Multiple myeloma classification: Difference between revisions
m (Bot: Removing from Primary care) |
|||
| (16 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
{{Multiple myeloma}} | {{Multiple myeloma}} | ||
{{CMG}} | {{CMG}} {{AE}} {{HMHJ}}; {{HL}}; {{shyam}}; {{SN}} | ||
==Overview== | ==Overview== | ||
Multiple myeloma can be classified into several subtypes based on the extent of organ involvement ([[medullary]] or extramedullary) and the disease clinical presentation (active symptomatic or smoldering asymptomatic). | Multiple myeloma can be classified into several subtypes based on the extent of organ involvement ([[medullary]] or extramedullary) and the disease clinical presentation (active symptomatic or smoldering asymptomatic). The four general categories include solitary [[plasmacytoma]], [[monoclonal gammopathy of undetermined significance]], smoldering multiple myeloma, and active multiple myeloma. Within the category of active multiple myeloma, the three risk groups include standard risk, intermediate risk, and high risk, based on the [[cytogenetic]] profile. | ||
==Classification== | ==Classification== | ||
Multiple myeloma is still believed to be a single disease entity but it is a group of different cytogenetically distinct plasma cell malignancies. It can be classified on several basis.<ref name="pmid27291302">{{cite journal |vauthors=Rajkumar SV |title=Multiple myeloma: 2016 update on diagnosis, risk-stratification, and management |journal=Am. J. Hematol. |volume=91 |issue=7 |pages=719–34 |date=July 2016 |pmid=27291302 |pmc=5291298 |doi=10.1002/ajh.24402 |url=}}</ref> | Multiple myeloma is still believed to be a single disease entity but it is a group of different cytogenetically distinct plasma cell malignancies. It can be classified on several basis.<ref name="pmid27291302">{{cite journal |vauthors=Rajkumar SV |title=Multiple myeloma: 2016 update on diagnosis, risk-stratification, and management |journal=Am. J. Hematol. |volume=91 |issue=7 |pages=719–34 |date=July 2016 |pmid=27291302 |pmc=5291298 |doi=10.1002/ajh.24402 |url=}}</ref><ref name="pmid10357398">{{cite journal| author=Alexiou C, Kau RJ, Dietzfelbinger H, Kremer M, Spiess JC, Schratzenstaller B et al.| title=Extramedullary plasmacytoma: tumor occurrence and therapeutic concepts. | journal=Cancer | year= 1999 | volume= 85 | issue= 11 | pages= 2305-14 | pmid=10357398 | doi= | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=10357398 }}</ref> | ||
===Biological genetic classification=== | ===Biological genetic classification=== | ||
{| | {| | ||
! colspan="3" align="center" style="background:#4479BA; color: #FFFFFF;" + |Biological Genetic Classification | |||
|- | |- | ||
!Hyperdiploid MM | ! colspan="2" align="center" style="background:#DCDCDC;" + |Hyperdiploid MM | ||
| align="left" style="background:#F5F5F5;" + | | |||
* Characterized by the presence of trisomies | |||
* Characterized by presence of trisomies | |||
* Relatively indolent form of the disease | * Relatively indolent form of the disease | ||
* Relatively more favorable outcome | * Relatively more favorable outcome | ||
| Line 23: | Line 21: | ||
* Higher incidence of MM bone disease | * Higher incidence of MM bone disease | ||
|- | |- | ||
! rowspan="4" align="center" style="background:#DCDCDC;" + |Non-hyperdiploid MM | |||
| | ! align="center" style="background:#DCDCDC;" + |t(11;14)(q13;q32) | ||
| | | align="left" style="background:#F5F5F5;" + | | ||
* 15% of all MM cases | * 15% of all MM cases | ||
* Subsequent upregulation of cyclin D1 | * Subsequent upregulation of cyclin D1 | ||
| Line 34: | Line 32: | ||
* Disease can be heterogeneous; global effect on prognosis neutral | * Disease can be heterogeneous; global effect on prognosis neutral | ||
|- | |- | ||
| | ! align="center" style="background:#DCDCDC;" + |t(4;14)(p16;q32) | ||
| | | align="left" style="background:#F5F5F5;" + | | ||
* Short remission duration after high-dose chemotherapy with stem cell support | * Short remission duration after high-dose chemotherapy with stem cell support | ||
* New treatments to target the translocations (ie TKI-258) are under-development | * New treatments to target the translocations (ie TKI-258) are under-development | ||
| Line 42: | Line 40: | ||
* Observed more frequently in patients with smoldering multiple myeloma | * Observed more frequently in patients with smoldering multiple myeloma | ||
|- | |- | ||
| | ! align="center" style="background:#DCDCDC;" + |t(14;16)(q32;q23) | ||
| | | align="left" style="background:#F5F5F5;" + | | ||
* 5-7% of all MM patients | * 5-7% of all MM patients | ||
* Association with higher frequency of chromosome 13 deletion | * Association with higher frequency of chromosome 13 deletion | ||
| Line 49: | Line 47: | ||
* Aggressive clinical outcome | * Aggressive clinical outcome | ||
|- | |- | ||
| | ! align="center" style="background:#DCDCDC;" + |Chromosome 13 deletion | ||
| | | align="left" style="background:#F5F5F5;" + | | ||
* Significant as a marker now thought to be as a surrogate of its association with nh-MM | * Significant as a marker now thought to be as a surrogate of its association with nh-MM | ||
* Detected in 50% of patients; 85% of chromosome 13 deletions are monosomy, and 15% interstitial deletions | * Detected in 50% of patients; 85% of chromosome 13 deletions are monosomy, and 15% interstitial deletions | ||
| Line 56: | Line 54: | ||
* Almost 90% of cases with t(4;14)(p16;q32) will have chromosome 13 deletion | * Almost 90% of cases with t(4;14)(p16;q32) will have chromosome 13 deletion | ||
|} | |} | ||
===Molecular Cytogenetic Classification=== | ===Molecular Cytogenetic Classification=== | ||
{| | {| | ||
| | ! colspan="4" style="background:#4479BA; color: #FFFFFF;" align="center" + |Molecular Cytogenetic Classification | ||
! style="background:# | |- | ||
! colspan="3" align="center" style="background:#DCDCDC;" + |Hyperdiploid | |||
| align="left" style="background:#F5F5F5;" + |More favorable, IgG-k, older patients | |||
|- | |- | ||
| | ! rowspan="7" align="center" style="background:#DCDCDC;" + |Non-hyperdiploid | ||
Aggressive, IgA-l, younger individuals | |||
! rowspan="3" align="center" style="background:#DCDCDC;" + |Cyclin D translocation | |||
! align="center" style="background:#DCDCDC;" + |t(11;14)(q13;q32) | |||
| align="left" style="background:#F5F5F5;" + |Upregulation of CCND1; favorable prognosis; bone lesions. Two subtypes by GEP | |||
|- | |||
! align="center" style="background:#DCDCDC;" + |t(6;14q)(p21;32) | |||
| align="left" style="background:#F5F5F5;" + |Probably same as CCND1 | |||
|- | |||
! align="center" style="background:#DCDCDC;" + |t(12;14)(p13;q32) | |||
| align="left" style="background:#F5F5F5;" + |Rare | |||
|- | |||
! align="center" style="background:#DCDCDC;" + |MMSET translocation | |||
! align="center" style="background:#DCDCDC;" + |t(4;14)(p16;q32) | |||
| align="left" style="background:#F5F5F5;" + |Upregulation of MMSET; upregulation of FGFR3 in 75%, unfavorable prognosis with conventional therapy; bone lesion less frequent | |||
|- | |||
! rowspan="3" align="center" style="background:#DCDCDC;" + |MAF translocation | |||
! align="center" style="background:#DCDCDC;" + |t(14;16)(q32;q23) | |||
| align="left" style="background:#F5F5F5;" + |Confirmed as aggressive by at least two series | |||
|- | |||
! align="center" style="background:#DCDCDC;" + |t(14;20)(q32;q11) | |||
| align="left" style="background:#F5F5F5;" + |One series shows more aggressive disease | |||
|- | |||
! align="center" style="background:#DCDCDC;" + |t(8;14)(q24;q32) | |||
| align="left" style="background:#F5F5F5;" + |Unknown effect on outcome but | |||
|- | |||
! colspan="3" align="center" style="background:#DCDCDC;" + |Unclassified (other) | |||
| align="left" style="background:#F5F5F5;" + |Various subtypes and some with overlap | |||
|} | |} | ||
* A more detailed version is given below: | * A more detailed version is given below: | ||
{| | {| | ||
! colspan="3" style="background:#4479BA; color: #FFFFFF;" align="center" + |Primary Molecular Cytogenetic Classification of Multiple Myeloma | ! colspan="3" style="background:#4479BA; color: #FFFFFF;" align="center" + |Primary Molecular Cytogenetic Classification of Multiple Myeloma | ||
|- | |- | ||
! align="center" style="background:#4479BA; color: #FFFFFF;" + |Subtype | |||
! align="center" style="background:#4479BA; color: #FFFFFF;" + |Gene(s)/chromosomes affected | |||
! align="center" style="background:#4479BA; color: #FFFFFF;" + |Percentage of myeloma patients | |||
|- | |- | ||
| | ! align="center" style="background:#DCDCDC;" + |Trisomic MM | ||
|Recurrent trisomies involving odd-numbered | | align="left" style="background:#F5F5F5;" + | | ||
chromosomes with the exception of chromosomes | * Recurrent trisomies involving odd-numbered chromosomes with the exception of chromosomes 1, 13, and 21 | ||
| align="center" style="background:#F5F5F5;" + |42 | |||
1, 13, and 21 | |||
| | |||
|- | |- | ||
| | ! align="center" style="background:#DCDCDC;" + |IgH translocated MM | ||
| | | align="left" style="background:#F5F5F5;" + | | ||
* '''t(11;14) (q13;q32)''' ''CCND1'' (cyclin D1) | * '''t(11;14) (q13;q32)''' ''CCND1'' (cyclin D1) | ||
* '''t(4;14) (p16;q32)''' ''FGFR-3'' and MMSET | * '''t(4;14) (p16;q32)''' ''FGFR-3'' and MMSET | ||
| Line 101: | Line 112: | ||
* '''t(14;20) (q32;q11)''' ''MAFB'' | * '''t(14;20) (q32;q11)''' ''MAFB'' | ||
* '''Other IgH translocations''' ''CCND3'' (cyclin D3) in t(6;14) MM | * '''Other IgH translocations''' ''CCND3'' (cyclin D3) in t(6;14) MM | ||
| | | align="center" style="background:#F5F5F5;" + |30 | ||
|- | |- | ||
| | ! align="center" style="background:#DCDCDC;" + |Combined IgH translocated/trisomic MM | ||
| align="left" style="background:#F5F5F5;" + | | |||
|Presence of trisomies and any one of the recurrent IgH translocations in the same patient | * Presence of trisomies and any one of the recurrent IgH translocations in the same patient | ||
| | | align="center" style="background:#F5F5F5;" + |15 | ||
|- | |- | ||
| | ! align="center" style="background:#DCDCDC;" + |Isolated Monosomy 14 | ||
|Few cases may represent 14q32 translocations | | align="left" style="background:#F5F5F5;" + | | ||
involving unknown partner chromosomes | * Few cases may represent 14q32 translocations involving unknown partner chromosomes | ||
| | | align="center" style="background:#F5F5F5;" + |4.5 | ||
|- | |- | ||
| | ! align="center" style="background:#DCDCDC;" + |Other cytogenetic abnormalities in absence of IgH translocations or trisomy or monosomy 14 | ||
| align="left" style="background:#F5F5F5;" + | | |||
| align="center" style="background:#F5F5F5;" + |5.5 | |||
| | |||
| | |||
|- | |- | ||
| | ! align="center" style="background:#DCDCDC;" + |Normal | ||
| | | align="left" style="background:#F5F5F5;" + |NA | ||
| | | align="center" style="background:#F5F5F5;" + |3 | ||
|} | |} | ||
===Mayo Clinic Risk Stratification for Multiple Myeloma (mSMART)=== | ===Mayo Clinic Risk Stratification for Multiple Myeloma (mSMART)=== | ||
* Multiple myeloma can be classified according to risk status. The risk stratification for multiple myeloma consists of three groups, as follows:<ref name="pmid27291302">{{cite journal| author=Rajkumar SV| title=Multiple myeloma: 2016 update on diagnosis, risk-stratification, and management. | journal=Am J Hematol | year= 2016 | volume= 91 | issue= 7 | pages= 719-34 | pmid=27291302 | doi=10.1002/ajh.24402 | pmc=5291298 | url=https://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=27291302 }} </ref><ref name="pmid27002115">{{cite journal| author=Sonneveld P, Avet-Loiseau H, Lonial S, Usmani S, Siegel D, Anderson KC et al.| title=Treatment of multiple myeloma with high-risk cytogenetics: a consensus of the International Myeloma Working Group. | journal=Blood | year= 2016 | volume= 127 | issue= 24 | pages= 2955-62 | pmid=27002115 | doi=10.1182/blood-2016-01-631200 | pmc=4920674 | url=https://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=27002115 }} </ref> | * Multiple myeloma can be classified according to risk status. The risk stratification for multiple myeloma consists of three groups, as follows:<ref name="pmid27291302">{{cite journal| author=Rajkumar SV| title=Multiple myeloma: 2016 update on diagnosis, risk-stratification, and management. | journal=Am J Hematol | year= 2016 | volume= 91 | issue= 7 | pages= 719-34 | pmid=27291302 | doi=10.1002/ajh.24402 | pmc=5291298 | url=https://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=27291302 }} </ref><ref name="pmid27002115">{{cite journal| author=Sonneveld P, Avet-Loiseau H, Lonial S, Usmani S, Siegel D, Anderson KC et al.| title=Treatment of multiple myeloma with high-risk cytogenetics: a consensus of the International Myeloma Working Group. | journal=Blood | year= 2016 | volume= 127 | issue= 24 | pages= 2955-62 | pmid=27002115 | doi=10.1182/blood-2016-01-631200 | pmc=4920674 | url=https://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=27002115 }} </ref> | ||
{| | {| | ||
! colspan="3" align="center" style="background:#4479BA; color: #FFFFFF;" + |Mayo Clinic Risk Stratification for Multiple Myeloma (mSMART) | |||
|- | |- | ||
| style=" | ! align="center" style="background:#4479BA; color: #FFFFFF;" + |Risk group | ||
Standard risk | ! align="center" style="background:#4479BA; color: #FFFFFF;" + |Chromosomal Abnormalities | ||
! align="center" style="background:#4479BA; color: #FFFFFF;" + |Frequency | |||
|- | |||
! align="center" style="background:#DCDCDC;" + |Standard risk | |||
| style="padding: 5px 5px; background: #F5F5F5;" | | | style="padding: 5px 5px; background: #F5F5F5;" | | ||
*'''t(11;14) (CCND1;IgH)''' | *'''t(11;14) (CCND1;IgH)''' | ||
| Line 141: | Line 148: | ||
*'''Trisomies''' (chromosomes 3, 5, 7, 9, 11, 15, 19, 21) | *'''Trisomies''' (chromosomes 3, 5, 7, 9, 11, 15, 19, 21) | ||
*'''Hyperdiploidy''' | *'''Hyperdiploidy''' | ||
| style=" | | align="center" style="background:#F5F5F5;" + |75% | ||
75% | |||
|- | |- | ||
! align="center" style="background:#DCDCDC;" + |Intermediate Risk | |||
Intermediate Risk | |||
| style="padding: 5px 5px; background: #F5F5F5;" | | | style="padding: 5px 5px; background: #F5F5F5;" | | ||
*'''t(4;14) (FGFR3;IgH)''' | *'''t(4;14) (FGFR3;IgH)''' | ||
*'''Gain(1q21)''' | *'''Gain(1q21)''' | ||
| style=" | | align="center" style="background:#F5F5F5;" + |10% | ||
10% | |||
|- | |- | ||
! align="center" style="background:#DCDCDC;" + |High risk | |||
High risk | |||
| style="padding: 5px 5px; background: #F5F5F5;" | | | style="padding: 5px 5px; background: #F5F5F5;" | | ||
*'''t(14;16) (IgH;MAF)''' | *'''t(14;16) (IgH;MAF)''' | ||
| Line 159: | Line 162: | ||
*'''del(17p) (''p53'' deletion)''' | *'''del(17p) (''p53'' deletion)''' | ||
*'''Non-hyperdiploidy''' | *'''Non-hyperdiploidy''' | ||
| style=" | | align="center" style="background:#F5F5F5;" + |15% | ||
15% | |||
|- | |- | ||
|} | |} | ||
==References== | ==References== | ||
{{Reflist|2}} | {{Reflist|2}} | ||
[[Category:Medicine]] | |||
[[Category: | |||
[[Category:Hematology]] | [[Category:Hematology]] | ||
[[Category:Neurology]] | [[Category:Neurology]] | ||
[[Category:Neurosurgery]] | [[Category:Neurosurgery]] | ||
[[Category:Oncology]] | |||
[[Category:Up-To-Date]] | |||
[[Category:Surgery]] | |||
Latest revision as of 22:46, 29 July 2020
|
Multiple myeloma Microchapters |
|
Diagnosis |
|---|
|
Treatment |
|
Case Studies |
|
Multiple myeloma classification On the Web |
|
American Roentgen Ray Society Images of Multiple myeloma classification |
|
Risk calculators and risk factors for Multiple myeloma classification |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1] Associate Editor(s)-in-Chief: Hannan Javed, M.D.[2]; Haytham Allaham, M.D. [3]; Shyam Patel [4]; "sandbox:SN"
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [5]; Associate Editor(s)-in-Chief:
Overview
Pernicious anemia (also called Addison's anemia) is a type of red blood cell disorder caused by impaired vitamin B12 metabolism. Vitamin B12 is primarily absorbed by the small intestine, after being bound to intrinsic factor secreted by parietal cells of gastric mucosa. When this process is disrupted by conditions like atrophic gastritis, celiac disease, small bowel resection etc, B12 deficiency ensues.
Historical perspective
- Pernicious anemia was first discovered by Thomas Addison, hence it is also known as addison's anemia.
- Loss of life from large volume blood loss in the people fighting in the first world war inspired George Whipple to investigate blood forming components such as arsenic, iron pills etc, but found liver to be the most effective. He bled dogs until they had clinical anemia and fed them cooked liver which showed an improvement in symptoms and hematopoeisis. [1]
- In 1948, Smith, Rickles et al., isolated the anti-pernicious factor from liver extract and named it Vitamin B12. They showed that even small amounts of this factor can be used to treat and to prevent pernicious anemia. [2]
Pathophysiology
Vitamin B12 is an essential vitamin for humans and animals because we cannot synthesise it on our own. B12 is a cofactor in DNA synthesis and other important biochemical reactions. Vitamin B12 deficiency manifests as anemia because hematopoetic stem cells in the bone marrow which are rapidly dividing need B12 for division and DNA production. This process is impaired leading to ineffective hematopoeisis. Vitamin B12 is also necessary for production of myelin which is an important component in the covering sheath of nerves. Deficiency results in improper nerve conduction due to nerve destabilisation. [3]
Physiology
- Vitamin B12 is also called cobalamin because it contains cobalt at the core of its structure. Dietary sources of vitamin B12 include meat, fish and eggs.[4]
- When consumed through its dietary source, B12 is bound to protein till it enters the stomach.
- In the stomach, B12 is uncoupled from its carrier protein due to the presence of gastric acid, which is why vitamin B12 deficiency is so commonly seen among those on chronic antacid medication. [5]
- Once in the stomach, it is then bound to gastric R binder, a glycoprotein secreted by the salivary glands till it reaches the duodenum.[6]
- In the duodenum and jejunum, the pancreatic enzymes digest the gastric R binder and cobalamin is bound to intrinsic factor (IF).
- Intrinsic factor is secreted by the gastric parietal cells. Once bound to IF, vitamin B12 travels up to the ileum where IF is removed and B12 binds with carrier proteins called transcobalamins and this complex is taken up by the liver and bone marrow, among other tissues.
- Inside the cells, the transcobalamin-B12 complex is dissolved and cobalamin is reduced to methylcobalamin which serves as a cofactor and coenzyme in many important biochemical reactions[7].
The two major reactions involving B12 in the human body are:
- Vitamin B12 in the from of cyanocobalamin is required in the synthesis of methionine. Methionine is produced from homocysteine and is catalysed by the enzyme methionine synthase. This enzyme utilises cyanocobalamin as a cofactor. Deficiency of vitamin B12 causes a decreased production of methionine and buildup of homocysteine. Hyperhomocysteinemia is implicated as a risk factor in cardiovascular disease.[8]
- The Kreb's cycle utilises vitamin B12 in the reaction converting methylmalonyl-CoA to succinyl-CoA. Thus vitamin B12 deficiency causes a buildup of methylmalonic acid, the substrate for the enzyme methylmalonyl coenzyme A mutase. Methylmalonic acid levels are elevated in the urine of people affected with pernicious anemia and other forms of B12 deficiency.
Storage
The human body can store anywhere from 2-5mg of vitamin B12. Most of this is stored in the liver and is recycled via enterohepatic circulation.
Pathogenesis
Pernicious anemia is a type of megaloblastic anemia caused due to improper vitamin B12 absorption by the body. Impaired absorption occurs because of deficiency of intrinsic factor which is produced by the parietal cells of the stomach. The etiology of pernicious anemia can be due to autoimmune causes or genetic disease. In autoimmune disease, the antibodies attack most of the gastric mucosa, but the antrum is spared.
Autoimmune causes of pernicious anemia
This is the most common cause of pernicious anemia. In autoimmune pernicious anemia, the body produces antibodies against parietal cells or intrinsic factor.
- Antibodies against parietal cells of the gastric mucosa work to inhibit the H+/K(+)-ATPase which is the proton pump present in the parietal cells. The proton pump serves as an auto antigen and activates the cytotoxic CD4+ T cells which proceed to destroy gastric mucosal cells.[9][10]
- Intrinsic factor antibodies are present in fewer cases of pernicious anaemia but are highly specific. There are 2 types of IF antibodies. They prevent the binding and absorption of cobalamin in the ileum via its receptor.[11]
Clinical features
- The symptoms of pernicious anemia take months, and often years to manifest. Patients most commonly present with symptoms of anemia like lightheadedness, dizziness, shortness of breath etc. The population affected with pernicious anemia is usually the elderly (>60 years) owing to its insidious onset.
- Pernicious anemia has hematological, gastrointestinal and neurological manifestations.
- Hematological signs are the earliest manifestation of the disease while neurological signs are seen much later.
- Patients with pernicious anemia usually have very low levels of hydrochloric acid in the stomach (achlorhydria) and high levels on gastrin (hypergastrinemia).
Differentiating pernicious anemia from other diseases
Pernicious anemia shares many similarities with other forms of megaloblastic anemia like B12 and folate deficiency.
- Vitamin B12 deficiency due to insufficient intake (eg veganism) has all the features of pernicious anemia like megaloblasts, hypersegmented neutrophils, neuropsychiatric manifestations. But atrophic gastritis is absent, so achlorhydria, parietal cell antibodies or IF antibodies are absent. Intrinsic factor levels are also normal.[6]
- Folic acid deficiency also results in megaloblastic anemia and similar hematological changes as pernicious anemia, but urinary excretion of methylmalonic acid is absent, so are features of pernicious anemia like achlorhydria, antibodies and normal IF levels.
- Ileal resection causes B12 deficiency due to decreased absorption.
- Certain drugs such as methotrexate, azathioprine cause folate deficiency and result in megaloblastic anemia. This is usually seen in patients taking chemotherapy or other chronic conditions such as rheumatoid arthritis. [12]
- Chronic proton pump inhibitor therapy also results in B12 deficiency as vitamin B12 cannot dissociate from its carrier protein in the absence of an acidic environment.[13]
- Long term use of metformin, such as in diabetics, is linked to vitamin B12 deficiency and symptoms similar to pernicious anemia, but this can be differentiated from pernicious anemia as it is seen in diabetics on chronic therapy.[14]
Associated Conditions
People affected with pernicious anemia might have other coexisting autoimmune conditions such as autoimmune thyroiditis, autoimmune diabetes, vitiligo etc. Autoimmune thyroiditis is most commonly seen in patients with pernicious anemia, particularly females. HLA DR3 has been implicated in the development of autoimmune diseases such as pernicious anemia[15].
Epidemiology and demographics
- Pernicious anemia is a disease of the elderly. The mean age of patients who are symptomatic is >60.[16]
- An exception is the genetic form of the disease which is a congenital deficiency of intrinsic factor and is seen in children <10 years of age.
- Men and women are equally affected
- Prevalence of pernicious anemia is estimated at 0.1% of the population.[17]
Genetics
- Some forms of pernicious anemia are congenital and a genetic link has been postulated because of a higher incidence in certain populations.
- Affected people have a complete or near total absence of intrinsic factor and the presence of antibodies against intrinsic factor.
- The genetic variant is transmitted through an autosomal recessive pattern.[18]
Risk factors
- People who have autoimmune conditions like diabetes mellitus, autoimmune thyroiditis are at higher risk of developing pernicious anemia.
Natural History, Complications and Prognosis
- In most cases, patients affected with pernicious anemia remain asymptomatic for many years.
- Early manifestations include fatigue, shortness of breath, pallor and weakness.
- Long standing untreated pernicious anemia results in irreversible neurological damage such as subacute combined degeneration of the spinal cord.
- Neurological changes are irreversible once they set in and do not resolve with cobalamin supplementation.
Diagnosis
A diagnosis of pernicious anemia is made by a history and physical examination, along with hematological and neurological examination.
Diagnostic criteria
- The only specific criteria to diagnose pernicious anemia is an intrinsic factor output of less than 200U/h after pentagastrin stimulation, where normal levels would be >2000U/h. [19]
Symptoms
Symptoms of pernicious anemia are summarised below
| Hematological symptoms | Gastrointestinal symptoms | Neurological symptoms |
|---|---|---|
| Fatigue | Loss of appetite | Parasthesias |
| Weakness | Weight loss
|
Depression |
| Shortness of breath | Nausea | Gait problems |
| Dizziness | Burning sensation on tongue | Weakness |
| Tachycardia | Diarrhea | Loss of balance |
| Lightheadedness | Vomiting | Confusion |
Physical examination findings
Most important physical examination findings are the neurological findings of long standing B12 deficiency which leads to subacute combined degeneration of the spinal cord.
- Hematological signs include pallor and icterus.[20]
- Neurological signs: Vitamin B12 deficiency causes nerve demyelination. B12 deficiency also causes a buildup of methylmalonic acid which is toxic to neuronal cells and causes apoptosis.[21].
The main neurological manifestation of pernicious anemia and vitamin B12 deficiency is subacute combined degeneration. The posterior and lateral columns of the spinal cord are affected. Lateral column demyelination manifests as hyperreflexia and spasticity, while posterior column defects are loss of proprioception and vibration sense. Ataxia and loss of tandem gait are also manifestations of posterior column demyelination. Recreational or accidental inhalation of nitrous oxide gas (laughing gas) can precipitate subacute combined degeneration in people with low levels of vitamin B12.[22]
- Gastrointestinal signs: Upto 25% of people affected with pernicious anemia develop glossitis. The tongue appears red, "beefy" and smooth due to atrophy and blunting of the lingual papillae.[23]
Subacute combined degeneration

Laboratory findings
- The first step in diagnosis is a blood vitamin B12 level. Blood levels less than 200 pg/ml are seen in pernicious anemia.
- Intrinsic factor antibodies and Parietal cell antibodies.
- Low intrinsic factor level.[24]
- Gastric mucosal sampling shows parietal cell atrophy with antral sparing.[25]
- Increased level of gastrin.
- Increased levels of homocysteine and methylmalonyl-CoA.
- Decreased folate levels are seen due to "folate trapping" in the form of methyltetrahydrofolate.
Shilling Test
The Shilling test is no longer done to detect an IF deficiency but has historical importance. After a vitamin B12 deficiency is noted, the patient is given radioactively tagged cobalamin to take orally. Soon after this step, the patient is injected with unlabelled cobalamin intramuscularly. Urine is checked for radioactive cobalamin for the next 24 hours. In pernicious anemia, there is an intrinsic factor deficiency, therefore the orally consumed radioactive cobalamin will not be absorbed and can be detected in the urine. In the next step, the patient is given radioactive cobalamin along with intrinsic factor and their urine is checked for traces of radioactive cobalamin. Absence of radioactive cobalamin in the urine points to the deficiency of intrinsic factor in the patients stomach which is the cause of vitamin B12 deficiency[26]. If the cobalamin absorption does not increase even with intrinsic factor supplementation, patient can be given a course of antibiotics as bacterial overgrowth may hinder absorption.
Peripheral smear findings
- The most obvious peripheral smear finding is megaloblasts and macrocytes.
Megaloblastic anemia results due to the lagging behind of nuclear development when compared to cytoplasmic development. This is known as nuclear-cytoplasmic asynchrony. Such defective cells are destroyed in the bone marrow (intramedullary hemolysis).
- Decreased number of RBCs (erythopenia)
- Macrocytosis- the RBCs in pernicious anemia are very large. Macrocytosis is defined as cells that have an MCV >100 femtolitres (normal :80-100fL)
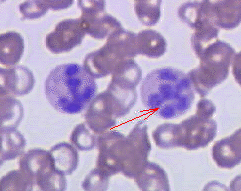
- Hypersegmented neutrophils : Neutrophils containing ≥ 6 lobes. [27]
- Poikilocytosis and anisocytosis
- Low reticulocyte count (reticulopenia)
- Howell-Jolly bodies
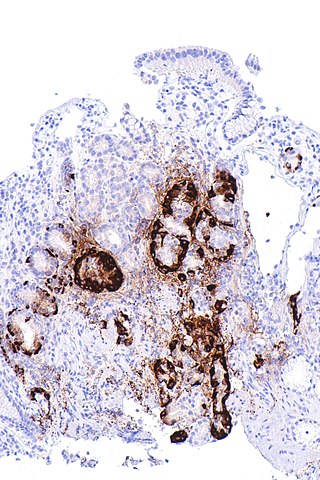
-
Atrophic gastritis
-
Hypersegmented neutrophil
Treatment
- Standard treatment for pernicious anemia is replacement of cobalamin via intramuscular injection. [28]
- 1000 mcg IM everyday for one week, followed by weekly injections the next month and then monthly once injections.
- Response to treatment is measured by an increase in reticulocyte count within 5 days of starting therapy.
- Patient also experience a sense of wellbeing shortly after beginning therapy.
- If reticulocytosis is not observed within the first week of therapy, other factors such as hypothyroidism, folate deficiency should be considered.
- Intramuscular therapy can be replaced by high dose oral therapy.[17]
- Neurological disease always warrants parenteral treatment.
- Within the first 3-4 weeks of treatment, marrow changes revert and there is resolution in macrocytosis.
- Most patients require lifelong monthly therapy.
- Routine follow up should be done with a CBC every few months.
- A small percentage of patients develop gastric carcinoma, particularly in the elderly. Regular surveillance helps in early detection and treatment. [29]
Prevention
- There is no primary preventive measure for pernicious anemia.
- Once sucessfully diagnosed and treated, patients with pernicious anemia are followed up every year for development of stomach cancer[30], or symptoms of anemia.
References
Overview
Multiple myeloma can be classified into several subtypes based on the extent of organ involvement (medullary or extramedullary) and the disease clinical presentation (active symptomatic or smoldering asymptomatic). The four general categories include solitary plasmacytoma, monoclonal gammopathy of undetermined significance, smoldering multiple myeloma, and active multiple myeloma. Within the category of active multiple myeloma, the three risk groups include standard risk, intermediate risk, and high risk, based on the cytogenetic profile.
Classification
Multiple myeloma is still believed to be a single disease entity but it is a group of different cytogenetically distinct plasma cell malignancies. It can be classified on several basis.[31][32]
Biological genetic classification
| Biological Genetic Classification | ||
|---|---|---|
| Hyperdiploid MM |
| |
| Non-hyperdiploid MM | t(11;14)(q13;q32) |
|
| t(4;14)(p16;q32) |
| |
| t(14;16)(q32;q23) |
| |
| Chromosome 13 deletion |
| |
Molecular Cytogenetic Classification
| Molecular Cytogenetic Classification | |||
|---|---|---|---|
| Hyperdiploid | More favorable, IgG-k, older patients | ||
| Non-hyperdiploid
Aggressive, IgA-l, younger individuals |
Cyclin D translocation | t(11;14)(q13;q32) | Upregulation of CCND1; favorable prognosis; bone lesions. Two subtypes by GEP |
| t(6;14q)(p21;32) | Probably same as CCND1 | ||
| t(12;14)(p13;q32) | Rare | ||
| MMSET translocation | t(4;14)(p16;q32) | Upregulation of MMSET; upregulation of FGFR3 in 75%, unfavorable prognosis with conventional therapy; bone lesion less frequent | |
| MAF translocation | t(14;16)(q32;q23) | Confirmed as aggressive by at least two series | |
| t(14;20)(q32;q11) | One series shows more aggressive disease | ||
| t(8;14)(q24;q32) | Unknown effect on outcome but | ||
| Unclassified (other) | Various subtypes and some with overlap | ||
- A more detailed version is given below:
| Primary Molecular Cytogenetic Classification of Multiple Myeloma | ||
|---|---|---|
| Subtype | Gene(s)/chromosomes affected | Percentage of myeloma patients |
| Trisomic MM |
|
42 |
| IgH translocated MM |
|
30 |
| Combined IgH translocated/trisomic MM |
|
15 |
| Isolated Monosomy 14 |
|
4.5 |
| Other cytogenetic abnormalities in absence of IgH translocations or trisomy or monosomy 14 | 5.5 | |
| Normal | NA | 3 |
Mayo Clinic Risk Stratification for Multiple Myeloma (mSMART)
- Multiple myeloma can be classified according to risk status. The risk stratification for multiple myeloma consists of three groups, as follows:[31][33]
| Mayo Clinic Risk Stratification for Multiple Myeloma (mSMART) | ||
|---|---|---|
| Risk group | Chromosomal Abnormalities | Frequency |
| Standard risk |
|
75% |
| Intermediate Risk |
|
10% |
| High risk |
|
15% |
References
- ↑ Sinclair L (2008). "Recognizing, treating and understanding pernicious anaemia". J R Soc Med. 101 (5): 262–4. doi:10.1258/jrsm.2008.081006. PMC 2376267. PMID 18463283.
- ↑ SMITH EL (1948). "Purification of anti-pernicious anaemia factors from liver". Nature. 161 (4095): 638. doi:10.1038/161638a0. PMID 18856623.
- ↑ Miles LM, Allen E, Clarke R, Mills K, Uauy R, Dangour AD (2017). "Impact of baseline vitamin B12 status on the effect of vitamin B12 supplementation on neurologic function in older people: secondary analysis of data from the OPEN randomised controlled trial". Eur J Clin Nutr. 71 (10): 1166–1172. doi:10.1038/ejcn.2017.7. PMID 28225050.
- ↑ Watanabe F (2007). "Vitamin B12 sources and bioavailability". Exp Biol Med (Maywood). 232 (10): 1266–74. doi:10.3181/0703-MR-67. PMID 17959839.
- ↑ Jung SB, Nagaraja V, Kapur A, Eslick GD (2015). "Association between vitamin B12 deficiency and long-term use of acid-lowering agents: a systematic review and meta-analysis". Intern Med J. 45 (4): 409–16. doi:10.1111/imj.12697. PMID 25583062.
- ↑ 6.0 6.1 Del Corral A, Carmel R (1990). "Transfer of cobalamin from the cobalamin-binding protein of egg yolk to R binder of human saliva and gastric juice". Gastroenterology. 98 (6): 1460–6. doi:10.1016/0016-5085(90)91076-i. PMID 2110915.
- ↑ Harrington DJ (2017). "Laboratory assessment of vitamin B12 status". J Clin Pathol. 70 (2): 168–173. doi:10.1136/jclinpath-2015-203502. PMID 27169753.
- ↑ Tinelli C, Di Pino A, Ficulle E, Marcelli S, Feligioni M (2019). "Hyperhomocysteinemia as a Risk Factor and Potential Nutraceutical Target for Certain Pathologies". Front Nutr. 6: 49. doi:10.3389/fnut.2019.00049. PMC 6491750. PMID 31069230.
- ↑ Callaghan JM, Khan MA, Alderuccio F, van Driel IR, Gleeson PA, Toh BH (1993). "Alpha and beta subunits of the gastric H+/K(+)-ATPase are concordantly targeted by parietal cell autoantibodies associated with autoimmune gastritis". Autoimmunity. 16 (4): 289–95. doi:10.3109/08916939309014648. PMID 7517707.
- ↑ Toh BH, Sentry JW, Alderuccio F (2000). "The causative H+/K+ ATPase antigen in the pathogenesis of autoimmune gastritis". Immunol Today. 21 (7): 348–54. doi:10.1016/s0167-5699(00)01653-4. PMID 10871877.
- ↑ Schade SG, Abels J, Schilling RF (1967). "Studies on antibody to intrinsic factor". J Clin Invest. 46 (4): 615–20. doi:10.1172/JCI105563. PMC 442045. PMID 6021209.
- ↑ Green R, Datta Mitra A (2017). "Megaloblastic Anemias: Nutritional and Other Causes". Med Clin North Am. 101 (2): 297–317. doi:10.1016/j.mcna.2016.09.013. PMID 28189172.
- ↑ Heidelbaugh JJ (2013). "Proton pump inhibitors and risk of vitamin and mineral deficiency: evidence and clinical implications". Ther Adv Drug Saf. 4 (3): 125–33. doi:10.1177/2042098613482484. PMC 4110863. PMID 25083257.
- ↑ Aroda VR, Edelstein SL, Goldberg RB, Knowler WC, Marcovina SM, Orchard TJ; et al. (2016). "Long-term Metformin Use and Vitamin B12 Deficiency in the Diabetes Prevention Program Outcomes Study". J Clin Endocrinol Metab. 101 (4): 1754–61. doi:10.1210/jc.2015-3754. PMC 4880159. PMID 26900641.
- ↑ Zulfiqar AA, Andres E (2017). "Association pernicious anemia and autoimmune polyendocrinopathy: a retrospective study". J Med Life. 10 (4): 250–253. PMC 5771255. PMID 29362601.
- ↑ Carmel R (1996). "Prevalence of undiagnosed pernicious anemia in the elderly". Arch Intern Med. 156 (10): 1097–100. PMID 8638997.
- ↑ 17.0 17.1 Andres E, Serraj K (2012). "Optimal management of pernicious anemia". J Blood Med. 3: 97–103. doi:10.2147/JBM.S25620. PMC 3441227. PMID 23028239.
- ↑ Gordon MM, Brada N, Remacha A, Badell I, del Río E, Baiget M; et al. (2004). "A genetic polymorphism in the coding region of the gastric intrinsic factor gene (GIF) is associated with congenital intrinsic factor deficiency". Hum Mutat. 23 (1): 85–91. doi:10.1002/humu.10297. PMID 14695536.
- ↑ Cattan D (2011). "Pernicious anemia: what are the actual diagnosis criteria?". World J Gastroenterol. 17 (4): 543–4. doi:10.3748/wjg.v17.i4.543. PMC 3027024. PMID 21274387.
- ↑ Seynabou F, Fatou Samba Diago N, Oulimata Diop D, Abibatou Fall S, Nafissatou D (2016). "Biermer anemia: Hematologic characteristics of 66 patients in a Clinical Hematology Unit at Senegal". Med Sante Trop. 26 (4): 402–407. doi:10.1684/mst.2016.0625. PMID 28073728.
- ↑ Han L, Wu S, Han F, Gu X (2015). "Insights into the molecular mechanisms of methylmalonic acidemia using microarray technology". Int J Clin Exp Med. 8 (6): 8866–79. PMC 4538064. PMID https://www.ncbi.nlm.nih.gov/pubmed/26309541 Check
|pmid=value (help). - ↑ Choi C, Kim T, Park KD, Lim OK, Lee JK (2019). "Subacute Combined Degeneration Caused by Nitrous Oxide Intoxication: A Report of Two Cases". Ann Rehabil Med. 43 (4): 530–534. doi:10.5535/arm.2019.43.4.530. PMC 6734019 Check
|pmc=value (help). PMID 31499607. - ↑ Stoopler ET, Kuperstein AS (2013). "Glossitis secondary to vitamin B12 deficiency anemia". CMAJ. 185 (12): E582. doi:10.1503/cmaj.120970. PMC 3761039. PMID 23359038.
- ↑ Lahner E, Annibale B (2009). "Pernicious anemia: new insights from a gastroenterological point of view". World J Gastroenterol. 15 (41): 5121–8. doi:10.3748/wjg.15.5121. PMC 2773890. PMID 19891010.
- ↑ Korman MG, Strickland RG, Hansky J (1972). "The functional 'G' cell mass in atrophic gastritis". Gut. 13 (5): 349–51. doi:10.1136/gut.13.5.349. PMC 1412218. PMID 5036089.
- ↑ "StatPearls". 2020. PMID 29939561.
- ↑ Farrelly SJ, O'Connor KA (2017). "Hypersegmented neutrophils and oval macrocytes in the setting of B12 deficiency and pancytopaenia". BMJ Case Rep. 2017. doi:10.1136/bcr-2016-218508. PMC 5612428. PMID 28821482.
- ↑ Annibale B, Lahner E, Fave GD (2011). "Diagnosis and management of pernicious anemia". Curr Gastroenterol Rep. 13 (6): 518–24. doi:10.1007/s11894-011-0225-5. PMID 21947876.
- ↑ Murphy G, Dawsey SM, Engels EA, Ricker W, Parsons R, Etemadi A; et al. (2015). "Cancer Risk After Pernicious Anemia in the US Elderly Population". Clin Gastroenterol Hepatol. 13 (13): 2282-9.e1-4. doi:10.1016/j.cgh.2015.05.040. PMC 4655146. PMID 26079040.
- ↑ Venerito M, Link A, Rokkas T, Malfertheiner P (2016). "Gastric cancer - clinical and epidemiological aspects". Helicobacter. 21 Suppl 1: 39–44. doi:10.1111/hel.12339. PMID 27531538.
- ↑ 31.0 31.1 Rajkumar SV (July 2016). "Multiple myeloma: 2016 update on diagnosis, risk-stratification, and management". Am. J. Hematol. 91 (7): 719–34. doi:10.1002/ajh.24402. PMC 5291298. PMID 27291302.
- ↑ Alexiou C, Kau RJ, Dietzfelbinger H, Kremer M, Spiess JC, Schratzenstaller B; et al. (1999). "Extramedullary plasmacytoma: tumor occurrence and therapeutic concepts". Cancer. 85 (11): 2305–14. PMID 10357398.
- ↑ Sonneveld P, Avet-Loiseau H, Lonial S, Usmani S, Siegel D, Anderson KC; et al. (2016). "Treatment of multiple myeloma with high-risk cytogenetics: a consensus of the International Myeloma Working Group". Blood. 127 (24): 2955–62. doi:10.1182/blood-2016-01-631200. PMC 4920674. PMID 27002115.