Epithelial ovarian tumors pathogenesis
|
Epithelial ovarian tumors Microchapters |
|
Differentiating Epithelial Ovarian Tumors from other Diseases |
|---|
|
Diagnosis |
|
Treatment |
|
Case Studies |
|
Epithelial ovarian tumors pathogenesis On the Web |
|
American Roentgen Ray Society Images of Epithelial ovarian tumors pathogenesis |
|
Risk calculators and risk factors for Epithelial ovarian tumors pathogenesis |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]Associate Editor(s)-in-Chief: Hannan Javed, M.D.[2]
Pathogenesis
Secondary Müllerian system
- Although ovarian surface epithelium is not a derivative of Müllerian ducts but ovarian epithelial cancers are characterized by presence of Müllerian lesions.[2]
- Serous carcinoma of ovary is though to originate from fallopian tubes while clear cell, endometrioid, and sero-mucinous carcinomas are thought to have their origin in endometriosis. Similarly Walthard nests potentially give rise to mucinous and Brenner malignant tumors, at least partially. All of these precursors are Müllerian system derivatives..[3][4]
- Secondary Müllerian system is a hypothesis that tries to explain this apparent enigma of existence of Müllerian epithelial lesions in locations not derived from Müllerian ducts such as ovaries and peritoneal cavity.[5][4]
- According to this hypothesis, Müllerian tissues, considered as vestigial, are found in locations such as para-tubal and para-ovarian locations and these tissues or cysts, not the ovarian epithelium itself, give rise to epithelial ovarian neoplasms.[3][4]
Hereditary ovarian carcinoma: An understanding of genome
- More than one fifth cases of ovarian epithelial cancers are found to have hereditary causes. These hereditary diseases/syndromes appear to possess heterogeneous, both in genetic anomalies and in clinical manifestations.[6][7]
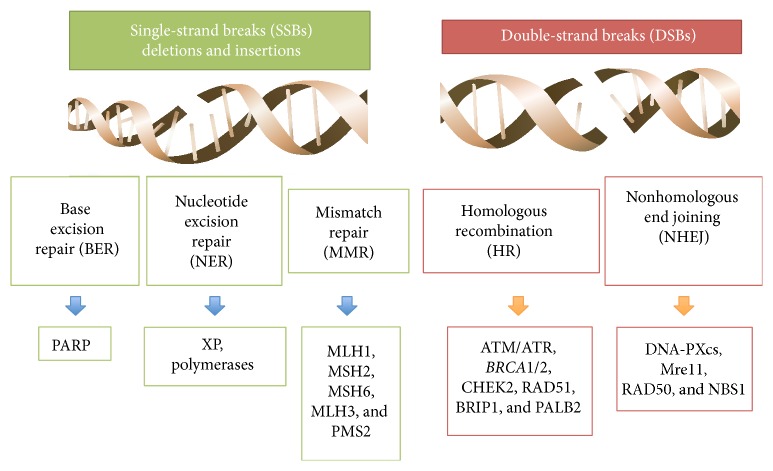
- Majority of these hereditary cancers are caused by two genetic anomalies: a defect in so-called mismatch repair genes named as MLH1, MSH2, MSH6 and PMS2, and in DNA defects repair genes named as BRCA1 and BRCA2.[6][7][1][8]
The role of BRCA1 gene in DNA repair
- BRCA1 is a protein that, through a complex interaction with other proteins such as tumor suppressors, regulators of cell cycle and other DNA repair genes, is involved in DNA repair pathways.[6][7] [9]
- This protein has two domains: amino-terminal RING domain and a BRCT domain. The former posses E3 ubiquitin ligase activity and the later facilitates phospho-protein binding.[9]
- Tumor suppressor role of both domains is highlighted by the fact that mutations in both domains have been found in breast and gynecological malignancies.[9]
- The major role of BRCA1 appears to sense and repair double stranded DNA breaks in homologous recombination.[9]
Binding of BRCA1 to double stranded DNA breaks through its association with the abraxas–RAP80 macro-complex → processing of double stranded DNA breaks through interaction of BRCA1 with CtIP (transcription factor) and the MRN complex → The BRCA1–CtIP complex → CtIP-mediated 5′-end resection of double stranded DNA breaks
- Another role of BRCA1 in Non-homologous end joining (NHEJ) pathway has also been proposed. Though still controversial, it has been suggested that BRCA1 plays a critical function by removal of Non-homologous end joining proteins such as p53-binding protein 1 (53BP1) from double stranded DNA breaks.[9]
- G1/S, S-phase and G2/M checkpoints activation during cell cycle has also been found defective in cells lacking or having mutated BRCA1. A brief interaction of BRCA1 with cell cycle is given below:[9]
Phosphorylation of BRCA1 by ataxia telangiectasia mutated (ATM) or ataxia telangiectasia and Rad3-related protein (ATR) → phosphorylation of p53 → transcriptional induction of the cyclin dependent kinase (CDK) inhibitor p21.

The role of BRCA2 gene in DNA repair
- BRCA2, as opposed to BRCA1 that functions in multiple pathways involving DNA repair, has its primary role in homologous recombination (HR).[6][7][9]
- DNA-binding domain (DBD) of BRCA2 binds single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) and eight BRC repeats. The eight BRC repeats bind RAD51 (a recombinase).[9][11]
- The binding of BRCA2 to RAD51 leads to recruitment of RAD51 to double stranded DNA breaks, an essential step in homologous recombination double stranded DNA repair.[9][11][12]
- After recruitment, BRCA2 helps RAD51 in displacement of replication protein A (RPA) in single stranded DNA. It then prevents nucleation of RAD51 at double stranded DNA and promotes RAD51 filament formation on single stranded DNA.[9][12]
The connection between BRCA1 and BRCA2
- The common pathway that seems to link both BRCA! and BRCA2 proteins is homologous recombination mediated repair.[9][13][14]
- Partner and localizer of BRCA2 (PALB2) physically connects BRCA1 and BRCA2 through N-terminal coiled-coil domain and the C terminus.[9][13][14]
- The interaction between BRCA2 and PALB2 is observed for two critical function in homologous recombination mediated repair: interaction of RAD51 with replication protein A (RPA) in single stranded DNA and recruitment of BRCA2 and RAD51 on the site of DNA damage.[9]
The role of mismatch repair genes
- Mismatch repair genes mutated in pathogenesis of hereditary epithelial ovarian cancer include human MutS homolog (MSH2 and 6), the human MutL homolog (MLH1 and 3), and post-meiotic segregation MutL homolog (PMS2) genes.[6][7][1][8]
- A simplified version of repair mechanism by mismatch repair genes products is described below:[8][15]
MutS homologs (MSHs) recognize the DNA mismatch → MutS homologs (MSHs) recruit MutL homologs (MLHs) → excision of mismatched DNA → DNA polymerase re-synthesizes DNA.
- Cells deficient in mismatch repair mechanism develop high rate of mutations including DNA sequences that include microsatellite repeats, resulting in microsatellite instability. This microsatellite instability has been implicated in impaired or defective signaling transduction, DNA repair and apoptosis, transcriptional regulation and protein translocation, and immune regulation.[1][8][15]
TP53 mutations and loss of tumor suppression
- TP53 is a tumor suppressor gene that encodes for a transcription factor. The transcription factor encoded by TP53, known as p53, is a major regulator of cell cycle.[16][17]
- Called by some as “Guardian of the Genome”, it is involved in variety of cellular functions such as cellular proliferation and cell cycle, apoptosis, and stability & integrity of the genome.[18][17]
Template:Epithelial ovarian cancer
An insight on molecular pathogenesis of epithelial ovarian cancer
Dualistic Model
- This model attempts to explain clinicopathological and molecular genetic features of epithelial tumors by diving them in two subgroups: type I and type II epithelial ovarian tumors.[21][22]
- Another advantage of this classification is that it tries to group precursor lesions with their putative malignant lesions.[21][22]
- Type I tumors generally arise from endometriosis or fallopian tubal related serous epithelium. They are clinically stable, exhibit less aggressive clinical course and a different genetic than that of Type II.[23][24]
- Type II tumors generally arise from fallopian tubal epithelium. They exhibit more aggressive clinical course and a different genetic profile relative to Type I.[23][24]
- Type I tumors are generally characterized by chromosomal stability and somatic mutations that may include KRAS, BRAF, PTEN, PIK3CA, CTNNB1, ARID1A and PPP2R1A. BRCA1 mutation, on the other hand, has not been observed and TP53 mutation is very rare.[21][22][25]
- Type II tumors are characterized by chromosomal instability. The mutations characteristic of high grade tumors, especially TP53 are common. TP53 has been reported in more than 90% of these tumors and a high proportion contains either BRCA mutations or BRCA related mutations such as RAD51, PALB2.[21][26][27]
- A simplified version of this classification is provided below:
| Epithelial Ovarian Cancer | |
|---|---|
| Type I | Type II |
|
|
Dualistic model for serous tumor
- Serous tumor provides, perhaps the most, evidence for the proposed model. Studies suggest that it exhibits distinct morphological and genetic types/stages that may explain the progression from benign tumor (cystadenoma) to low grade serous tumor.[21][22]
- This idea is supported by advances in discovery and understanding of so-called borderline serous tumors. These advances demonstrated that one type of these borderline tumors resembled benign serous tumors in their cinicopathological behavior and were named as “atypical proliferative serous tumor (APST)”. The other type behaved in way closer to low grade serous cancer and were termed as “micropapillary serous carcinoma (MPSC)”.[21][22][28]
- The absence of KRAS and BRAF mutation in serous cystadenoma but presence of these mutations in atypical proliferative serous tumor indicates that these mutations occur somewhat early in transformation of serous cystadenoma into atypical proliferative serous tumor.[21][29]
- More support was provided by studies that showed that genes involved in MAPK pathway were expressed more in micropapillary serous carcinoma than in atypical proliferative serous tumor. In addition, micropapillary serous carcinoma exhibited more chromosomal instability than atypical proliferative serous tumor.[21][28]
- This indicates the step-wise development of low grade serous carcinoma from benign cystadenoma with developemnet of abnormalities in KRAS, BRAF and MAPK pathways. A simplistic version is given below:[21][30][31][32]
ERRB2 (mutation) → PI3K → AKT → mTOR → Cyclin D1 → cell cycle control and cellular survival → Tumor initiation and progression
↓
KRAS → BRAF → MEK → ERK → Cell cycle control and cellular survival → Tumor initiation and progression
PI3K (mutation) → AKT → Tumor initiation and progression
KRAS (mutation) → BRAF → MEK → ERK → Tumor initiation and progression
PI3K (mutation) → Tumor initiation and progression
BRAF (mutation) → MEK → ERK → Cell cycle control and cellular survival → Tumor initiation and progression
*ERK can directly promote tumor initiation, and cellular growth and survival or can promote these through activation of glucose transporter-1 and cyclin D1.[21]
- High grade serous carcinoma, on the other hand, is characterized by mutations rarely found in either of low grade serous carcinoma, micropapillary serous carcinoma and atypical prolferative serous tumor. Of these mutations, TP53 is the most common mutation and is found in >90% of the cases.[21][27]
- While BRCA1 and BRCA2 mutations occur in majority of familial high grade serous carcinoma, inactivation of BRCA1 and/or BRCA2 by indirect mechanisms such as mutation and/or inactivation of promoter occur more frequently in sporadic high grade serous cancer and have been observed in about half of these cancers.[21][33]
- The most noteworthy feature in molecular pathogenesis of high grade serous carcinoma is high level of DNA copy number gains or losses. These gains or losses are diffuse and include foci such as CCNE1 (cyclin E1), NOTCH3, AKT2, RSF1, and PIK3CA.[21][34]
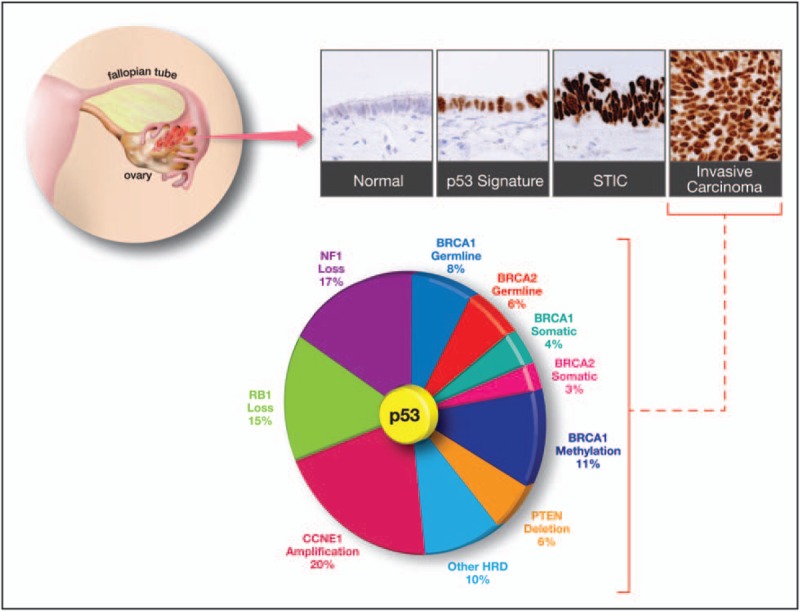
Genetic alterations in clear Cell
- Inactivating mutation of ARID1A. ARID1A encodes for a product that functions in tumor suppression and is observed in half of clear cell cancers.[21][25][37]
- Activating mutation of PIK3CA, also observed in about half of these tumors, results in actiavtion of PI3k pathway.[21][38]
- Deletion of PTEN, observed in about 20% of the cases, results in loss of tumor suppressor gene.[21][39]
- These alterations indicate the importance of PI3K/PTEN pathway in development of clear cell carcinoma of ovary.[21][39]

Genetic alterations in endometrioid tumors
- Low grade endometrioid cancer also exhibits dysregulated either PI3K/PTEN pathway or Wnt/b-catenin signaling pathway. Later has been observed in about 40% of the low grade endometrioid tumors.[21][41][42][43]
- PI3K/PTEN pathway is deregulated either by activating mutations in PIK3CA or inactivation/deletion of PTEN, a tumor suppressor gene. Activating mutations of CTNNB1, that encodes β-catenin, are usually the cause for deregulated Wnt/b-catenin signaling pathway.[21][41][42][43]
- High grade endometrioid carcinoma, on the other hand, dooes not exhibit dysregulated PI3K/PTEN pathway or Wnt/b-catenin signaling pathway but frequently has TP53 mutations present.[21][43]
Genetic alterations in mucinous tumors
- KRAS mutations are present in up to two thirds of these tumors and have also been used as molecular marker.[21][44][45]
References
- ↑ 1.0 1.1 1.2 1.3 Toss A, Tomasello C, Razzaboni E, Contu G, Grandi G, Cagnacci A, Schilder RJ, Cortesi L (2015). "Hereditary ovarian cancer: not only BRCA 1 and 2 genes". Biomed Res Int. 2015: 341723. doi:10.1155/2015/341723. PMC 4449870. PMID 26075229.
- ↑ Devouassoux-Shisheboran M, Genestie C (January 2015). "Pathobiology of ovarian carcinomas". Chin J Cancer. 34 (1): 50–5. doi:10.5732/cjc.014.10273. PMC 4302089. PMID 25556618.
- ↑ 3.0 3.1 Devouassoux-Shisheboran M, Genestie C (January 2015). "Pathobiology of ovarian carcinomas". Chin J Cancer. 34 (1): 50–5. doi:10.5732/cjc.014.10273. PMC 4302089. PMID 25556618.
- ↑ 4.0 4.1 4.2 Lauchlan SC (July 1984). "Metaplasias and neoplasias of Müllerian epithelium". Histopathology. 8 (4): 543–57. PMID 6090303.
- ↑ Devouassoux-Shisheboran M, Genestie C (January 2015). "Pathobiology of ovarian carcinomas". Chin J Cancer. 34 (1): 50–5. doi:10.5732/cjc.014.10273. PMC 4302089. PMID 25556618.
- ↑ 6.0 6.1 6.2 6.3 6.4 Lynch HT, Casey MJ, Snyder CL, Bewtra C, Lynch JF, Butts M, Godwin AK (April 2009). "Hereditary ovarian carcinoma: heterogeneity, molecular genetics, pathology, and management". Mol Oncol. 3 (2): 97–137. doi:10.1016/j.molonc.2009.02.004. PMID 19383374.
- ↑ 7.0 7.1 7.2 7.3 7.4 Neff RT, Senter L, Salani R (August 2017). "BRCA mutation in ovarian cancer: testing, implications and treatment considerations". Ther Adv Med Oncol. 9 (8): 519–531. doi:10.1177/1758834017714993. PMID 28794804.
- ↑ 8.0 8.1 8.2 8.3 Martín-López JV, Fishel R (June 2013). "The mechanism of mismatch repair and the functional analysis of mismatch repair defects in Lynch syndrome". Fam. Cancer. 12 (2): 159–68. doi:10.1007/s10689-013-9635-x. PMC 4235668. PMID 23572416.
- ↑ 9.00 9.01 9.02 9.03 9.04 9.05 9.06 9.07 9.08 9.09 9.10 9.11 9.12 Roy R, Chun J, Powell SN (December 2011). "BRCA1 and BRCA2: different roles in a common pathway of genome protection". Nat. Rev. Cancer. 12 (1): 68–78. doi:10.1038/nrc3181. PMC 4972490. PMID 22193408.
- ↑ Wu W, Koike A, Takeshita T, Ohta T (January 2008). "The ubiquitin E3 ligase activity of BRCA1 and its biological functions". Cell Div. 3: 1. doi:10.1186/1747-1028-3-1. PMC 2254412. PMID 18179693.
- ↑ 11.0 11.1 Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC (October 2010). "The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA". Nat. Struct. Mol. Biol. 17 (10): 1263–5. doi:10.1038/nsmb.1905. PMC 4041013. PMID 20729858.
- ↑ 12.0 12.1 Carreira A, Hilario J, Amitani I, Baskin RJ, Shivji MK, Venkitaraman AR, Kowalczykowski SC (March 2009). "The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51". Cell. 136 (6): 1032–43. doi:10.1016/j.cell.2009.02.019. PMC 2669112. PMID 19303847.
- ↑ 13.0 13.1 Sy SM, Huen MS, Chen J (April 2009). "PALB2 is an integral component of the BRCA complex required for homologous recombination repair". Proc. Natl. Acad. Sci. U.S.A. 106 (17): 7155–60. doi:10.1073/pnas.0811159106. PMC 2678481. PMID 19369211.
- ↑ 14.0 14.1 Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM (June 2006). "Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2". Mol. Cell. 22 (6): 719–29. doi:10.1016/j.molcel.2006.05.022. PMID 16793542.
- ↑ 15.0 15.1 Hsieh P, Yamane K (2008). "DNA mismatch repair: molecular mechanism, cancer, and ageing". Mech. Ageing Dev. 129 (7–8): 391–407. doi:10.1016/j.mad.2008.02.012. PMC 2574955. PMID 18406444.
- ↑ Toss A, Tomasello C, Razzaboni E, Contu G, Grandi G, Cagnacci A, Schilder RJ, Cortesi L (2015). "Hereditary ovarian cancer: not only BRCA 1 and 2 genes". Biomed Res Int. 2015: 341723. doi:10.1155/2015/341723. PMC 4449870. PMID 26075229.
- ↑ 17.0 17.1 Miller M, Shirole N, Tian R, Pal D, Sordella R (2016). "The Evolution of TP53 Mutations: From Loss-of-Function to Separation-of-Function Mutants". J Cancer Biol Res. 4 (4). PMC 5298884. PMID 28191499.
- ↑ Toss A, Tomasello C, Razzaboni E, Contu G, Grandi G, Cagnacci A, Schilder RJ, Cortesi L (2015). "Hereditary ovarian cancer: not only BRCA 1 and 2 genes". Biomed Res Int. 2015: 341723. doi:10.1155/2015/341723. PMC 4449870. PMID 26075229.
- ↑ Pantziarka P (2015). "Primed for cancer: Li Fraumeni Syndrome and the pre-cancerous niche". Ecancermedicalscience. 9: 541. doi:10.3332/ecancer.2015.541. PMC 4462886. PMID 26082798.
- ↑ D'Andrilli G, Giordano A, Bovicelli A (February 2008). "Epithelial ovarian cancer: the role of cell cycle genes in the different histotypes". Open Clin Cancer J. 2: 7–12. doi:10.2174/1874189400802010007. PMC 2490600. PMID 18665245.
- ↑ 21.00 21.01 21.02 21.03 21.04 21.05 21.06 21.07 21.08 21.09 21.10 21.11 21.12 21.13 21.14 21.15 21.16 21.17 21.18 21.19 21.20 Kurman RJ, Shih I (July 2011). "Molecular pathogenesis and extraovarian origin of epithelial ovarian cancer--shifting the paradigm". Hum. Pathol. 42 (7): 918–31. doi:10.1016/j.humpath.2011.03.003. PMID 21683865. Vancouver style error: initials (help)
- ↑ 22.0 22.1 22.2 22.3 22.4 Shih I, Kurman RJ (May 2004). "Ovarian tumorigenesis: a proposed model based on morphological and molecular genetic analysis". Am. J. Pathol. 164 (5): 1511–8. PMID 15111296. Vancouver style error: initials (help)
- ↑ 23.0 23.1 Rojas V, Hirshfield KM, Ganesan S, Rodriguez-Rodriguez L (December 2016). "Molecular Characterization of Epithelial Ovarian Cancer: Implications for Diagnosis and Treatment". Int J Mol Sci. 17 (12). doi:10.3390/ijms17122113. PMC 5187913. PMID 27983698.
- ↑ 24.0 24.1 McCluggage WG (August 2011). "Morphological subtypes of ovarian carcinoma: a review with emphasis on new developments and pathogenesis". Pathology. 43 (5): 420–32. doi:10.1097/PAT.0b013e328348a6e7. PMID 21716157.
- ↑ 25.0 25.1 Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, Yang W, Heravi-Moussavi A, Giuliany R, Chow C, Fee J, Zayed A, Prentice L, Melnyk N, Turashvili G, Delaney AD, Madore J, Yip S, McPherson AW, Ha G, Bell L, Fereday S, Tam A, Galletta L, Tonin PN, Provencher D, Miller D, Jones SJ, Moore RA, Morin GB, Oloumi A, Boyd N, Aparicio SA, Shih I, Mes-Masson AM, Bowtell DD, Hirst M, Gilks B, Marra MA, Huntsman DG (October 2010). "ARID1A mutations in endometriosis-associated ovarian carcinomas". N. Engl. J. Med. 363 (16): 1532–43. doi:10.1056/NEJMoa1008433. PMC 2976679. PMID 20942669. Vancouver style error: initials (help)
- ↑ Ahmed AA, Etemadmoghadam D, Temple J, Lynch AG, Riad M, Sharma R, Stewart C, Fereday S, Caldas C, Defazio A, Bowtell D, Brenton JD (May 2010). "Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary". J. Pathol. 221 (1): 49–56. doi:10.1002/path.2696. PMC 3262968. PMID 20229506.
- ↑ 27.0 27.1 Senturk E, Cohen S, Dottino PR, Martignetti JA (November 2010). "A critical re-appraisal of BRCA1 methylation studies in ovarian cancer". Gynecol. Oncol. 119 (2): 376–83. doi:10.1016/j.ygyno.2010.07.026. PMID 20797776.
- ↑ 28.0 28.1 Burks RT, Sherman ME, Kurman RJ (November 1996). "Micropapillary serous carcinoma of the ovary. A distinctive low-grade carcinoma related to serous borderline tumors". Am. J. Surg. Pathol. 20 (11): 1319–30. PMID 8898836.
- ↑ Ho CL, Kurman RJ, Dehari R, Wang TL, Shih I (October 2004). "Mutations of BRAF and KRAS precede the development of ovarian serous borderline tumors". Cancer Res. 64 (19): 6915–8. doi:10.1158/0008-5472.CAN-04-2067. PMID 15466181. Vancouver style error: initials (help)
- ↑ Singer G, Oldt R, Cohen Y, Wang BG, Sidransky D, Kurman RJ, Shih I (March 2003). "Mutations in BRAF and KRAS characterize the development of low-grade ovarian serous carcinoma". J. Natl. Cancer Inst. 95 (6): 484–6. PMID 12644542. Vancouver style error: initials (help)
- ↑ Mayr D, Hirschmann A, Löhrs U, Diebold J (December 2006). "KRAS and BRAF mutations in ovarian tumors: a comprehensive study of invasive carcinomas, borderline tumors and extraovarian implants". Gynecol. Oncol. 103 (3): 883–7. doi:10.1016/j.ygyno.2006.05.029. PMID 16806438.
- ↑ Hsu CY, Bristow R, Cha MS, Wang BG, Ho CL, Kurman RJ, Wang TL, Shih I (October 2004). "Characterization of active mitogen-activated protein kinase in ovarian serous carcinomas". Clin. Cancer Res. 10 (19): 6432–6. doi:10.1158/1078-0432.CCR-04-0893. PMID 15475429. Vancouver style error: initials (help)
- ↑ May T, Virtanen C, Sharma M, Milea A, Begley H, Rosen B, Murphy KJ, Brown TJ, Shaw PA (April 2010). "Low malignant potential tumors with micropapillary features are molecularly similar to low-grade serous carcinoma of the ovary". Gynecol. Oncol. 117 (1): 9–17. doi:10.1016/j.ygyno.2010.01.006. PMID 20117829.
- ↑ Nakayama K, Nakayama N, Jinawath N, Salani R, Kurman RJ, Shih I, Wang TL (June 2007). "Amplicon profiles in ovarian serous carcinomas". Int. J. Cancer. 120 (12): 2613–7. doi:10.1002/ijc.22609. PMID 17351921. Vancouver style error: initials (help)
- ↑ 35.0 35.1 Kroeger PT, Drapkin R (February 2017). "Pathogenesis and heterogeneity of ovarian cancer". Curr. Opin. Obstet. Gynecol. 29 (1): 26–34. doi:10.1097/GCO.0000000000000340. PMC 5201412. PMID 27898521.
- ↑ Karst AM, Drapkin R (2010). "Ovarian cancer pathogenesis: a model in evolution". J Oncol. 2010: 932371. doi:10.1155/2010/932371. PMC 2739011. PMID 19746182.
- ↑ Jones S, Wang TL, Shih I, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA, Vogelstein B, Kinzler KW, Velculescu VE, Papadopoulos N (October 2010). "Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma". Science. 330 (6001): 228–31. doi:10.1126/science.1196333. PMC 3076894. PMID 20826764. Vancouver style error: initials (help)
- ↑ Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, Hooi CS, Cristiano BE, Pearson RB, Phillips WA (November 2004). "Mutation of the PIK3CA gene in ovarian and breast cancer". Cancer Res. 64 (21): 7678–81. doi:10.1158/0008-5472.CAN-04-2933. PMID 15520168.
- ↑ 39.0 39.1 Sato N, Tsunoda H, Nishida M, Morishita Y, Takimoto Y, Kubo T, Noguchi M (December 2000). "Loss of heterozygosity on 10q23.3 and mutation of the tumor suppressor gene PTEN in benign endometrial cyst of the ovary: possible sequence progression from benign endometrial cyst to endometrioid carcinoma and clear cell carcinoma of the ovary". Cancer Res. 60 (24): 7052–6. PMID 11156411.
- ↑ Chandler RL, Damrauer JS, Raab JR, Schisler JC, Wilkerson MD, Didion JP, Starmer J, Serber D, Yee D, Xiong J, Darr DB, Pardo-Manuel de Villena F, Kim WY, Magnuson T (January 2015). "Coexistent ARID1A-PIK3CA mutations promote ovarian clear-cell tumorigenesis through pro-tumorigenic inflammatory cytokine signalling". Nat Commun. 6: 6118. doi:10.1038/ncomms7118. PMC 4308813. PMID 25625625.
- ↑ 41.0 41.1 Obata K, Morland SJ, Watson RH, Hitchcock A, Chenevix-Trench G, Thomas EJ, Campbell IG (May 1998). "Frequent PTEN/MMAC mutations in endometrioid but not serous or mucinous epithelial ovarian tumors". Cancer Res. 58 (10): 2095–7. PMID 9605750.
- ↑ 42.0 42.1 Catasús L, Bussaglia E, Rodrguez I, Gallardo A, Pons C, Irving JA, Prat J (November 2004). "Molecular genetic alterations in endometrioid carcinomas of the ovary: similar frequency of beta-catenin abnormalities but lower rate of microsatellite instability and PTEN alterations than in uterine endometrioid carcinomas". Hum. Pathol. 35 (11): 1360–8. doi:10.1016/j.humpath.2004.07.019. PMID 15668893.
- ↑ 43.0 43.1 43.2 Wu R, Hendrix-Lucas N, Kuick R, Zhai Y, Schwartz DR, Akyol A, Hanash S, Misek DE, Katabuchi H, Williams BO, Fearon ER, Cho KR (April 2007). "Mouse model of human ovarian endometrioid adenocarcinoma based on somatic defects in the Wnt/beta-catenin and PI3K/Pten signaling pathways". Cancer Cell. 11 (4): 321–33. doi:10.1016/j.ccr.2007.02.016. PMID 17418409.
- ↑ Ichikawa Y, Nishida M, Suzuki H, Yoshida S, Tsunoda H, Kubo T, Uchida K, Miwa M (January 1994). "Mutation of K-ras protooncogene is associated with histological subtypes in human mucinous ovarian tumors". Cancer Res. 54 (1): 33–5. PMID 8261457.
- ↑ Gemignani ML, Schlaerth AC, Bogomolniy F, Barakat RR, Lin O, Soslow R, Venkatraman E, Boyd J (August 2003). "Role of KRAS and BRAF gene mutations in mucinous ovarian carcinoma". Gynecol. Oncol. 90 (2): 378–81. PMID 12893203.