Hantavirus infection
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]
Please Take Over This Page and Apply to be Editor-In-Chief for this topic: There can be one or more than one Editor-In-Chief. You may also apply to be an Associate Editor-In-Chief of one of the subtopics below. Please mail us [2] to indicate your interest in serving either as an Editor-In-Chief of the entire topic or as an Associate Editor-In-Chief for a subtopic. Please be sure to attach your CV and or biographical sketch.
Overview
Hantavirus pulmonary syndrome (HPS) is a deadly disease from rodents.
Also known as: Sin nombre virus, HPS
References
http://www.cdc.gov/ncidod/diseases/hanta/hps/index.htm
Epidemiology and Demographics
HPS an old disease, newly recognized
Although the high-profile investigation of the HPS syndrome emphasized public health authorities' warnings about new and emerging infectious diseases, HPS has turned out to be a newly identified, but not a "new," disease. In fact, the earliest case of a serologically confirmed SNV infection was in a person who developed an HPS-compatible illness in July 1959 and was found to have IgG antibodies in September 1994. The earliest case of HPS to be confirmed by IHC with direct visualization of hantaviral antigens in postmortem tissue involved a patient who died in 1978.
Hantavirus infection is apparently not deleterious to its rodent reservoir host and is associated with a brisk antibody response against the virion envelope and core proteins and chronic, probably lifelong infection. In natural populations, most infections occur through age-dependent horizontal route(s). The highest antibody prevalence is observed in large (mature) animals. A striking male predilection for hantavirus infection is observed in some rodent species such as harvest mice and deer mice, but not in urban rats (Rattus norvegicus). Horizontal transmission among cage-mates was experimentally demonstrated, but vertical transmission from dam to pup is negligible or absent both in wild and experimental settings.
Outbreaks of hantaviral disease have been associated with changes in rodent population densities, which may vary greatly across time, both seasonally and from year to year. Cycles respond to such extrinsic factors as interspecific competition, climatic changes, and predation. Spring and summer outbreaks of HFRS in agricultural settings in Asia and Europe are linked to human contact with field rodents through the planting and harvesting of crops. PUU outbreaks in Scandinavia and the HPS outbreak in the Four Corners region of the United States were associated with natural rodent population increases, followed by invasion of buildings by rodents. The ecologic events that led to 1994 and 1996 outbreaks of Andes virus-HPS in Patagonia, a region in southern South America, are being investigated. Human interventions, such as the introduction of Old World plant species (e.g., rosas mosquetas and Scottish brougham) to Patagonia, with associated alteration in rodent population dynamics, have been suggested as possible factors. Recent fires and a mild winter in Argentina's Rio Negro and Chubut Provinces may also have had a positive effect on the carrier rodent, the colilargo, Oligoryzomys longicaudatus (M. Christie and O. Pearson, pers. comm.).
Although the aerosol route of infection is undoubtedly the most common means of transmission among rodents and to humans, virus transmission by bite may occur among certain rodents and may also occasionally result in human infection (often inside a closed space, such as a rodent-infested grain silo, garage, or outbuilding used for food storage). Epidemiologic investigations have linked virus exposure to such activities as heavy farm work, threshing, sleeping on the ground, and military exercises. Indoor exposure was linked to invasion of homes by field rodents during cold weather or to nesting of rodents in or near dwellings. Genetic sequencing of rodent- and patient-associated viruses has been used to pinpoint the precise locations of human infections, which has supported the role of indoor exposure in hantavirus transmission (32,33). Many hantavirus infections have occurred in persons of lower socioeconomic status because poorer housing conditions and agricultural activities favor closer contact between humans and rodents. However, suburbanization, wilderness camping, and other outdoor recreational activities have spread infection to persons of middle and upper incomes.
Nosocomial transmission of hantaviruses has not been documented until very recently and must be regarded as rare. However, viruses have been isolated from blood and urine of HFRS patients, so exposure to bodily fluids of infected persons could result in secondary transmission. Only rarely have multiple North American HPS cases been associated with particular households or buildings. During recent outbreaks of HPS in South America, however, clustering of cases in households and among personal contacts appeared to be more common (M. Christie, pers. comm.). During a recent outbreak of Andes-virus-associated HPS in Patagonia, a Buenos Aires physician apparently contracted the infection after minimal exposure to infected patient blood (34; D.A. Pirola, pers. comm.). An adolescent patient in Buenos Aires apparently contracted hantavirus infection from her parents, who were infected in Patagonia. This unprecedented observation of apparent person-to-person spread of a hantavirus clearly requires laboratory confirmation, especially by careful comparative analysis of the viral sequences.
Hantaviruses have also caused several laboratory-associated outbreaks of HFRS. Laboratory-acquired infections were traced to persistently infected rats obtained from breeders, to wild-caught, naturally infected rodents, or to experimentally infected rodents. No illnesses due to laboratory infections have been reported among workers using cell-culture adapted viruses, although asymptomatic seroconversions have been documented.
Hantavirus Distribution and Disease-causing Potential
The worldwide distribution of rodents known to harbor hantaviruses suggests great disease-causing potential. Each hantavirus appears to have a single predominant natural reservoir. With rare exception, the phylogenetic interrelationships among the viruses and those of their predominant host show remarkable concordance. These observations suggest that hantaviruses do not adapt readily to new hosts and that they are closely adapted for success in their host, possibly because of thousands of years of coexistence. As many as three hantaviruses can be found in a particular geographic site, each circulating in its own rodent reservoir, with no apparent evolutionary influence on one another.
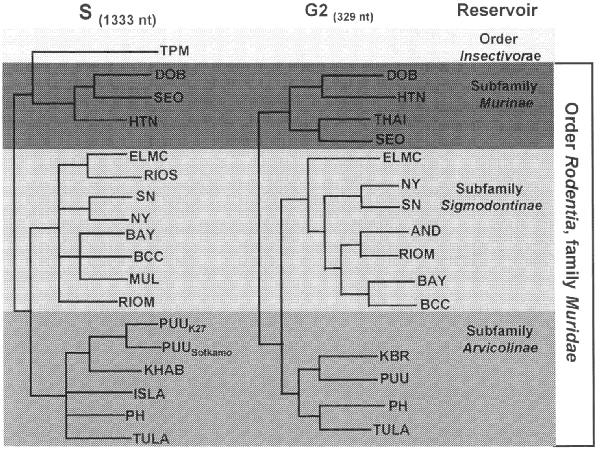
Figure. Phylogeny of hantaviruses and their relationships to natural reservoirs. The trees were constructed by comparing the complete coding regions of the S segments of hantaviruses or of 330 nucleotides corresponding to those of the M segment of Hantaan virus (strain 76118) from nucleotides 1987 to 2315. Abrreviations for viruses are as in Table 1. For each analysis, a single most parsimonious tree was derived by using PAUP 3.1.1 software. For the S segment tree, boostrap values resulting from 100 replications were all greater than 87% except for the branch leading to BCC (78%) and the branch leading to DOB (52%). The next most common placing of DOB was on a branch with HTN.
All known hantaviruses, except Thotta-palayam (TPM) virus, have been isolated or detected in murid rodents. Because only one isolate of TPM virus was made from a shrew (Order Insectivora), it is not clear if Suncus is the true primary reservoir or an example of a "spillover" host, i.e., a secondary host infected through contact with the primary host. Spillover is common in sympatric murid rodents, including those identified as the predominant carrier of another hantavirus; thus, the opportunity for genetic exchange among hantaviruses is present in nature. Spillover hosts are believed to have little or no impact on hantaviral distribution or associated disease. However, rodents other than the primary reservoirs can play an important carrier role. For example, Microtus rossiaemeri-dionalis may play a role in maintenance of Tula virus in some settings, and Peromyscus leucopus and Peromyscus boylii can be important reservoirs for SN virus in the western United States (T. Yates and B. Hjelle, unpub. data). Apparent spillover may also be the result of laboratory errors such as polymerase chain reaction (PCR) contamination or misidentification of rodent species. However, spillover is probably under-appreciated in many studies that rely on reverse transcriptase PCR for identifying specific viruses because many primer pairs may not detect an unexpected spillover virus. In either case, because mistaken identities and cell culture contaminations with other hantaviruses have occasionally been reported, investigators should verify unusual findings to prevent further confusion.
Epidemiology in the Virology Laboratory
During the outbreak in 1993, definitive proof that the agent causing HPS was a novel hantavirus was obtained using a genetic detection assay. Oligonucleotide primers were designed on the basis of regions of the M segment (G2 coding region) conserved among hantaviruses and were used in a nested RT-PCR assay to amplify hantavirus-specific DNA fragments from RNA extracted from the tissues of patients. The amplified DNA fragments were then sequenced. Comparative and phylogenetic analyses of derived sequence data demonstrated that the hantavirus associated with the HPS outbreak (SNV) was a novel virus most closely related to Prospect Hill virus (PHV). In addition, a direct genetic link was made between the human HPS cases and the virus harbored by peridomestic P. maniculatus rodents. Characterization of hantaviral genetic sequences recovered from human tissues demonstrated that these sequences were identical to those from rodents captured at the site of the patient's presumed infection. This characterization has continued to facilitate identification of the site of infection when more than one such site exists and therefore focus the public health response. These techniques also allow implication of a specific rodent host in areas of overlapping hosts.
Hantavirus Pulmonary Syndrome Case Count and Descriptive Statistics in the U.S.
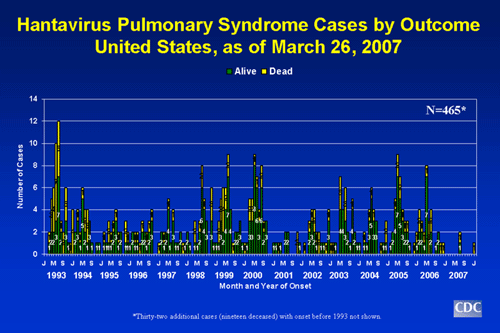
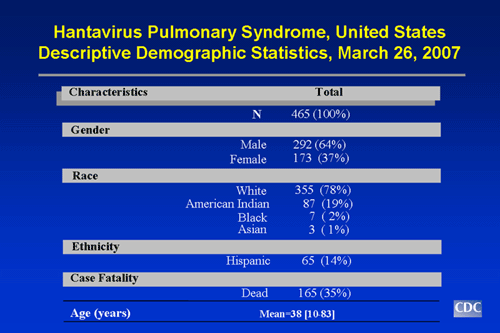
- Through March 26, 2007, a total of 465 cases of hantavirus pulmonary syndrome have been reported in the United States. The case count started when the disease was first recognized in May 1993. Thirty-five percent of all reported cases have resulted in death.
- Of persons ill with HPS, 64% have been male, 37% female.
- The mean age of confirmed case patients is 38 years (range: 10 to 83 years).
- HPS can strike anyone; however, whites currently account for 78% of all cases. American Indians account for about 19% of cases, African Americans for 2% of cases, and Asians for 1% of cases. About 14% of HPS cases have been reported among Hispanics (ethnicity considered separately from race).
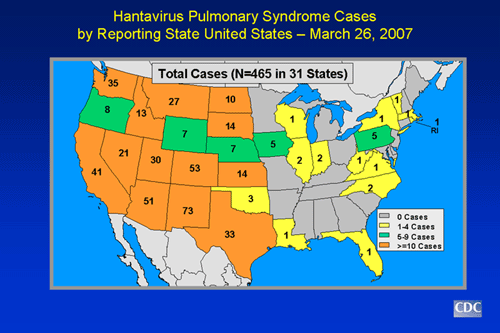
- Cases have been reported in 30 states, including most of the western half of the country and some eastern states as well. Over half of the confirmed cases have been reported from areas outside the Four Corners area.
- About three-quarters of patients with HPS have been residents of rural areas.
Slide 1: Hantavirus pulmonary syndrome cases by outcome
Slide 2: Characteristics of HPS Case-Patients, United States
Slide 3: Hantavirus pulmonary syndrome cases by state of residence
Outbreaks of HPS in South and Central America
Argentina
An outbreak of 18 cases of hantavirus pulmonary syndrome (HPS) occurred in the southern Andean city of El Bolson, in the Rio Negro Province of Argentina, between September 22 and December 5, 1996. All of these cases displayed the common characteristics of HPS. However, some patients displayed a flushed facial appearance reminiscent of some viral hemorrhagic fevers.
Research performed on the lung and liver tissues of a patient who had died of the same disease in this region March of 1995, identified a new hantavirus, named Andes virus, by polymerase chain reaction.
All of the people who became sick during the outbreak were either permanent residents of the El Bolson area, or had been visitors to it between two and five weeks before their admission to the hospital. In addition, three doctors who treated patients with the disease became ill themselves.
One feature of this outbreak that makes it highly unusual for hantaviral-associated illnesses, is that the available data strongly suggests person-to-person transmission. Specifically, the majority of cases had had contact with another cases two to three weeks before becoming ill, and cases tended to become ill within two weeks of each other. This was especially clear in the case of a physician in Buenos Aires (well outside the El Bolson area) who became ill 27 days after taking care of an HPS patient who had been transferred from the outbreak area to Buenos Aires. Person-to-person transmission has never before been observed with any other type of hantavirus, either those causing HPS in North, Central and South America, or those causing HFRS in Europe and Asia.
Chile
Between August 1 and October 8, 1997, an outbreak of HPS occurred in Chile. A total of 25 cases were reported, including three family case clusters. Officials in Chile requested epidemiological assistance from Special Pathogens Branch at CDC, and a team left for the region on September 24 to investigate the outbreak. Outbreak investigation findings have been published in "An Outbreak of Hantavirus Pulmonary Syndrome, Chile, 1997". The article is available in the EID October - December 1998 issue. In addition, the initial report made by the team is available in the CDC journal Morbidity and Mortality Weekly Report, Vo. 46, No. 40, and may be read online in Adobe Acrobat Reader (pdf) format.
CDC is only able to offer the information as listed above. As a U.S government agency, all of the data we have gathered during investigations is reported and available. However, we are limited by the fact that our disease control and surveillance assistance is provided outside our national borders only if formally requested by another nation, and only to the degree that assistance is actually provided. Therefore, our information on HPS outside of the U.S. is somewhat limited. If you are looking for more information on hantaviruses in Chile, please refer to the Ministerio de Salud, Chile (Chilean Ministry of Health).
Panama
In mid-January 1999, an outbreak of HPS occurred in Panama. The Special Pathogens Branch of the CDC and the Pan-American Health Organization (PAHO) collaborated with health authorities in Panama to investigate the outbreak. For more information on the outbreak investigation, please read the article "Hantavirus Pulmonary Syndrome -- Panama, 1999-2000", published in the Morbidity and Mortality Weekly Report, March 17, 2000.
For specific information about this outbreak, please contact the Panama Ministry of Health at: hantapanama@hotmail.com
References
http://www.cdc.gov/ncidod/diseases/hanta/hps/noframes/phys/epi.htm
http://www.cdc.gov/ncidod/diseases/hanta/hps/noframes/phys/virology.htm
http://www.cdc.gov/ncidod/EID/vol3no2/schmaljo.htm
http://www.cdc.gov/ncidod/diseases/hanta/hps/noframes/argtina.htm
http://www.cdc.gov/ncidod/diseases/hanta/hps/noframes/caseinfo.htm
http://www.cdc.gov/ncidod/diseases/hanta/hps/noframes/epislides/episl2.htm
Risk Factors
Little is known about activities that lead to a greater risk of infection. However, an early case-control study suggests that increased numbers of rodents in the household is the strongest risk factor for infection. Entering rarely opened or seasonally closed buildings may also contribute to infection. Among the confirmed cases of HPS for which exposure information is available, 70% of the patients in the case control study had exposures closely associated with peridomestic activities, such as cleaning, in homes that showed signs of rodent infestation. Four clusters of HPS cases involving 2-4 persons have been documented; for each cluster, exposure probably occurred within a shared, enclosed structure. Taken together, these observations suggest that disturbing or inhabiting closed, actively rodent-infested structures may constitute an important risk factor for contracting HPS.
Potentially occupationally acquired SNV infections have been recognized but are infrequent. Among documented U.S. cases of HPS, patients with potential occupational exposures have included grain farmers, an extension livestock specialist, field biologists, and agricultural, mill, construction, utility and feedlot workers. Many of these individuals had concurrent peridomestic exposures. Among U.S. mammalogists and rodent workers with varying degrees of rodent exposure, the seroprevalence of SNV antibodies was 1.14%. In contrast, a recent HPS seroprevalence study focused on selected occupational groups with frequent contact with rodents and their excreta (e.g., farm workers, laborers, professionals, home repairers, service industry and park service workers, heating and plumbing contractors, utility workers, and technicians) found no evidence of SNV infection.
Travel to and within all areas where hantavirus infection has been reported is not considered a risk factor for infection with HPS. The possibility of exposure to hantavirus for campers, hikers, and tourists is very small and is reduced further if steps are taken to reduce rodent contact.
References
http://www.cdc.gov/ncidod/diseases/hanta/hps/noframes/phys/epi.htm
Pathophysiology & Etiology
Hantaviruses belong to the bunyavirus family of viruses. There are 5 genera within the family: bunyavirus, phlebovirus, nairovirus, tospovirus, and hantavirus. Each is made up of negative-sensed, single-stranded RNA viruses. All these genera include arthropod-borne viruses, with the exception of hantavirus, which is rodent-borne.
Like other members of the bunyavirus family, hantaviruses are enveloped viruses with a genome that consists of three single-stranded RNA segments designated S (small), M (medium), and L (large). All hantaviral genes are encoded in the negative (genome complementary) sense. The S RNA encodes the nucleocapsid (N) protein. The M RNA encodes a polyprotein that is cotranslationally cleaved to yield the envelope glycoproteins G1 and G2. The L RNA encodes the L protein, which functions as the viral transcriptase/replicase. Within virions, the genomic RNAs of hantaviruses are thought to complex with the N protein to form helical nucleocapsids, which circularize due to sequence complementarity between the 5' and 3' terminal sequences of each genomic segment.
Hantaviruses replicate exclusively in the host cell cytoplasm. Entry into host cells is thought to occur by attachment of virions to cellular receptors and subsequent endocytosis. Nucleocapsids are introduced into the cytoplasm by pH-dependent fusion of the virion with the endosomal membrane. Transcription of viral genes is initiated by association of the L protein with the three nucleocapsid species. In addition to transcriptase and replicase functions, the viral L protein is also thought to have an endonuclease activity that cleaves cellular messenger RNAs (mRNAs) for the production of capped primers used to initiate transcription of viral mRNAs. As a result of this "cap snatching," the mRNAs of hantaviruses are believed to be capped and contain nontemplated 5' terminal extensions. The viral N and L mRNAs are thought to undergo translation at free ribosomes, whereas the M mRNA is translated in the endoplasmic reticulum. G1 and G2 glycoproteins form heterodimers and are then transported from the endoplasmic reticulum to the Golgi complex, where glycosylation is completed. The L protein produces nascent genomes by replication via a positive-sense RNA intermediate. Hantavirions are believed to form by association of nucleocapsids with glycoproteins embedded in the membranes of the Golgi, followed by budding into the Golgi cisternae. Nascent virions are then transported in secretory vesicles to the plasma membrane and released by exocytosis.
Disease development
Within 24 hours of initial evaluation, most patients develop some degree of hypotension and progressive evidence of pulmonary edema and hypoxia, usually requiring mechanical ventilation. The patients with fatal infections appear to have severe myocardial depression which can progress to sinus bradycardia with subsequent electromechanical dissociation, ventricular tachycardia or fibrillation.
Hemodynamic compromise occurs a median of 5 days after symptom onset--usually dramatically within the first day of hospitalization. In contrast to HFRS, overt hemorrhage occurs rarely in HPS, although hemorrhage is occasionally seen in association with disseminated intravascular coagulation. In contrast to septic shock, HPS patients have a low cardiac output with a raised systemic vascular resistance. Poor prognostic indicators include a plasma lactate of greater than 4.0 mmol/L or a cardiac index of less than 2.2 L/min/m2 Whilst pulmonary edema and pleural effusions are common, multiorgan dysfunction syndrome is rarely seen. However, HPS patients sometimes have mildly impaired renal function. Survivors frequently become polyuric during convalescence and improve almost as rapidly as they decompensated.
Other Hantaviruses
Several members of the hantavirus genus cause different forms of hemorrhagic fever with renal syndrome (HFRS), an ancient disease first described in Russia in 1913. The four viruses that are associated with HFRS, each named for the region from where they were first isolated, have different primary rodent hosts: Apodemus agrarius (the striped field mouse) for Hantaan virus, Rattus norvegicus (the Norway rat) and Rattus rattus (the black rat) for Seoul virus, Clethrionomys glareolus (the bank vole) for Puumala virus, and Apodemus flavicollis (the yellow-necked field mouse) for Dobrava virus. Hantaan virus from Korea and Dobrava virus from Slovenia are associated with a severe form of HFRS characterized by renal failure that can precede pulmonary edema and disseminated intravascular coagulation (DIC), with estimated mortality rates of 5% to 15%. A moderate form of HFRS caused by Seoul virus (which, along with its host, is distributed worldwide) is responsible for thousands of Eurasian cases annually. Serologic evidence for infection with Seoul-like hantaviruses has been found in rodents in major cities of the United States, and this virus was recently implicated in human cases of HFRS in Baltimore. One report has also associated Seoul virus with chronic renal disease. A mild form of HFRS, caused by Puumala virus, is responsible for nephropathia epidemica in Scandinavia, with an estimated mortality rate of 1% to 3%.
References
http://www.cdc.gov/ncidod/diseases/hanta/hps/noframes/phys/virology.htm
http://www.cdc.gov/ncidod/diseases/hanta/hps/noframes/phys/clinical.htm
Genetics
Sin nombre virus sequencing
The entire genomic sequence of SNV has subsequently been determined by using RNA extracted from autopsy material as well as RNA extracted from cell culture-adapted virus. The L RNA is 6562 nucleotides (nt) in length; the M RNA is 3696 nt long; and the S RNA is 2059 to 2060 nt long. Interestingly, when the prototype sequence (NMH10) of SNV detected in tissues from an HPS case was compared with the sequence of the SNV isolate (NMR11; isolated in Vero E6 cells from P. maniculatus trapped in the residence of the same case), only 16 nucleotide changes were found, and none of these changes resulted in alterations in amino acid sequences of viral proteins. It had been assumed that in the process of adaptation to cell culture, selection of SNV variants which grow optimally in cell culture would occur, and selected variants would differ genetically from the parental virus. Though NMH10 and NMR11 are identical in protein sequence, nucleotide substitutions in nontranslated regions of the genome could be responsible for altered viral phenotypes, as could changes in protein glycosylation or virus membrane components.
The nested RT-PCR assay developed during the initial HPS outbreak provided a rapid method for the genetic characterization of novel hantaviruses that did not require a virus isolate. Numerous new hantaviruses have been detected by RT-PCR in rodent tissues but have yet to be associated with human disease. These include El Moro Canyon virus associated with the western harvest mouse (Reithrodontomys megalotis), Tula virus with Microtus arvalis and M. rossiaemeridionalis, Rio Segundo virus with the Mexican harvest mouse (R. mexicanus), Isla Vista virus with the California vole (M. californicus), and Prospect Hill-like viruses in Microtus species.
Phylogenetic Analysis
Phylogenetic analysis of Old World and American hantaviruses indicates that the relationship among hantaviruses corresponds with the phylogeny of their rodent hosts. Viruses of rodents belonging to the subfamily Murinae are monophyletic as are hantaviruses of arvicoline and sigmodontine rodents, suggesting that long-term virus-rodent coevolution is taking place. Hantavirus evolution is best understood as co-evolution within specific lineages in the rodent family Muridae. The apparent coupling between hantaviruses and their rodent hosts suggests that viruses of sigmodontine rodents share a common ancestor, as do viruses of the subfamily Arvicolinae and Murinae. This coupling also has a geographic and clinical correlate: viruses found in Old World murine rodents, including Hantaan virus (HTNV), Seoul virus (SEOV) and Dobrava virus, are associated with HFRS in Eurasia. By contrast, viruses carried by New World sigmodontine rodents, including SNV Black Creek Canal virus (BCCV) and Bayou virus (BAYV), are associated with HPS in the Americas. This distinction can narrow the search for a rodent host for newly discovered HPS-like diseases and suggest disease implications for the various new viruses being genetically amplified from rodents.
Sequence Divergence
Detection and characterization of Sin Nombre-like viruses in P. maniculatus and P. leucopus populations have shown that multiple phylogenetic lineages of SNV exist in North America, and in some cases similar viruses are detected in both Peromyscus species. Sequence divergence among SNV genes has been shown to be as high as 23% nucleotide dissimilarity and 7% amino acid dissimilarity. Comparison of SNV sequences with those of other hantaviruses provides no obvious explanation as to why SNV and related viruses cause HPS while other hantaviruses are associated with HFRS. The development of a reverse genetics system for manipulation of virus genomes and an animal model for studying pathogenesis will be necessary to define the molecular mechanism(s) of SNV pathogenicity.
Genomic reassortment by RNA viruses with segmented genomes is well documented and has the potential to produce viruses with altered biological activity, host range, and disease potential. For example, segment reassortment among influenza virus strains (antigenic shift) is thought to be responsible for influenza pandemics. Genomic reassortment among SNV variants is known to occur in nature, but the precise role of genomic reassortment in the epidemiology of HPS and HFRS is unknown.
References
http://www.cdc.gov/ncidod/diseases/hanta/hps/noframes/phys/virology.htm
Natural History
The "First" Outbreak

In May 1993, an outbreak of an unexplained pulmonary illness occurred in the southwestern United States, in an area shared by Arizona, New Mexico, Colorado and Utah known as "The Four Corners." A young, physically fit Navajo man suffering from shortness of breath was rushed to a hospital in New Mexico and died very rapidly.
While reviewing the results of the case, medical personnel discovered that the young man's fiancee had died a few days before after showing similar symptoms, a piece of information that proved key to discovering the disease. As Dr. James Cheek of the Indian Health Service (IHS) noted, "I think if it hadn't been for that initial pair of people that became sick within a week of each other, we never would have discovered the illness at all."
An investigation combing the entire Four Corners region was launched by the New Mexico Office of Medical Investigations (OMI) to find any other people who had a similar case history. Within a few hours, Dr. Bruce Tempest of IHS, working with OMI, had located five young, healthy people who had all died after acute respiratory failure.
A series of laboratory tests had failed to identify any of the deaths as caused by a known disease, such as bubonic plague. At this point, the CDC Special Pathogens Branch was notified. CDC, the state health departments of New Mexico, Colorado and Utah, the Indian Health Service, the Navajo Nation, and the University of New Mexico all joined together to confront the outbreak.
During the next few weeks, as additional cases of the disease were reported in the Four Corners area, physicians and other scientific experts worked intensively to narrow down the list of possible causes. The particular mixture of symptoms and clinical findings pointed researchers away from possible causes, such as exposure to a herbicide or a new type of influenza, and toward some type of virus. Samples of tissue from patients who had gotten the disease were sent to CDC for exhaustive analysis. Virologists at CDC used several tests, including new methods to pinpoint virus genes at the molecular level, and were able to link the pulmonary syndrome with a virus, in particular a previously unknown type of hantavirus.
References
http://www.cdc.gov/ncidod/diseases/hanta/hps/index.htm
Diagnosis
A positive serological test result, evidence of viral antigen in tissue by immunohistochemistry, or the presence of amplifiable viral RNA sequences in blood or tissue, with compatible history of HPS, is considered diagnostic for HPS.
Serologic assays
At the time of the 1993 outbreak in the Four Corners area, cross-reactive antibodies to the previously known hantaviruses (e.g., Hantaan, Seoul, Puumala, and Prospect Hill viruses) were found in the acute- and convalescent-phase sera of some of the initial HPS patients. Tests based on specific viral antigens from SNV have since been developed and are now widely used for the routine diagnosis of HPS. CDC uses an enzyme-linked immunosorbent assay (ELISA) to detect IgM antibodies to SNV and to diagnose acute infections with other hantaviruses. This assay is also available in some state health laboratories.
An IgG test is used in conjunction with the IgM-capture test. Acute- and convalescent-phase sera should reflect a four-fold rise in IgG antibody titer or the presence of IgM in acute-phase sera to be diagnostic for hantaviral disease. Note that acute-phase serum sent as an initial diagnostic specimen may not yet have IgG. IgG antibody is long lasting, and sera of patients retrospectively identified appear to have retained antibody for many years. The SNV IgG ELISA has therefore been used in serologic investigations of the epidemiology of the disease and appears to be appropriate for this purpose. Investigations of selected populations using this assay have confirmed that infections with the virus are not common and that mild or inapparent infections are rare.
A Western blot assay using recombinant antigens and isotype-specific conjugates for IgM-IgG differentiation has also been developed and its results are generally in agreement with those of the IgM-capture format.
Also in use is a rapid immunoblot strip assay (RIBA), an investigational prototype assay to identify serum antibody to recombinant proteins and peptides specific for SNV and other hantaviruses.
Serologic confirmation of hantaviral infections has traditionally been done with neutralizing plaque assays, which have been recently described for SNV. However, these specific assays are also not commercially available.
Isolation
Isolation of hantaviruses (see below) from human sources is difficult, and the viruses causing HPS seem to be no exception to this rule. To date, no isolates of SNV-like viruses have been recovered from humans, and therefore virus isolation is not a consideration for diagnostic purposes.
Immunohistochemistry (IHC)
IHC testing of formalin-fixed tissues with specific monoclonal and polyclonal antibodies can be used to detect hantavirus antigens and has proven to be a sensitive method for laboratory confirmation of hantaviral infections. IHC has an important role in the diagnosis of HPS in patients from whom serum samples and frozen tissues are unavailable for diagnostic testing and in the retrospective assessment of disease prevalence in a defined geographic region.
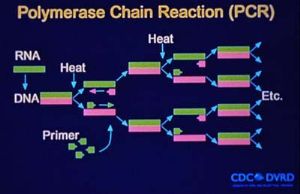
Polymerase Chain Reaction (PCR)
Reverse transcriptase-PCR (RT-PCR) can be used to detect hantaviral RNA in fresh frozen lung tissue, blood clots, or nucleated blood cells. However, RT-PCR is very prone to cross-contamination and should be considered an experimental technique. Differences in viruses in the United States complicate the use and sensitivity of RT-PCR for the routine diagnosis of hantaviral infections.
Differential Diagnosis
The prodromal phase of HPS is indistinguishable clinically from numerous other viral infections. Often the only guide to the etiology of the patient's illness is the blood picture, which may show circulating immunoblasts, which appear as large atypical lymphocytes, and thrombocytopenia. However, unlike other viral infections, HPS patients usually have concurrent left-shifted neutrophilia with circulating myelocytes.
In the cardiopulmonary stage of the disease, the patients have a diffuse pulmonary edema. The most frequent cause for such a picture is silent myocardial infarction so it is important to obtain an ECG and echocardiogram early to aid in the assessment. Intensivists at the University of New Mexico, where many of the patients have been managed, have found that a echocardiogram also helps to distinguish these patients from patients with ARDS as cardiac function is depressed to a much greater degree in the HPS patients and cardiac output does not respond to fluid challenge as it tends to with ARDS.
Infections in the immunocompetent which might present with a non-specific prodrome leading to acute cardiopulmonary deterioration as in HPS include leptospirosis, Legionnaire's disease, mycoplasma, Q fever, chlamydia, and in regions where the organisms are present, septicemic plague, tularemia, coccidioidomycosis and histoplasmosis. Non-infectious conditions such as Goodpasture's syndrome should also be considered. Lack of coryza aids the clinical distinction between HPS and Influenza A infection.
It must be remembered that HPS is relatively uncommon and in the immunocompromised PCP, CMV, cryptococcus, aspergillus and graft vs. host disease are far more likely to be the cause of diffuse pulmonary infiltrates than a hantaviral infection.
References
http://www.cdc.gov/ncidod/diseases/hanta/hps/noframes/phys/diag.htm
http://www.cdc.gov/ncidod/diseases/hanta/hps/noframes/phys/clinical.htm
History and Symptoms
Early symptoms
Early symptoms include fatigue, fever and muscle aches, especially in the large muscle groups-thighs, hips, back, and sometimes shoulders. These symptoms are universal.
There may also be headaches, dizziness, chills, and abdominal problems, such as nausea, vomiting, diarrhea, and abdominal pain. About half of all HPS patients experience these symptoms.
Late symptoms
Four to 10 days after the initial phase of illness, the late symptoms of HPS appear. These include coughing and shortness of breath, with the sensation of, as one survivor put it, a "...tight band around my chest and a pillow over my face" as the lungs fill with fluid.
Uncommon symptoms
Earache, sore throat, runny nose, and rash are very uncommon symptoms of HPS.
How long after contracting the virus do symptoms appear?
Due to the small number of HPS cases, the "incubation time" is not positively known. However, on the basis of limited information, it appears that symptoms may develop between 1 and 5 weeks after exposure to urine, droppings, or saliva of infected rodents.
Another important point to remember from the data that the CDC Special Pathogens Branch keeps on all reported cases of HPS, is that it appears many people who have become ill were in a situation where they did not see rodents or rodent droppings. Other people have had frequent contact with rodents and their droppings before becoming ill. This apparent inconsistency makes it very difficult to pin down the precise time when the virus was transmitted.
References
http://www.cdc.gov/ncidod/diseases/hanta/hps/noframes/symptoms.htm
Treatment
There is no specific treatment, cure, or vaccine for hantavirus infection. However, we do know that if infected individuals are recognized early and receive medical care in an intensive care unit, they may do better. In intensive care, patients are intubated and given oxygen therapy to help them through the period of severe respiratory distress.
The earlier the patient is brought in to intensive care, the better. If a patient is experiencing full distress, it is less likely the treatment will be effective.
Therefore, if you have been around rodents and have symptoms of fever, deep muscle aches, and severe shortness of breath, see your doctor immediately. Be sure to tell your doctor that you have been around rodents-this will alert your physician to look closely for any rodent-carried disease, such as HPS.
References
http://www.cdc.gov/ncidod/diseases/hanta/hps/noframes/treating.htm
Primary Prevention
Eliminate or minimize contact with rodents in your home, workplace, or campsite. If rodents don't find that where you are is a good place for them to be, then you're less likely to come into contact with them. Seal up holes and gaps in your home or garage. Place traps in and around your home to decrease rodent infestation. Clean up any easy-to-get food.
Recent research results show that many people who became ill with HPS developed the disease after having been in frequent contact with rodents and/or their droppings around a home or a workplace. On the other hand, many people who became ill reported that they had not seen rodents or rodent droppings at all. Therefore, if you live in an area where the carrier rodents are known to live, try to keep your home, vacation place, workplace, or campsite clean.
Prevention indoors and outdoors
Indoors

- Keep a clean home, especially kitchen (wash dishes, clean counters and floor, keep food covered in rodent-proof containers).
- Keep a tight-fitting lid on garbage, discard uneaten pet food at the end of the day.
- Set and keep spring-loaded rodent traps. Set traps near baseboards because rodents tend to run along walls and in tight spaces rather than out in the open.
- Set Environmental Protection Agency-approved rodenticide with bait under plywood or plastic shelter along baseboards. These are sometimes known as "covered bait stations." Remember to follow product use instructions carefully, since rodenticides are poisonous to pets and people, too.
- Seal all entry holes 1/4 inch wide or wider with lath screen or lath metal, cement, wire screening or other patching materials, inside and out.
- If bubonic plague is a problem in your area, spray flea killer or spread flea powder in the area before setting traps. This is important. If you control rodents but do not control fleas as well, you may increase the risk of infection with bubonic plague, since fleas will leave rodents once the rodents die and will seek out other food sources, including humans.
Outdoors
- Clear brush, grass and junk from around house foundations to eliminate a source of nesting materials.
- Use metal flashing around the base of wooden, earthen or adobe homes to provide a strong metal barrier. Install so that the flashing reaches 12 inches above the ground and six inches down into the ground
- Elevate hay, woodpiles and garbage cans to eliminate possible nesting sites. If possible, locate them 100 feet or more from your house.
- Trap rodents outside, too. Poisons or rodenticides may be used as well, but be sure to keep them out of the reach of children or pets.
- Encourage the presence of natural predators, such as non-poisonous snakes, owls and hawks.
- Remember, getting rid of all rodents isn't feasible, but with ongoing effort you can keep the population very low.
Clean Up Infested Areas, Using Safety Precautions:
- Put on latex rubber gloves before cleaning up.
- Do not stir up dust by sweeping up or vacuuming up droppings, urine or nesting materials. Instead, thoroughly wet contaminated areas with detergent or liquid to deactivate the virus. Most general purpose disinfectants and household detergents are effective. However, a hypochlorite solution prepared by mixing 1 and 1/2 cups of household bleach in 1 gallon of water may be used in place of commercial disinfectant. When using the chlorine solution, avoid spilling the mixture on clothing or other items that may be damaged.
- Once everything is wet, take up contaminated materials with a damp towel, then mop or sponge the area with disinfectant.
- Spray dead rodents with disinfectant, then double-bag along with all cleaning materials and bury or burn—or throw out in appropriate waste disposal system. If burning or burying isn't feasible, contact your local or state health department about other disposal methods.
- Finally, disinfect gloves before taking them off with disinfectant or soap and water. After taking off the clean gloves, thoroughly wash hands with soap and warm water.
- When going into cabins or outbuildings (or work areas) that have been closed for awhile, open them up and air out before cleaning.
Hantaviruses are surrounded by a lipid (fatty) envelope, so they are somewhat fragile. The lipid envelope can be destroyed and the virus killed by fat solvents, such as alcohol, ordinary disinfectants and household bleach. That is why one of the most important ways to prevent transmitting the disease is to carefully wet down dead rodents and areas where rodents have been with disinfectant and/or bleach. When you do this, you are killing the virus itself and reducing the chance that the virus will get into the air.
Special Precautions for Homes of Persons with Confirmed Hantavirus Infection or Buildings with Heavy Rodent Infestations
- Persons involved in the clean-up should wear coveralls (disposable, if possible), rubber boots or disposable shoe covers, rubber or plastic gloves, protective goggles, and an appropriate respiratory protection device, such as a half-mask air-purifying (or negative-pressure) respirator with a high-efficiency particulate air (HEPA) filter or a powered air-purifying respirator (PAPR) with HEPA filters.
- Personal protective gear should be decontaminated upon removal at the end of the day. If the coveralls are not disposable, they should be laundered on site. If no laundry facilities are available, the coveralls should be immersed in liquid disinfectant until they can be washed.
- All potentially infective waste material (including respirator filters) from clean-up operations that cannot be burned or deep buried on site should be double bagged in appropriate plastic bags. The bagged material should then be labeled as infectious (if it is to be transported) and disposed of in accordance with local requirements for infectious waste.
- Workers who develop symptoms suggestive of HPS within 45 days of the last potential exposure should immediately seek medical attention. The physician should contact local health authorities promptly if hantavirus-associated illness is suspected. A blood sample should be obtained and forwarded through the state health department to CDC for hantavirus antibody testing.
Precautions for Campers and Hikers in the Affected Areas

- Avoid coming into contact with rodents and rodent burrows or disturbing dens (such as pack rat nests).
- Air out, then disinfect cabins or shelters before using them. These places often shelter rodents.
- Do not pitch tents or place sleeping bags in areas in proximity to rodent droppings or burrows or near areas that may shelter rodents or provide food for them (e.g., garbage dumps or woodpiles).
- If possible, do not sleep on the bare ground. In shelters, use a cot with the sleeping surface at least 12 inches above the ground. Use tents with floors or a ground cloth if sleeping in the open air.
- Keep food in rodent-proof containers!
- Promptly bury (or--preferably--burn followed by burying, when in accordance with local requirements) all garbage and trash, or discard in covered trash containers.
- Use only bottled water or water that has been disinfected by filtration, boiling, chlorination, or iodination for drinking, cooking, washing dishes, and brushing teeth.
- And last but not least, do not play with or handle any rodents that show up at the camping or hiking site, even if they appear friendly.
References
http://www.cdc.gov/ncidod/diseases/hanta/hps/noframes/prevent.htm
http://www.cdc.gov/ncidod/diseases/hanta/hps/noframes/prevent2.htm
http://www.cdc.gov/ncidod/diseases/hanta/hps/noframes/prevent3.htm
http://www.cdc.gov/ncidod/diseases/hanta/hps/noframes/prevent4.htm
http://www.cdc.gov/ncidod/diseases/hanta/hps/noframes/prevent6.htm
Acknowledgements
The content on this page was first contributed by: C. Michael Gibson, M.S., M.D.
List of contributors:
Pilar Almonacid Template:SIB Template:WH Template:WikiDoc Sources