commercial identification systems or inoculated into screening media such as triple-sugar–iron and lysine-iron agar. Direct detection of Salmonella from stool and food specimens using polymerase chain reaction (PCR) and rapid serologic diagnosis using anti-Salmonella 09 IgM antibodies are under development.20-22 | |||
Isolates with typical biochemical profiles for Salmonella should be serogrouped with commercially available polyvalent antisera or sent to a reference or public health laboratory for complete serogrouping. Salmonellae are serogrouped according to their polysaccharide O (somatic) antigens, Vi (capsular) antigens, and H (flagellar) antigens according to the Kauffman-White scheme.23 The Vi antigen is a heat- labile capsular homopolymer of N-acetylgalactosaminouronic acid that is used for the identification of S. Typhi strains and occasionally other Salmonella serotypes by slide agglutination.24 In S. Typhi and S. Paratyphi C, the polysaccharide Vi antigen can inhibit O antigen agglutination because it is so abundant, and boiling is required to inactive Vi antigen and to detect O antigen. Most antigenic variability occurs in the O antigen, which is composed of chains of oligosaccha- ride attached to a core oligosaccharide linked covalently to lipid A. | |||
--> | |||
In a clinical laboratory, it is usually isolated on [[MacConkey agar]], [[XLD agar]], [[XLT agar]], [[DCA agar]], or Önöz agar. Numbers of salmonella may be so low in clinical samples that stools are routinely also subjected to "enrichment culture", where a small volume of stool is incubated in a selective broth medium, such as [[selenite broth]] or [[Rappaport Vassiliadis soya peptone broth]], overnight. These media are inhibitory to the growth of the microbes normally found in the healthy human bowel, while allowing salmonellae to become enriched in numbers. Salmonellae may then be recovered by inoculating the enrichment broth on one or more of the primary selective media. On [[blood agar]], they form moist colonies about 2 to 3 mm in diameter. When the cells are grown for a prolonged time at a range of 25—28°C, some strains produce a biofilm, which is a matrix of [[complex carbohydrate]]s, [[cellulose]] and [[proteins]]. The ability to produce biofilm (a.k.a. "rugose", "lacy", or "wrinkled") can be an indicator of dimorphism, which is the ability of a single [[genome]] to produce multiple [[phenotypes]] in response to environmental conditions. Salmonellae usually do not ferment lactose; most of them produce hydrogen sulfide which, in media containing ferric ammonium citrate, reacts to form a black spot in the centre of the creamy colonies. | |||
==Structure== | ==Structure== | ||
Revision as of 02:38, 21 August 2014
|
Salmonellosis Microchapters |
|
Diagnosis |
|---|
|
Treatment |
|
Case Studies |
|
Salmonella On the Web |
|
American Roentgen Ray Society Images of Salmonella |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]; Associate Editor(s)-in-Chief: João André Alves Silva, M.D. [2] Jolanta Marszalek, M.D. [3]
Overview
Salmonella is a genus of rod-shaped Gram-negative enterobacteria that causes typhoid fever, paratyphoid fever, and foodborne illness.[1] Salmonella species are motile and produce hydrogen sulfide.[2]
Taxonomy
Cellular organism; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae[3]
Biology
 |
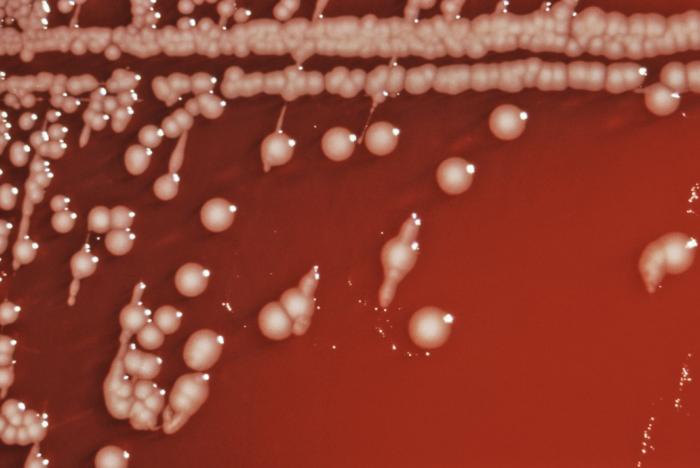 |
Salmonella is a Gram-negative bacterium, facultatively anaerobic, non-spore-forming bacilli. It measures 2 to 3 by 0.4 to 0.6 μm. Salmonella reduce nitrates, produce acid on glucose fermentation and are non producers of cytochrome oxidase.
In a clinical laboratory, it is usually isolated on MacConkey agar, XLD agar, XLT agar, DCA agar, or Önöz agar. Numbers of salmonella may be so low in clinical samples that stools are routinely also subjected to "enrichment culture", where a small volume of stool is incubated in a selective broth medium, such as selenite broth or Rappaport Vassiliadis soya peptone broth, overnight. These media are inhibitory to the growth of the microbes normally found in the healthy human bowel, while allowing salmonellae to become enriched in numbers. Salmonellae may then be recovered by inoculating the enrichment broth on one or more of the primary selective media. On blood agar, they form moist colonies about 2 to 3 mm in diameter. When the cells are grown for a prolonged time at a range of 25—28°C, some strains produce a biofilm, which is a matrix of complex carbohydrates, cellulose and proteins. The ability to produce biofilm (a.k.a. "rugose", "lacy", or "wrinkled") can be an indicator of dimorphism, which is the ability of a single genome to produce multiple phenotypes in response to environmental conditions. Salmonellae usually do not ferment lactose; most of them produce hydrogen sulfide which, in media containing ferric ammonium citrate, reacts to form a black spot in the centre of the creamy colonies.
Structure
Classification
Salmonella taxonomy is complicated.[5][6] As of December 7, 2005, there are two species within the genus: S. bongori (previously subspecies V) and S. enterica (formerly called S. choleraesuis), which is divided into six subspecies:
- I—enterica
- II—salamae
- IIIa—arizonae
- IIIb—diarizonae
- IV—houtenae
- V—obsolete (now designated S. bongori)
- VI—indica
There are also numerous (over 2500) serovars within both species, which are found in a disparate variety of environments and which are associated with many different diseases. The vast majority of human isolates (>99.5%) are subspecies S. enterica. For the sake of simplicity, the CDC recommends that Salmonella species be referred to only by their genus and serovar, e.g.,
- Salmonella Typhi
instead of the more technically correct designation,
- Salmonella enterica subspecies enterica serovar Typhi.
Salmonella isolates are most commonly classified according to serology (Kauffman-White classification).[5] The main division is first by the somatic O antigen, then by flagellar H antigens. H antigens are further divided into phase 1 and phase 2. Both phase 1 and phase 2 H antigens are required for the full identification of an isolate but in practise, routine labs will leave this to Reference Laboratories.
Note that, with the exception of typhoid and paratyphoid, salmonellosis is not a blood-related infection, as is commonly believed.
Examples:
- Salmonella Enteritidis (1,9,12:g,m) - where the O antigens present are 1, 9 and 12; the H antigens are g and m.
- Salmonella Typhi (9,12,Vi:d:−) - where the O antigens are 9, 12,; the H antigen is d: The Vi antigen is associated with the bacterial capsule, which acts as a Virulence factor, hence its name.
In a clinical laboratory, only a small number of serovars are looked for (the remainder being rare or not clinically significant). The Health Protection Agency recommend testing for the following antigens routinely:
- O antigens: 2 4 6.7 8 9 and 3.10
- phase 1 H antigens: a b d E G i r Vi
- phase 2 H antigens: 1,2 1,5 1,6 1,7
Isolates that cannot be identified using this panel are sent to the reference laboratory for identification.
Tropism
Natural Reservoir
Related Chapters
- 1984 Rajneeshee bioterror attack
- List of foodborne illness outbreaks
- Food Testing Strips
References
- ↑ Ryan KJ, Ray CG (editors) (2004). Sherris Medical Microbiology (4th ed. ed.). McGraw Hill. ISBN 0-8385-8529-9.
- ↑ Giannella RA (1996). "Salmonella". In Baron S et al (eds.). Baron's Medical Microbiology (4th ed. ed.). Univ of Texas Medical Branch. ISBN 0-9631172-1-1.
- ↑ "Salmonella (Taxonomy)".
- ↑ 4.0 4.1 "Public Health Image Library (PHIL), Centers for Disease Control and Prevention".
- ↑ 5.0 5.1 "The type species of the genus Salmonella Lignieres 1900 is Salmonella enterica (ex Kauffmann and Edwards 1952) Le Minor and Popoff 1987, with the type strain LT2T, and conservation of the epithet enterica in Salmonella enterica over all earlier epithets that may be applied to this species. Opinion 80". Int J Syst Evol Microbiol. 55 (Pt 1): 519–20. 2005. PMID 15653929.
- ↑ Tindall BJ; Grimont PAD, Garrity GM; Euzéby JP (2005). "Nomenclature and taxonomy of the genus Salmonella". Int J Syst Evol Microbiol. 55: 521&ndash, 524. PMID 15653930.