Poliovirus: Difference between revisions
Joao Silva (talk | contribs) No edit summary |
m (Bot: Removing from Primary care) |
||
| (18 intermediate revisions by 3 users not shown) | |||
| Line 5: | Line 5: | ||
==Overview== | ==Overview== | ||
Poliovirus is a small, nonenveloped, positive stranded RNA virus, that belongs to the family of Picornaviridae. It is a transient inhabitant of the GI tract, where it replicates, to further infect distant regions, however, [[poliovirus]] rarely causes symptoms. Three [[serotype]]s of [[poliovirus]], P1, P2 and P3, may be identified. Tissue tropism is dictated by | Poliovirus is a small, nonenveloped, positive stranded [[RNA]] virus, that belongs to the family of [[Picornaviridae]]. It is a transient inhabitant of the GI tract, where it replicates, to further infect distant regions, however, [[poliovirus]] rarely causes [[symptoms]]. Three [[serotype]]s of [[poliovirus]], P1, P2 and P3, may be identified. Tissue [[tropism]] is dictated by extracellular and intracellular factors. The [[cellular]] receptor [[CD155]] is the extracellular receptor for [[poliovirus]]. It may be identified in organs, such as the [[brain]], [[heart]], [[skeletal muscle]] and [[liver]]. Intracellular factors that influence viral [[replication]] include: polypyrimidine tract binding protein (PTB), which binds to IRES; the proteolytic processing of [[poliovirus]] [[proteins]]; and lack of an host factor for [[viral replication]]. Humans are the only [[natural reservoir]]s for [[poliovirus]]. | ||
==Taxonomy== | ==Taxonomy== | ||
[[Viruses]]; ssRNA viruses; ssRNA positive-strand viruses, no DNA stage; Picornavirales; [[Picornaviridae]]; [[Enterovirus]]; [[Poliovirus]]<ref name=NCBI>{{cite web | title = | [[Viruses]]; ssRNA viruses; ssRNA positive-strand viruses, no DNA stage; Picornavirales; [[Picornaviridae]]; [[Enterovirus]]; [[Poliovirus]]<ref name=NCBI>{{cite web | title = Poliovirus | url = http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=138950 }}</ref> | ||
==Biology== | ==Biology== | ||
| Line 15: | Line 15: | ||
|} | |} | ||
[[Poliovirus]] is a member of the genus [[enterovirus]], family [[Picornaviridae]]. Enteroviruses are small, nonenveloped, positive stranded RNA viruses. Other members of the family include: [[Rhinovirus]], [[Hepatovirus]], [[Cardiovirus]] and Apthovirus. Poliovirus is a transient inhabitant of the [[gastrointestinal tract]], stable at an acid pH.<ref name=CDC>{{cite web | title = Polyomavirus | url = http://www.cdc.gov/vaccines/pubs/pinkbook/downloads/polio.pdf }}</ref><ref name="pmid15885840">{{cite journal| author=Mueller S, Wimmer E, Cello J| title=Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event. | journal=Virus Res | year= 2005 | volume= 111 | issue= 2 | pages= 175-93 | pmid=15885840 | doi=10.1016/j.virusres.2005.04.008 | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=15885840 }} </ref> | [[Poliovirus]] is a member of the genus [[enterovirus]], family [[Picornaviridae]]. Enteroviruses are small, nonenveloped, positive stranded RNA viruses. Other members of the family include: [[Rhinovirus]], [[Hepatovirus]], [[Cardiovirus]] and Apthovirus. Poliovirus is a transient inhabitant of the [[gastrointestinal tract]], stable at an acid pH.<ref name=CDC>{{cite web | title = Polyomavirus | url = http://www.cdc.gov/vaccines/pubs/pinkbook/downloads/polio.pdf }}</ref><ref name="pmid15885840">{{cite journal| author=Mueller S, Wimmer E, Cello J| title=Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event. | journal=Virus Res | year= 2005 | volume= 111 | issue= 2 | pages= 175-93 | pmid=15885840 | doi=10.1016/j.virusres.2005.04.008 | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=15885840 }} </ref> Disease syndromes resulting from viral spread to other secondary organs are rare. Despite this fact, these syndromes lead to severe disease complications, seldom with fatal outcomes. | ||
There are three | There are three [[serotype]]s of poliovirus (P1, P2, and P3) that replicate efficiently in the [[gastrointestinal]] tract. There is minimal heterotypic [[immunity]] between the three [[serotype]]s. That is, immunity to one [[serotype]] does not produce significant immunity to any of the other [[serotype]]s. The poliovirus is rapidly inactivated by heat, formaldehyde, chlorine, and ultraviolet light.<ref name=CDC>{{cite web | title = Polyomavirus | url = http://www.cdc.gov/vaccines/pubs/pinkbook/downloads/polio.pdf }}</ref> The characteristics of [[poliovirus]] make it a good model for [[viral]] study, namely: high viral titers, stable [[capsid]] and ease of purification, along with a low bio-safety requirement.<ref name="pmid15885840">{{cite journal| author=Mueller S, Wimmer E, Cello J| title=Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event. | journal=Virus Res | year= 2005 | volume= 111 | issue= 2 | pages= 175-93 | pmid=15885840 | doi=10.1016/j.virusres.2005.04.008 | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=15885840 }} </ref> | ||
The characteristics of [[poliovirus]] make it a good model for [[viral]] study, | |||
==Structure== | ==Structure== | ||
The genome of poliovirus consists of a single positive-sense RNA molecule, of approximately 7740 nucleotides. At the 5' end of the RNA molecule are coded 88 nucleotides that interact to form a ''clover leaf structure'', which is involved in the replication process.<ref name="pmid15885840">{{cite journal| author=Mueller S, Wimmer E, Cello J| title=Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event. | journal=Virus Res | year= 2005 | volume= 111 | issue= 2 | pages= 175-93 | pmid=15885840 | doi=10.1016/j.virusres.2005.04.008 | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=15885840 }} </ref> At the 3' end of the genome is encoded a ''poly Adenine'' sequence, which varies about 60 adenylate residues in length.<ref name="pmid15885840">{{cite journal| author=Mueller S, Wimmer E, Cello J| title=Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event. | journal=Virus Res | year= 2005 | volume= 111 | issue= 2 | pages= 175-93 | pmid=15885840 | doi=10.1016/j.viruses.2005.04.008 | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=15885840 }} </ref> The translation of the genome is initiated by the attachment of | The [[genome]] of poliovirus consists of a single positive-sense [[RNA]] molecule, of approximately 7740 nucleotides. At the 5' end of the [[RNA]] molecule are coded 88 [[nucleotides]] that interact, to form a ''clover leaf structure'', which is involved in the [[replication]] process.<ref name="pmid15885840">{{cite journal| author=Mueller S, Wimmer E, Cello J| title=Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event. | journal=Virus Res | year= 2005 | volume= 111 | issue= 2 | pages= 175-93 | pmid=15885840 | doi=10.1016/j.virusres.2005.04.008 | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=15885840 }} </ref> At the 3' end of the [[genome]] is encoded a ''poly Adenine'' sequence, which varies about 60 adenylate residues in length.<ref name="pmid15885840">{{cite journal| author=Mueller S, Wimmer E, Cello J| title=Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event. | journal=Virus Res | year= 2005 | volume= 111 | issue= 2 | pages= 175-93 | pmid=15885840 | doi=10.1016/j.viruses.2005.04.008 | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=15885840 }} </ref> The [[translation]] of the [[genome]] is initiated by the attachment of host cell's ribosomes to the often called ''internal ribosomal entry site'' (IRES). This is a specific [[RNA]] segment in the 5' end region of the RNA (not translated), where the host cell's translational [[ribosomes]] first attach in order to initiate viral genome replication. The understanding of this mechanism has led to the establishment of a new mechanism of protein synthesis in [[eukaryotes]].<ref name="pmid15885840">{{cite journal| author=Mueller S, Wimmer E, Cello J| title=Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event. | journal=Virus Res | year= 2005 | volume= 111 | issue= 2 | pages= 175-93 | pmid=15885840 | doi=10.1016/j.virusres.2005.04.008 | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=15885840 }} </ref> | ||
==Tropism== | ==Tropism== | ||
=== | ===Extracellular Factors=== | ||
The cellular receptor for poliovirus was discovered after the transformation of mouse L-cells. These cells were altered with HeLa cell DNA, which led to susceptibility to poliovirus, of previously unsusceptible mice. The cDNA of the cellular receptor for poliovirus was later isolated and named CD155, or PVR. This receptor is a member of the immunoglobulin family, containing 3 Ig domains. CD155 is expressed in the following organs:<ref name="pmid15885840">{{cite journal| author=Mueller S, Wimmer E, Cello J| title=Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event. | journal=Virus Res | year= 2005 | volume= 111 | issue= 2 | pages= 175-93 | pmid=15885840 | doi=10.1016/j.virusres.2005.04.008 | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=15885840 }} </ref> | The cellular receptor for poliovirus was discovered after the transformation of mouse L-cells. These cells were altered with HeLa cell DNA, which led to susceptibility to poliovirus, of previously unsusceptible mice. The cDNA of the cellular receptor for poliovirus was later isolated and named CD155, or PVR. This receptor is a member of the immunoglobulin family, containing 3 Ig domains. CD155 is expressed in the following organs:<ref name="pmid15885840">{{cite journal| author=Mueller S, Wimmer E, Cello J| title=Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event. | journal=Virus Res | year= 2005 | volume= 111 | issue= 2 | pages= 175-93 | pmid=15885840 | doi=10.1016/j.virusres.2005.04.008 | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=15885840 }} </ref> | ||
* [[Brain]] | * [[Brain]] | ||
| Line 39: | Line 37: | ||
However, [[viral replication]] does not occur on all [[CD155]]-expressing cells. Possible explanations include:<ref name="pmid15885840">{{cite journal| author=Mueller S, Wimmer E, Cello J| title=Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event. | journal=Virus Res | year= 2005 | volume= 111 | issue= 2 | pages= 175-93 | pmid=15885840 | doi=10.1016/j.virusres.2005.04.008 | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=15885840 }} </ref> | However, [[viral replication]] does not occur on all [[CD155]]-expressing cells. Possible explanations include:<ref name="pmid15885840">{{cite journal| author=Mueller S, Wimmer E, Cello J| title=Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event. | journal=Virus Res | year= 2005 | volume= 111 | issue= 2 | pages= 175-93 | pmid=15885840 | doi=10.1016/j.virusres.2005.04.008 | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=15885840 }} </ref> | ||
* The detection method does not differentiate variants of the [[receptor]]. Some variants, despite detected, may not serve as | * The detection method does not differentiate variants of the [[receptor]]. Some variants, despite being detected, may not serve as receptors. | ||
* Excess [[secretion]] of non-receptor isoforms of [[CD155]] may compete for the virus, thereby inactivating | * Excess [[secretion]] of non-receptor isoforms of [[CD155]] may compete for the virus, thereby inactivating it. | ||
* Other ligands may compete with [[poliovirus]] for [[CD155]]. | * Other ligands may compete with [[poliovirus]] for [[CD155]]. | ||
* Physical barriers may block [[poliovirus]] access to [[CD155]]. | * Physical barriers may block [[poliovirus]] access to [[CD155]]. | ||
* [[Cytoplasm]] of certain cells may be inadequate for poliovirus [[viral replication|replication]]. | * [[Cytoplasm]] of certain cells may be inadequate for poliovirus [[viral replication|replication]]. | ||
CD155 positive tissues involved in the [[pathogenesis]] of the [[virus]], include:<ref name="pmid15885840">{{cite journal| author=Mueller S, Wimmer E, Cello J| title=Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event. | journal=Virus Res | year= 2005 | volume= 111 | issue= 2 | pages= 175-93 | pmid=15885840 | doi=10.1016/j.virusres.2005.04.008 | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=15885840 }} </ref> | [[CD155]] positive tissues involved in the [[pathogenesis]] of the [[virus]], include:<ref name="pmid15885840">{{cite journal| author=Mueller S, Wimmer E, Cello J| title=Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event. | journal=Virus Res | year= 2005 | volume= 111 | issue= 2 | pages= 175-93 | pmid=15885840 | doi=10.1016/j.virusres.2005.04.008 | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=15885840 }} </ref> | ||
* Germinal centers of [[tonsils]] | * Germinal centers of [[tonsils]] | ||
* Germinal centers of [[Peyer's patches]] | * Germinal centers of [[Peyer's patches]] | ||
| Line 52: | Line 50: | ||
* [[Colon]] [[enterocytes]] | * [[Colon]] [[enterocytes]] | ||
=== | ===Intracellular Factors=== | ||
Extracellular viral receptors are not the only determinants of tissue tropism. Genetic properties of the virus, which dictate the ability of poliovirus to replicate within a certain cell environment, are also important contributors to tissue [[tropism]]. [[Cellular]] host factors also interact with viral [[RNA]], influencing [[replication]]. An example is polypyrimidine tract binding protein (PTB), which binds to IRES. This binding initiates a cap-independent [[translation]] of the [[virus]], and has also been implicated in [[alternative splicing]] mechanisms.<ref name="pmid15885840">{{cite journal| author=Mueller S, Wimmer E, Cello J| title=Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event. | journal=Virus Res | year= 2005 | volume= 111 | issue= 2 | pages= 175-93 | pmid=15885840 | doi=10.1016/j.virusres.2005.04.008 | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=15885840 }} </ref> Other factors within the host cell may alter the poliovirus [[replication]] cycle: | |||
* Proteolytic processing of poliovirus proteins | * Proteolytic processing of poliovirus proteins | ||
* Lack of an host | * Lack of an host factors for viral replication | ||
* | * Cessation of protein synthesis within the host cell | ||
==Natural Reservoir== | ==Natural Reservoir== | ||
Only human cells, and certain primate species, show [[receptors]] for [[poliovirus]]. Therefore humans are considered the only [[natural reservoir]] for [[poliovirus]].<ref name="pmid12943679">{{cite journal| author=Baury B, Masson D, McDermott BM, Jarry A, Blottière HM, Blanchardie P et al.| title=Identification of secreted CD155 isoforms. | journal=Biochem Biophys Res Commun | year= 2003 | volume= 309 | issue= 1 | pages= 175-82 | pmid=12943679 | doi= | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=12943679 }} </ref><ref name="pmid10618373">{{cite journal| author=Belnap DM, McDermott BM, Filman DJ, Cheng N, Trus BL, Zuccola HJ et al.| title=Three-dimensional structure of poliovirus receptor bound to poliovirus. | journal=Proc Natl Acad Sci U S A | year= 2000 | volume= 97 | issue= 1 | pages= 73-8 | pmid=10618373 | doi= | pmc=PMC26618 | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=10618373 }} </ref> | Only human cells, and certain primate species, show [[receptors]] for [[poliovirus]]. Therefore humans are considered the only [[natural reservoir]] for [[poliovirus]].<ref name="pmid12943679">{{cite journal| author=Baury B, Masson D, McDermott BM, Jarry A, Blottière HM, Blanchardie P et al.| title=Identification of secreted CD155 isoforms. | journal=Biochem Biophys Res Commun | year= 2003 | volume= 309 | issue= 1 | pages= 175-82 | pmid=12943679 | doi= | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=12943679 }} </ref><ref name="pmid10618373">{{cite journal| author=Belnap DM, McDermott BM, Filman DJ, Cheng N, Trus BL, Zuccola HJ et al.| title=Three-dimensional structure of poliovirus receptor bound to poliovirus. | journal=Proc Natl Acad Sci U S A | year= 2000 | volume= 97 | issue= 1 | pages= 73-8 | pmid=10618373 | doi= | pmc=PMC26618 | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=10618373 }} </ref> There is no [[asymptomatic]] carrier state, except in the case of immunodeficient patients.<ref name=CDC>{{cite web | title = Polyomavirus | url = http://www.cdc.gov/vaccines/pubs/pinkbook/downloads/polio.pdf }}</ref> | ||
==References== | ==References== | ||
{{Reflist|2}} | {{Reflist|2}} | ||
{{WH}} | {{WH}} | ||
{{WS}} | {{WS}} | ||
[[Category:Disease]] | |||
Latest revision as of 23:46, 29 July 2020
|
Polio Microchapters |
|
Causes |
|---|
|
Diagnosis |
|
Treatment |
|
Case Studies |
|
Poliovirus On the Web |
|
American Roentgen Ray Society Images of Poliovirus |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]; Associate Editor(s)-in-Chief: João André Alves Silva, M.D. [2]
Overview
Poliovirus is a small, nonenveloped, positive stranded RNA virus, that belongs to the family of Picornaviridae. It is a transient inhabitant of the GI tract, where it replicates, to further infect distant regions, however, poliovirus rarely causes symptoms. Three serotypes of poliovirus, P1, P2 and P3, may be identified. Tissue tropism is dictated by extracellular and intracellular factors. The cellular receptor CD155 is the extracellular receptor for poliovirus. It may be identified in organs, such as the brain, heart, skeletal muscle and liver. Intracellular factors that influence viral replication include: polypyrimidine tract binding protein (PTB), which binds to IRES; the proteolytic processing of poliovirus proteins; and lack of an host factor for viral replication. Humans are the only natural reservoirs for poliovirus.
Taxonomy
Viruses; ssRNA viruses; ssRNA positive-strand viruses, no DNA stage; Picornavirales; Picornaviridae; Enterovirus; Poliovirus[1]
Biology
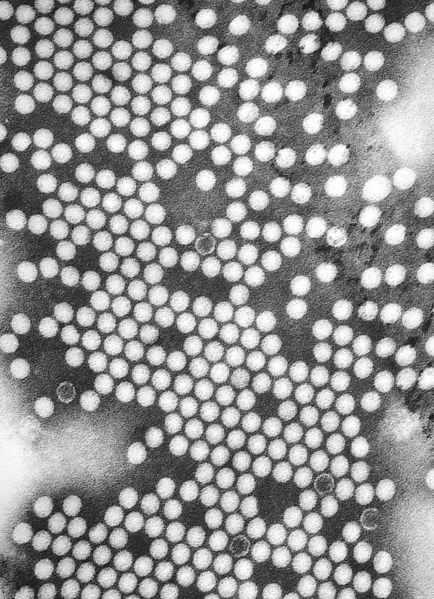 |
Poliovirus is a member of the genus enterovirus, family Picornaviridae. Enteroviruses are small, nonenveloped, positive stranded RNA viruses. Other members of the family include: Rhinovirus, Hepatovirus, Cardiovirus and Apthovirus. Poliovirus is a transient inhabitant of the gastrointestinal tract, stable at an acid pH.[3][4] Disease syndromes resulting from viral spread to other secondary organs are rare. Despite this fact, these syndromes lead to severe disease complications, seldom with fatal outcomes.
There are three serotypes of poliovirus (P1, P2, and P3) that replicate efficiently in the gastrointestinal tract. There is minimal heterotypic immunity between the three serotypes. That is, immunity to one serotype does not produce significant immunity to any of the other serotypes. The poliovirus is rapidly inactivated by heat, formaldehyde, chlorine, and ultraviolet light.[3] The characteristics of poliovirus make it a good model for viral study, namely: high viral titers, stable capsid and ease of purification, along with a low bio-safety requirement.[4]
Structure
The genome of poliovirus consists of a single positive-sense RNA molecule, of approximately 7740 nucleotides. At the 5' end of the RNA molecule are coded 88 nucleotides that interact, to form a clover leaf structure, which is involved in the replication process.[4] At the 3' end of the genome is encoded a poly Adenine sequence, which varies about 60 adenylate residues in length.[4] The translation of the genome is initiated by the attachment of host cell's ribosomes to the often called internal ribosomal entry site (IRES). This is a specific RNA segment in the 5' end region of the RNA (not translated), where the host cell's translational ribosomes first attach in order to initiate viral genome replication. The understanding of this mechanism has led to the establishment of a new mechanism of protein synthesis in eukaryotes.[4]
Tropism
Extracellular Factors
The cellular receptor for poliovirus was discovered after the transformation of mouse L-cells. These cells were altered with HeLa cell DNA, which led to susceptibility to poliovirus, of previously unsusceptible mice. The cDNA of the cellular receptor for poliovirus was later isolated and named CD155, or PVR. This receptor is a member of the immunoglobulin family, containing 3 Ig domains. CD155 is expressed in the following organs:[4]
However, viral replication does not occur on all CD155-expressing cells. Possible explanations include:[4]
- The detection method does not differentiate variants of the receptor. Some variants, despite being detected, may not serve as receptors.
- Excess secretion of non-receptor isoforms of CD155 may compete for the virus, thereby inactivating it.
- Other ligands may compete with poliovirus for CD155.
- Physical barriers may block poliovirus access to CD155.
- Cytoplasm of certain cells may be inadequate for poliovirus replication.
CD155 positive tissues involved in the pathogenesis of the virus, include:[4]
- Germinal centers of tonsils
- Germinal centers of Peyer's patches
- Epithelium of Peyer's patches
- Small intestine enterocytes
- Colon enterocytes
Intracellular Factors
Extracellular viral receptors are not the only determinants of tissue tropism. Genetic properties of the virus, which dictate the ability of poliovirus to replicate within a certain cell environment, are also important contributors to tissue tropism. Cellular host factors also interact with viral RNA, influencing replication. An example is polypyrimidine tract binding protein (PTB), which binds to IRES. This binding initiates a cap-independent translation of the virus, and has also been implicated in alternative splicing mechanisms.[4] Other factors within the host cell may alter the poliovirus replication cycle:
- Proteolytic processing of poliovirus proteins
- Lack of an host factors for viral replication
- Cessation of protein synthesis within the host cell
Natural Reservoir
Only human cells, and certain primate species, show receptors for poliovirus. Therefore humans are considered the only natural reservoir for poliovirus.[5][6] There is no asymptomatic carrier state, except in the case of immunodeficient patients.[3]
References
- ↑ "Poliovirus".
- ↑ "http://phil.cdc.gov/phil/details.asp". External link in
|title=(help) - ↑ 3.0 3.1 3.2 "Polyomavirus" (PDF).
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 4.8 Mueller S, Wimmer E, Cello J (2005). "Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event". Virus Res. 111 (2): 175–93. doi:10.1016/j.virusres.2005.04.008. PMID 15885840.
- ↑ Baury B, Masson D, McDermott BM, Jarry A, Blottière HM, Blanchardie P; et al. (2003). "Identification of secreted CD155 isoforms". Biochem Biophys Res Commun. 309 (1): 175–82. PMID 12943679.
- ↑ Belnap DM, McDermott BM, Filman DJ, Cheng N, Trus BL, Zuccola HJ; et al. (2000). "Three-dimensional structure of poliovirus receptor bound to poliovirus". Proc Natl Acad Sci U S A. 97 (1): 73–8. PMC 26618. PMID 10618373.