Bacteria
| Bacteria Fossil range: Archean or earlier - Recent | ||
|---|---|---|
 Escherichia coli cells magnified 25,000 times
| ||
| Scientific classification | ||
| ||
| Phyla | ||
|
Acidobacteria |
|
WikiDoc Resources for Bacteria |
|
Articles |
|---|
|
Most recent articles on Bacteria |
|
Media |
|
Evidence Based Medicine |
|
Clinical Trials |
|
Ongoing Trials on Bacteria at Clinical Trials.gov Clinical Trials on Bacteria at Google
|
|
Guidelines / Policies / Govt |
|
US National Guidelines Clearinghouse on Bacteria
|
|
Books |
|
News |
|
Commentary |
|
Definitions |
|
Patient Resources / Community |
|
Directions to Hospitals Treating Bacteria Risk calculators and risk factors for Bacteria
|
|
Healthcare Provider Resources |
|
Causes & Risk Factors for Bacteria |
|
Continuing Medical Education (CME) |
|
International |
|
|
|
Business |
|
Experimental / Informatics |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]
Bacteria (singular: bacterium) are unicellular microorganisms. Typically a few micrometres in length, bacteria have a wide range of shapes, ranging from spheres to rods to spirals. Bacteria are ubiquitous in every habitat on Earth, growing in soil, Hot spring|acidic hot springs, radioactive waste,[1] seawater, and deep in the Earth's crust. There are typically 40 million bacterial cells in a gram of soil and a million bacterial cells in a millilitre of fresh water; in all, there are approximately five nonillion (5×1030) bacteria on Earth,[2] forming much of the world's biomass.[3] Bacteria are vital in recycling nutrients, and many important steps in nutrient cycles depend on bacteria, such as the fixation of nitrogen from the atmosphere. However, most of these bacteria have not been characterized, and only about half of the phyla of bacteria have species that can be cultured in the laboratory.[4] The study of bacteria is known as bacteriology, a branch of microbiology.
There are approximately ten times as many bacterial cells as human cells in the human body, with large numbers of bacteria on the skin and in the digestive tract.[5] Although the vast majority of these bacteria are rendered harmless by the protective effects of the immune system, and a few are beneficial, some are pathogenic bacteria and cause infectious diseases, including cholera, syphilis, anthrax, leprosy and bubonic plague. The most common fatal bacterial diseases are respiratory infections, with tuberculosis alone killing about 2 million people a year, mostly in sub-Saharan Africa.[6] In developed countries, antibiotics are used to treat bacterial infections and in various agricultural processes, so antibiotic resistance is becoming common. In industry, bacteria are important in processes such as sewage treatment, the production of cheese and yoghurt, and the manufacture of antibiotics and other chemicals.[7]
Bacteria are prokaryotes. Unlike cells of animals and other eukaryotes, bacterial cells do not contain a nucleus and rarely harbour membrane-bound organelles. Although the term bacteria traditionally included all prokaryotes, the scientific classification changed after the discovery in the 1990s that prokaryotic life consists of two very different groups of organisms that evolved independently from an ancient common ancestor. These evolutionary domains are called Bacteria and Archaea.[8]
History of bacteriology

Bacteria were first observed by Antonie van Leeuwenhoek in 1676, using a single-lens microscope of his own design.[9] He called them "animalcules" and published his observations in a series of letters to the Royal Society.[10][11][12] The name bacterium was introduced much later, by Christian Gottfried Ehrenberg in 1838, and is derived from the Greek word βακτήριον -α , bacterion -a , meaning "small staff".[13]
Louis Pasteur demonstrated in 1859 that the fermentation process is caused by the growth of microorganisms, and that this growth is not due to spontaneous generation. (Yeasts and molds, commonly associated with fermentation, are not bacteria, but rather fungi.) Along with his contemporary, Robert Koch, Pasteur was an early advocate of the germ theory of disease.[14] Robert Koch was a pioneer in medical microbiology and worked on cholera, anthrax and tuberculosis. In his research into tuberculosis, Koch finally proved the germ theory, for which he was awarded a Nobel Prize in 1905.[15] In Koch's postulates, he set out criteria to test if an organism is the cause of a disease; these postulates are still used today.[16]
Though it was known in the nineteenth century that bacteria are the cause of many diseases, no effective antibacterial treatments were available.[17] In 1910, Paul Ehrlich developed the first antibiotic, by changing dyes that selectively stained Treponema pallidum—the spirochaete that causes syphilis—into compounds that selectively killed the pathogen.[18] Ehrlich had been awarded a 1908 Nobel Prize for his work on immunology, and pioneered the use of stains to detect and identify bacteria, with his work being the basis of the Gram stain and the Ziehl-Neelsen stain.[19]
A major step forward in the study of bacteria was the recognition in 1977 by Carl Woese that archaea have a separate line of evolutionary descent from bacteria.[20] This new phylogenetic taxonomy was based on the sequencing of 16S ribosomal RNA, and divided prokaryotes into two evolutionary domains, as part of the three-domain system.[21]
Origin and early evolution
The ancestors of modern bacteria were single-celled microorganisms that were the first forms of life to develop on earth, about 4 billion years ago. For about 3 billion years, all organisms were microscopic, and bacteria and archaea were the dominant forms of life.[22][23] Although bacterial fossils exist, such as stromatolites, their lack of distinctive morphology prevents them from being used to examine the past history of bacterial evolution, or to date the time of origin of a particular bacterial species. However, gene sequences can be used to reconstruct the bacterial phylogeny, and these studies indicate that bacteria diverged first from the archaeal/eukaryotic lineage.[24] The most recent common ancestor of bacteria and archaea was probably a hyperthermophile that lived about 2.5 billion–3.2 billion years ago.[25][26]
Bacteria were also involved in the second great evolutionary divergence, that of the archaea and eukaryotes. Here, eukaryotes resulted from ancient bacteria entering into endosymbiotic associations with the ancestors of eukaryotic cells, which were themselves possibly related to the Archaea.[27][28] This involved the engulfment by proto-eukaryotic cells of alpha-proteobacterial symbionts to form either mitochondria or hydrogenosomes, which are still being found in all known Eukarya (sometimes in highly reduced form, e.g. in ancient "amitochondrial" protozoa). Later on, an independent second engulfment by some mitochondria-containing eukaryotes of cyanobacterial-like organisms led to the formation of chloroplasts in algae and plants. There are even some algal groups known that clearly originated from subsequent events of endosymbiosis by heterotrophic eukaryotic hosts engulfing a eukaryotic algae that developed into "second-generation" plastids.[29][30]
Morphology
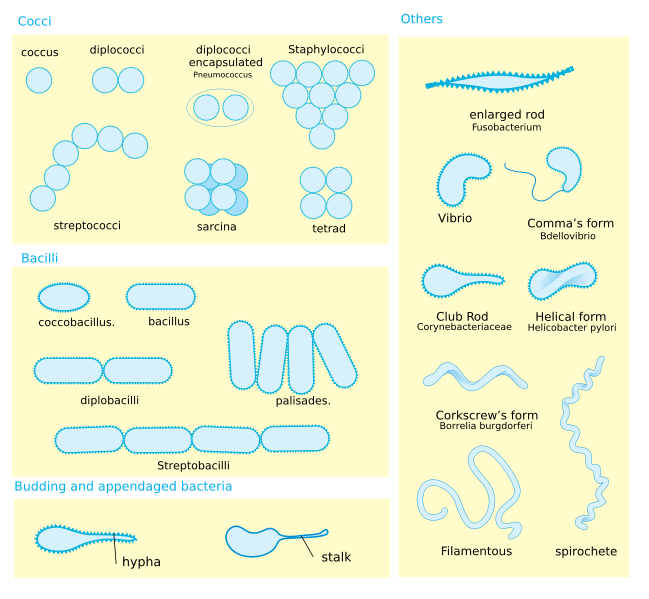
Bacteria display a wide diversity of shapes and sizes, called morphologies. Bacterial cells are about one tenth the size of eukaryotic cells and are typically 0.5–5.0 micrometres in length. However, a few species–for example Thiomargarita namibiensis and Epulopiscium fishelsoni–are up to half a millimetre long and are visible to the unaided eye.[31] Among the smallest bacteria are members of the genus Mycoplasma, which measure only 0.3 micrometres, as small as the largest viruses.[32]
Most bacterial species are either spherical, called cocci (sing. coccus, from Greek kókkos, grain, seed) or rod-shaped, called bacilli (sing. bacillus, from Latin baculus, stick). Some rod-shaped bacteria, called vibrio, are slightly curved or comma-shaped; others, can be spiral-shaped, called spirilla, or tightly coiled, called spirochaetes. A small number of species even have tetrahedral or cuboidal shapes.[33] This wide variety of shapes is determined by the bacterial cell wall and cytoskeleton, and is important because it can influence the ability of bacteria to acquire nutrients, attach to surfaces, swim through liquids and escape predators.[34][35]
Many bacterial species exist simply as single cells, others associate in characteristic patterns: Neisseria form diploids (pairs), Streptococcus form chains, and Staphylococcus group together in "bunch of grapes" clusters. Bacteria can also be elongated to form filaments, for example the Actinobacteria. Filamentous bacteria are often surrounded by a sheath that contains many individual cells; certain types, such as species of the genus Nocardia, even form complex, branched filaments, similar in appearance to fungal mycelia.[36]
Bacteria often attach to surfaces and form dense aggregations called biofilms or bacterial mats. These films can range from a few micrometers in thickness to up to half a meter in depth, and may contain multiple species of bacteria, protists and archaea. Bacteria living in biofilms display a complex arrangement of cells and extracellular components, forming secondary structures such as microcolonies, through which there are networks of channels to enable better diffusion of nutrients.[37][38] In natural environments, such as soil or the surfaces of plants, the majority of bacteria are bound to surfaces in biofilms.[39] Biofilms are also important for chronic bacterial infections and infections of implanted medical devices, as bacteria protected within these structures are much harder to kill than individual bacteria.[40]
Even more complex morphological changes are sometimes possible. For example, when starved of amino acids, Myxobacteria detect surrounding cells in a process known as quorum sensing, migrate towards each other, and aggregate to form fruiting bodies up to 500 micrometres long and containing approximately 100,000 bacterial cells.[41] In these fruiting bodies, the bacteria perform separate tasks; this type of cooperation is a simple type of multicellular organisation. For example, about one in 10 cells migrate to the top of these fruiting bodies and differentiate into a specialised dormant state called myxospores, which are more resistant to desiccation and other adverse environmental conditions than are ordinary cells.[42]
Cellular structure
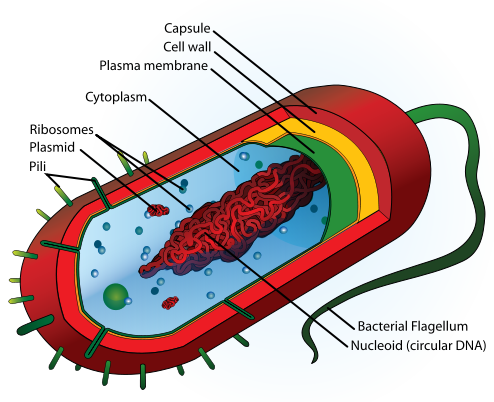
Intracellular structures
The bacterial cell is surrounded by a lipid membrane, or cell membrane, which encompasses the contents of the cell and acts as a barrier to hold nutrients, proteins and other essential components of the cytoplasm within the cell. As they are prokaryotes, bacteria do not tend to have membrane-bound organelles in their cytoplasm and thus contain few intracellular structures. They consequently lack a nucleus, mitochondria, chloroplasts and the other organelles present in eukaryotic cells, such as the Golgi apparatus and endoplasmic reticulum.[43] However, recent research is identifying increasing amounts of structural complexity in bacteria, such as the discovery of the prokaryotic cytoskeleton.[44][45]
Many important biochemical reactions, such as energy generation, occur due to concentration gradients across membranes, creating a potential difference analogous to a battery. The absence of internal membranes in bacteria means these reactions, such as electron transport, occur across the cell membrane, between the cytoplasm and the periplasmic space.[46] Additionally, while some transporter proteins consume chemical energy, others harness concentration gradients to import nutrients across the cell membrane or to expel undesired molecules from the cytoplasm.
Bacteria do not have a membrane-bound nucleus, and their genetic material is typically a single circular chromosome located in the cytoplasm in an irregularly shaped body called the nucleoid.[47] The nucleoid contains the chromosome with associated proteins and RNA. Like all living organisms, bacteria contain ribosomes for the production of proteins, but the structure of the bacterial ribosome is different from those of eukaryotes and Archaea.[48] The order Planctomycetes are an exception to the general absence of internal membranes in bacteria, because they have a membrane around their nucleoid and contain other membrane-bound cellular structures.[49]
Some bacteria produce intracellular nutrient storage granules, such as glycogen,[50] polyphosphate,[51] sulfur[52] or polyhydroxyalkanoates.[53] These granules enable bacteria to store compounds for later use. Certain bacterial species, such as the photosynthetic Cyanobacteria, produce internal gas vesicles, which they use to regulate their buoyancy - allowing them to move up or down into water layers with different light intensities and nutrient levels.[54]
Extracellular structures
Around the outside of the cell membrane is the bacterial cell wall. Bacterial cell walls are made of peptidoglycan (called murein in older sources), which is made from polysaccharide chains cross-linked by unusual peptides containing D-amino acids.[55] Bacterial cell walls are different from the cell walls of plants and fungi, which are made of cellulose and chitin, respectively.[56] The cell wall of bacteria is also distinct from that of Archaea, which do not contain peptidoglycan. The cell wall is essential to the survival of many bacteria, and the antibiotic penicillin is able to kill bacteria by inhibiting a step in the synthesis of peptidoglycan.[56]
There are broadly speaking two different types of cell wall in bacteria, called Gram-positive and Gram-negative. The names originate from the reaction of cells to the Gram stain, a test long-employed for the classification of bacterial species.[57]
Gram-positive bacteria possess a thick cell wall containing many layers of peptidoglycan and teichoic acids. In contrast, Gram-negative bacteria have a relatively thin cell wall consisting of a few layers of peptidoglycan surrounded by a second lipid membrane containing lipopolysaccharides and lipoproteins. Most bacteria have the Gram-negative cell wall, and only the Firmicutes and Actinobacteria (previously known as the low G+C and high G+C Gram-positive bacteria, respectively) have the alternative Gram-positive arrangement.[58] These differences in structure can produce differences in antibiotic susceptibility; for instance, vancomycin can kill only Gram-positive bacteria and is ineffective against Gram-negative pathogens, such as Haemophilus influenzae or Pseudomonas aeruginosa.[59]
In many bacteria an S-layer of rigidly arrayed protein molecules covers the outside of the cell.[60] This layer provides chemical and physical protection for the cell surface and can act as a macromolecular diffusion barrier. S-layers have diverse but mostly poorly understood functions, but are known to act as virulence factors in Campylobacter and contain surface enzymes in Bacillus stearothermophilus.[61]

Flagella are rigid protein structures, about 20 nanometres in diameter and up to 20 micrometres in length, that are used for motility. Flagella are driven by the energy released by the transfer of ions down an electrochemical gradient across the cell membrane.[62]
Fimbriae are fine filaments of protein, just 2–10 nanometres in diameter and up to several micrometers in length. They are distributed over the surface of the cell, and resemble fine hairs when seen under the electron microscope. Fimbriae are believed to be involved in attachment to solid surfaces or to other cells and are essential for the virulence of some bacterial pathogens.[63] Pili (sing. pilus) are cellular appendages, slightly larger than fimbriae, that can transfer genetic material between bacterial cells in a process called conjugation (see bacterial genetics, below).[64]
Capsules or slime layers are produced by many bacteria to surround their cells, and vary in structural complexity: ranging from a disorganised slime layer of extra-cellular polymer, to a highly structured capsule or glycocalyx. These structures can protect cells from engulfment by eukaryotic cells, such as macrophages.[65] They can also act as antigens and be involved in cell recognition, as well as aiding attachment to surfaces and the formation of biofilms.[66]
The assembly of these extracellular structures is dependent on bacterial secretion systems. These transfer proteins from the cytoplasm into the periplasm or into the environment around the cell. Many types of secretion systems are known and these structures are often essential for the virulence of pathogens, so are intensively studied.[67]
Endospores
Certain genera of Gram-positive bacteria, such as Bacillus, Clostridium, Sporohalobacter, Anaerobacter and Heliobacterium, can form highly resistant, dormant structures called endospores.[68] In almost all cases, one endospore is formed and this is not a reproductive process, although Anaerobacter can make up to seven endospores in a single cell.[69] Endospores have a central core of cytoplasm containing DNA and ribosomes surrounded by a cortex layer and protected by an impermeable and rigid coat.
Endospores show no detectable metabolism and can survive extreme physical and chemical stresses, such as high levels of UV light, gamma radiation, detergents, disinfectants, heat, pressure and desiccation.[70] In this dormant state, these organisms may remain viable for millions of years,[71][72] and endospores even allow bacteria to survive exposure to the vacuum and radiation in space.[73] Endospore-forming bacteria can also cause disease: for example, anthrax can be contracted by the inhalation of Bacillus anthracis endospores, and contamination of deep puncture wounds with Clostridium tetani endospores causes tetanus.[74]
Metabolism

In contrast to higher organisms, bacteria exhibit an extremely wide variety of metabolic types.[75] The distribution of metabolic traits within a group of bacteria has traditionally been used to define their taxonomy, but these traits often do not correspond with modern genetic classifications.[76] Bacterial metabolism is classified on the basis of three major criteria: the kind of energy used for growth, the source of carbon, and the electron donors used for growth. An additional criterion of respiratory microorganisms are the electron acceptors used for aerobic or anaerobic respiration.[77]
Carbon metabolism in bacteria is either heterotrophic, where organic carbon compounds are used as carbon sources, or autotrophic, meaning that cellular carbon is obtained by fixing carbon dioxide. Typical autotrophic bacteria are phototrophic cyanobacteria, green sulfur-bacteria and some purple bacteria, but also many chemolithotrophic species, such as nitrifying or sulfur-oxidising bacteria.[78] Energy metabolism of bacteria is either based on phototrophy, the use of light through photosynthesis, or on chemotrophy, the use of chemical substances for energy, which are mostly oxidised at the expense of oxygen or alternative electron acceptors (aerobic/anaerobic respiration).
Finally, bacteria are further divided into lithotrophs that use inorganic electron donors and organotrophs that use organic compounds as electron donors. Chemotrophic organisms use the respective electron donors for energy conservation (by aerobic/anaerobic respiration or fermentation) and biosynthetic reactions (e.g. carbon dioxide fixation), whereas phototrophic organisms use them only for biosynthetic purposes. Respiratory organisms use chemical compounds as a source of energy by taking electrons from the reduced substrate and transferring them to a terminal electron acceptor in a redox reaction. This reaction releases energy that can be used to synthesise ATP and drive metabolism. In aerobic organisms, oxygen is used as the electron acceptor. In anaerobic organisms other inorganic compounds, such as nitrate, sulfate or carbon dioxide are used as electron acceptors. This leads to the ecologically important processes of denitrification, sulfate reduction and acetogenesis, respectively.
Another way of life of chemotrophs in the absence of possible electron acceptors is fermentation, where the electrons taken from the reduced substrates are transferred to oxidised intermediates to generate reduced fermentation products (e.g. lactate, ethanol, hydrogen, butyric acid). Fermentation is possible, because the energy content of the substrates is higher than that of the products, which allows the organisms to synthesise ATP and drive their metabolism.[79][80]
These processes are also important in biological responses to pollution; for example, sulfate-reducing bacteria are largely responsible for the production of the highly toxic forms of mercury (methyl- and dimethylmercury) in the environment.[81] Non-respiratory anaerobes use fermentation to generate energy and reducing power, secreting metabolic by-products (such as ethanol in brewing) as waste. Facultative anaerobes can switch between fermentation and different terminal electron acceptors depending on the environmental conditions in which they find themselves.
Lithotrophic bacteria can use inorganic compounds as a source of energy. Common inorganic electron donors are hydrogen, carbon monoxide, ammonia (leading to nitrification), ferrous iron and other reduced metal ions, and several reduced sulfur compounds. Unusually, the gas methane can be used by methanotrophic bacteria as both a source of electrons and a substrate for carbon anabolism.[82] In both aerobic phototrophy and chemolithotrophy, oxygen is used as a terminal electron acceptor, while under anaerobic conditions inorganic compounds are used instead. Most lithotrophic organisms are autotrophic, whereas organotrophic organisms are heterotrophic.
In addition to fixing carbon dioxide in photosynthesis, some bacteria also fix nitrogen gas (nitrogen fixation) using the enzyme nitrogenase. This environmentally important trait can be found in bacteria of nearly all the metabolic types listed above, but is not universal.[83]
Growth and reproduction
Unlike multicellular organisms, increases in the size of bacteria (cell growth) and their reproduction by cell division are tightly linked in unicellular organisms. Bacteria grow to a fixed size and then reproduce through binary fission, a form of asexual reproduction.[84] Under optimal conditions, bacteria can grow and divide extremely rapidly, and bacterial populations can double as quickly as every 9.8 minutes.[85] In cell division, two identical clone daughter cells are produced. Some bacteria, while still reproducing asexually, form more complex reproductive structures that help disperse the newly-formed daughter cells. Examples include fruiting body formation by Myxobacteria and arial hyphae formation by Streptomyces, or budding. Budding involves a cell forming a protrusion that breaks away and produces a daughter cell.

In the laboratory, bacteria are usually grown using solid or liquid media. Solid growth media such as agar plates are used to isolate pure cultures of a bacterial strain. However, liquid growth media are used when measurement of growth or large volumes of cells are required. Growth in stirred liquid media occurs as an even cell suspension, making the cultures easy to divide and transfer, although isolating single bacteria from liquid media is difficult. The use of selective media (media with specific nutrients added or deficient, or with antibiotics added) can help identify specific organisms.[87]
Most laboratory techniques for growing bacteria use high levels of nutrients to produce large amounts of cells cheaply and quickly. However, in natural environments nutrients are limited, meaning that bacteria cannot continue to reproduce indefinitely. This nutrient limitation has led the evolution of different growth strategies (see r/K selection theory). Some organisms can grow extremely rapidly when nutrients become available, such as the formation of algal (and cyanobacterial) blooms that often occur in lakes during the summer.[88] Other organisms have adaptations to harsh environments, such as the production of multiple antibiotics by Streptomyces that inhibit the growth of competing microorganisms.[89] In nature, many organisms live in communities (e.g. biofilms) which may allow for increased supply of nutrients and protection from environmental stresses.[39] These relationships can be essential for growth of a particular organism or group of organisms (syntrophy).[90]
Bacterial growth follows three phases. When a population of bacteria first enter a high-nutrient environment that allows growth, the cells need to adapt to their new environment. The first phase of growth is the lag phase, a period of slow growth when the cells are adapting to the high-nutrient environment and preparing for fast growth. The lag phase has high biosynthesis rates, as proteins necessary for rapid growth are produced.[91] The second phase of growth is the logarithmic phase (log phase), also known as the exponential phase. The log phase is marked by rapid exponential growth. The rate at which cells grow during this phase is known as the growth rate (k), and the time it takes the cells to double is known as the generation time (g). During log phase, nutrients are metabolised at maximum speed until one of the nutrients is depleted and starts limiting growth. The final phase of growth is the stationary phase and is caused by depleted nutrients. The cells reduce their metabolic activity and consume non-essential cellular proteins. The stationary phase is a transition from rapid growth to a stress response state and there is increased expression of genes involved in DNA repair, antioxidant metabolism and nutrient transport.[92]
Genetics
Most bacteria have a single circular chromosome that can range in size from only 160,000 base pairs in the endosymbiotic bacteria Candidatus Carsonella ruddii,[93] to 12,200,000 base pairs in the soil-dwelling bacteria Sorangium cellulosum.[94] Spirochaetes of the genus Borrelia are a notable exception to this arrangement, with bacteria such as Borrelia burgdorferi, the cause of Lyme disease, containing a single linear chromosome.[95] The genes in bacterial genomes are usually a single continuous stretch of DNA and although several different types of introns do exist in bacteria, these are much more rare than in eukaryotes.[96]
Bacteria may also contain plasmids, which are small extra-chromosomal DNAs that may contain genes for antibiotic resistance or virulence factors. Another type of bacterial DNA are integrated viruses (bacteriophages). Many types of bacteriophage exist, some simply infect and lyse their host bacteria, while others insert into the bacterial chromosome. A bacteriophage can contain genes that contribute to its host's phenotype: for example, in the evolution of Escherichia coli O157:H7 and Clostridium botulinum, the toxin genes in an integrated phage converted a harmless ancestral bacteria into a lethal pathogen.[97]
Bacteria, as asexual organisms, inherit identical copies of their parent's genes (i.e., they are clonal). However, all bacteria can evolve by selection on changes to their genetic material DNA caused by genetic recombination or mutations. Mutations come from errors made during the replication of DNA or from exposure to mutagens. Mutation rates vary widely among different species of bacteria and even among different clones of a single species of bacteria.[98] Genetic changes in bacterial genomes come from either random mutation during replication or "stress-directed mutation", where genes involved in a particular growth-limiting process have an increased mutation rate.[99]
Some bacteria also transfer genetic material between cells. This can occur in three main ways. Firstly, bacteria can take up exogenous DNA from their environment, in a process called transformation. Genes can also be transferred by the process of transduction, when the integration of a bacteriophage introduces foreign DNA into the chromosome. The third method of gene transfer is bacterial conjugation, where DNA is transferred through direct cell contact. This gene acquisition from other bacteria or the environment is called horizontal gene transfer and may be common under natural conditions.[100] Gene transfer is particularly important in antibiotic resistance as it allows the rapid transfer of resistance genes between different pathogens.[101]
Movement

Motile bacteria can move using flagella, bacterial gliding, twitching motility or changes of buoyancy.[102] In twitching motility, bacterial use their type IV pili as a grappling hook, repeatedly extending it, anchoring it and then retracting it with remarkable force (>80 pN).[103]
Bacterial species differ in the number and arrangement of flagella on their surface; some have a single flagellum (monotrichous), a flagellum at each end (amphitrichous), clusters of flagella at the poles of the cell (lophotrichous), while others have flagella distributed over the entire surface of the cell (peritrichous). The bacterial flagella is the best-understood motility structure in any organism and is made of about 20 proteins, with approximately another 30 proteins required for its regulation and assembly.[102] The flagellum is a rotating structure driven by a motor at the base that uses the electrochemical gradient across the membrane for power. This motor drives the motion of the filament, which acts as a propeller. Many bacteria (such as E. coli) have two distinct modes of movement: forward movement (swimming) and tumbling. The tumbling allows them to reorient and makes their movement a three-dimensional random walk.[104] (See external links below for link to videos.) The flagella of a unique group of bacteria, the spirochaetes, are found between two membranes in the periplasmic space. They have a distinctive helical body that twists about as it moves.[102]
Motile bacteria are attracted or repelled by certain stimuli in behaviors called taxes: these include chemotaxis, phototaxis and magnetotaxis.[105][106] In one peculiar group, the myxobacteria, individual bacteria move together to form waves of cells that then differentiate to form fruiting bodies containing spores.[107] The myxobacteria move only when on solid surfaces, unlike E. coli which is motile in liquid or solid media.
Several Listeria and Shigella species move inside host cells by usurping the cytoskeleton, which is normally used to move organelles inside the cell. By promoting actin polymerization at one pole of their cells, they can form a kind of tail that pushes them through the host cell's cytoplasm.[108]
Classification and identification

Classification seeks to describe the diversity of bacterial species by naming and grouping organisms based on similarities. Bacteria can be classified on the basis of cell structure, cellular metabolism or on differences in cell components such as DNA, fatty acids, pigments, antigens and quinones.[87] While these schemes allowed the identification and classification of bacterial strains, it was unclear whether these differences represented variation between distinct species or between strains of the same species. This uncertainty was due to the lack of distinctive structures in most bacteria, as well as lateral gene transfer between unrelated species.[109] Due to lateral gene transfer, some closely related bacteria can have very different morphologies and metabolisms. To overcome this uncertainty, modern bacterial classification emphasizes molecular systematics, using genetic techniques such as guanine cytosine ratio determination, genome-genome hybridization, as well as sequencing genes that have not undergone extensive lateral gene transfer, such as the rRNA gene.[110] Classification of bacteria is determined by publication in the International Journal of Systematic Bacteriology,[111] and Bergey's Manual of Systematic Bacteriology.[112]
The term "bacteria" was traditionally applied to all microscopic, single-celled prokaryotes. However, molecular systematics showed prokaryotic life to consist of two separate domains, originally called Eubacteria and Archaebacteria, but now called Bacteria and Archaea that evolved independently from an ancient common ancestor.[113] The archaea and eukaryotes are more closely-related to each other than either is to the bacteria. These two domains, along with Eukarya, are the basis of the three-domain system, which is currently the most widely used classification system in microbiolology.[114] However, due to the relatively recent introduction of molecular systematics and a rapid increase in the number of genome sequences that are available, bacterial classification remains a changing and expanding field.[4][115] For example, a few biologists argue that the Archaea and Eukaryotes evolved from Gram-positive bacteria.[116]
Identification of bacteria in the laboratory is particularly relevant in medicine, where the correct treatment is determined by the bacterial species causing an infection. Consequently, the need to identify human pathogens was a major impetus for the development of techniques to identify bacteria.

The Gram stain, developed in 1884 by Hans Christian Gram, characterises bacteria based on the structural characteristics of their cell walls.[57] The thick layers of peptidoglycan in the "Gram-positive" cell wall stain purple, while the thin "Gram-negative" cell wall appears pink. By combining morphology and Gram-staining, most bacteria can be classified as belonging to one of four groups (Gram-positive cocci, Gram-positive bacilli, Gram-negative cocci and Gram-negative bacilli). Some organisms are best identified by stains other than the Gram stain, particularly mycobacteria or Nocardia, which show acid-fastness on Ziehl–Neelsen or similar stains.[118] Other organisms may need to be identified by their growth in special media, or by other techniques, such as serology.
Culture techniques are designed to promote the growth and identify particular bacteria, while restricting the growth of the other bacteria in the sample. Often these techniques are designed for specific specimens; for example, a sputum sample will be treated to identify organisms that cause pneumonia, while stool specimens are cultured on selective media to identify organisms that cause diarrhoea, while preventing growth of non-pathogenic bacteria. Specimens that are normally sterile, such as blood, urine or spinal fluid, are cultured under conditions designed to grow all possible organisms.[119][87] Once a pathogenic organism has been isolated, it can be further characterised by its morphology, growth patterns such as (aerobic or anaerobic growth, patterns of hemolysis) and staining.
As with bacterial classification, identification of bacteria is increasingly using molecular methods. Diagnostics using such DNA-based tools, such as polymerase chain reaction, are increasingly popular due to their specificity and speed, compared to culture-based methods.[120] These methods also allow the detection and identification of "viable but nonculturable" cells that are metabolically active but non-dividing.[121] However, even using these improved methods, the total number of bacterial species is not known and cannot even be estimated with any certainty. Attempts to quantify bacterial diversity have ranged from 107 to 109 total species, but even these diverse estimates may be out by many orders of magnitude.[122][123]
Interactions with other organisms
Despite their apparent simplicity, bacteria can form complex associations with other organisms. These symbiotic associations can be divided into parasitism, mutualism and commensalism. Due to their small size, commensal bacteria are ubiquitous and grow on animals and plants exactly as they will grow on any other surface. However, their growth can be increased by warmth and sweat, and large populations of these organisms in humans are the cause of body odor.
Mutualists
Certain bacteria form close spatial associations that are essential for their survival. One such mutualistic association, called interspecies hydrogen transfer, occurs between clusters of anaerobic bacteria that consume organic acids such as butyric acid or propionic acid and produce hydrogen, and methanogenic Archaea that consume hydrogen.[124] The bacteria in this association are unable to consume the organic acids as this reaction produces hydrogen that accumulates in their surroundings. Only the intimate association with the hydrogen-consuming Archaea keeps the hydrogen concentration low enough to allow the bacteria to grow.
In soil, microorganisms which reside in the rhizosphere (a zone that includes the root surface and the soil that adheres to the root after gentle shaking) carry out nitrogen fixation, converting nitrogen gas to nitrogenous compounds.[125] This serves to provide an easily absorbable form of nitrogen for many plants, which cannot fix nitrogen themselves. Many other bacteria are found as symbionts in humans and other organisms. For example, the presence of over 1,000 bacterial species in the normal human gut flora of the intestines can contribute to gut immunity, synthesise vitamins such as folic acid, vitamin K and biotin, convert milk protein to lactic acid (see Lactobacillus), as well as fermenting complex undigestible carbohydrates.[126][127][128] The presence of this gut flora also inhibits the growth of potentially pathogenic bacteria (usually through competitive exclusion) and these beneficial bacteria are consequently sold as probiotic dietary supplements.[129]
Pathogens

If bacteria form a parasitic association with other organisms, they are classed as pathogens. Pathogenic bacteria are a major cause of human death and disease and cause infections such as tetanus, typhoid fever, diphtheria, syphilis, cholera, foodborne illness, leprosy and tuberculosis. A pathogenic cause for a known medical disease may only be discovered many years after, as was the case with Helicobacter pylori and peptic ulcer disease. Bacterial diseases are also important in agriculture, with bacteria causing leaf spot, fire blight and wilts in plants, as well as Johne's disease, mastitis, salmonella and anthrax in farm animals.
Each species of pathogen has a characteristic spectrum of interactions with its human hosts. Some organisms, such as Staphylococcus or Streptococcus, can cause skin infections, pneumonia, meningitis and even overwhelming sepsis, a systemic inflammatory response producing shock, massive vasodilation and death.[130] Yet these organisms are also part of the normal human flora and usually exist on the skin or in the nose without causing any disease at all. Other organisms invariably cause disease in humans, such as the Rickettsia, which are obligate intracellular parasites able to grow and reproduce only within the cells of other organisms. One species of Rickettsia causes typhus, while another causes Rocky Mountain spotted fever. Chlamydia, another phylum of obligate intracellular parasites, contains species that can cause pneumonia, or urinary tract infection and may be involved in coronary heart disease.[131] Finally, some species such as Pseudomonas aeruginosa, Burkholderia cenocepacia, and Mycobacterium avium are opportunistic pathogens and cause disease mainly in people suffering from immunosuppression or cystic fibrosis.[132][133]
Bacterial infections may be treated with antibiotics, which are classified as bacteriocidal if they kill bacteria, or bacteriostatic if they just prevent bacterial growth. There are many types of antibiotics and each class inhibits a process that is different in the pathogen from that found in the host. An example of how antibiotics produce selective toxicity are chloramphenicol and puromycin, which inhibit the bacterial ribosome, but not the structurally different eukaryotic ribosome.[134] Antibiotics are used both in treating human disease and in intensive farming to promote animal growth, where they may be contributing to the rapid development of antibiotic resistance in bacterial populations.[135] Infections can be prevented by antiseptic measures such as sterilizating the skin prior to piercing it with the needle of a syringe, and by proper care of indwelling catheters. Surgical and dental instruments are also sterilized to prevent contamination and infection by bacteria. Disinfectants such as bleach are used to kill bacteria or other pathogens on surfaces to prevent contamination and further reduce the risk of infection.
Significance in technology and industry
Bacteria, often Lactobacillus in combination with yeasts and molds, have been used for thousands of years in the preparation of fermented foods such as cheese, pickles, soy sauce, sauerkraut, vinegar, wine and yoghurt.[136][137]
The ability of bacteria to degrade a variety of organic compounds is remarkable and has been used in waste processing and bioremediation. Bacteria capable of digesting the hydrocarbons in petroleum are often used to clean up oil spills.[138] Fertilizer was added to some of the beaches in Prince William Sound in an attempt to promote the growth of these naturally occurring bacteria after the infamous 1989 Exxon Valdez oil spill. These efforts were effective on beaches that were not too thickly covered in oil. Bacteria are also used for the bioremediation of industrial toxic wastes.[139] In the chemical industry, bacteria are most important in the production of enantiomerically pure chemicals for use as pharmaceuticals or agrichemicals.[140]
Bacteria can also be used in the place of pesticides in the biological pest control. This commonly involves Bacillus thuringiensis (also called BT), a Gram-positive, soil dwelling bacterium. Subspecies of this bacteria are used as a Lepidopteran-specific insecticides under trade names such as Dipel and Thuricide.[141] Because of their specificity, these pesticides are regarded as environmentally friendly, with little or no effect on humans, wildlife, pollinators and most other beneficial insects.[142][143]
Because of their ability to quickly grow and the relative ease with which they can be manipulated, bacteria are the workhorses for the fields of molecular biology, genetics and biochemistry. By making mutations in bacterial DNA and examining the resulting phenotypes, scientists can determine the function of genes, enzymes and metabolic pathways in bacteria, then apply this knowledge to more complex organisms.[144] This aim of understanding the biochemistry of a cell reaches its most complex expression in the synthesis of huge amounts of enzyme kinetic and gene expression data into mathematical models of entire organisms. This is achievable in some well-studied bacteria, with models of Escherichia coli metabolism now being produced and tested.[145][146] This understanding of bacterial metabolism and genetics allows the use of biotechnology to bioengineer bacteria for the production of therapeutic proteins, such as insulin, growth factors, or antibodies.[147][148]
See also
- Human flora
- Bioaerosol
- Biotechnology
- Contamination control
- Denitrification
- Desulforudis audaxviator
- Extremophiles
- Transgenic bacteria
- Microorganism
References
- ↑ Fredrickson J, Zachara J, Balkwill D; et al. (2004). "Geomicrobiology of high-level nuclear waste-contaminated vadose sediments at the Hanford site, Washington state". Appl Environ Microbiol. 70 (7): 4230&ndash, 41. PMID 15240306.
- ↑ Whitman W, Coleman D, Wiebe W (1998). "Prokaryotes: the unseen majority". Proc Natl Acad Sci U S A. 95 (12): 6578&ndash, 83. PMID 9618454.
- ↑ Whitman W, Coleman D, Wiebe W (1998). "Prokaryotes: the unseen majority". Proc Natl Acad Sci U S A. 95 (12): 6578–83. PMID 9618454.
- ↑ 4.0 4.1 Rappé MS, Giovannoni SJ (2003). "The uncultured microbial majority". Annu. Rev. Microbiol. 57: 369–94. doi:10.1146/annurev.micro.57.030502.090759. PMID 14527284.
- ↑ Sears CL (2005). "A dynamic partnership: celebrating our gut flora". Anaerobe. 11 (5): 247–51. doi:10.1016/j.anaerobe.2005.05.001. PMID 16701579.
- ↑ "2002 WHO mortality data". Retrieved 2007-01-20.
- ↑ Ishige T, Honda K, Shimizu S (2005). "Whole organism biocatalysis". Curr Opin Chem Biol. 9 (2): 174&ndash, 80. PMID 15811802.
- ↑ Woese C, Kandler O, Wheelis M (1990). "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya". Proc Natl Acad Sci U S A. 87 (12): 4576&ndash, 9. PMID 2112744.
- ↑ Porter JR (1976). "Antony van Leeuwenhoek: Tercentenary of his discovery of bacteria". Bacteriological reviews. 40 (2): 260–269. PMID 786250. Retrieved 2007-08-19.
- ↑ van Leeuwenhoek A (1684). "An abstract of a letter from Mr. Anthony Leevvenhoek at Delft, dated Sep. 17, 1683, Containing Some Microscopical Observations, about Animals in the Scurf of the Teeth, the Substance Call'd Worms in the Nose, the Cuticula Consisting of Scales". Philosophical Transactions (1683–1775). 14: 568–574. Retrieved 2007-08-19.
- ↑ van Leeuwenhoek A (1700). "Part of a Letter from Mr Antony van Leeuwenhoek, concerning the Worms in Sheeps Livers, Gnats, and Animalcula in the Excrements of Frogs". Philosophical Transactions (1683–1775). 22: 509–518. Retrieved 2007-08-19.
- ↑ van Leeuwenhoek A (1702). "Part of a Letter from Mr Antony van Leeuwenhoek, F. R. S. concerning Green Weeds Growing in Water, and Some Animalcula Found about Them". Philosophical Transactions (1683-1775). 23: 1304–11. Retrieved 2007-08-19.
- ↑ "Etymology of the word "bacteria"". Online Etymology dictionary. Retrieved 2006-11-23.
- ↑ "Pasteur's Papers on the Germ Theory". LSU Law Center's Medical and Public Health Law Site, Historic Public Health Articles. Retrieved 2006-11-23.
- ↑ "The Nobel Prize in Physiology or Medicine 1905". Nobelprize.org. Retrieved 2006-11-22.
- ↑ O'Brien S, Goedert J (1996). "HIV causes AIDS: Koch's postulates fulfilled". Curr Opin Immunol. 8 (5): 613–618. PMID 8902385.
- ↑ Thurston A (2000). "Of blood, inflammation and gunshot wounds: the history of the control of sepsis". Aust N Z J Surg. 70 (12): 855–61. PMID 11167573.
- ↑ Schwartz R (2004). "Paul Ehrlich's magic bullets". N Engl J Med. 350 (11): 1079–80. PMID 15014180.
- ↑ "Biography of Paul Ehrlich". Nobelprize.org. Retrieved 2006-11-26.
- ↑ Woese C, Fox G (1977). "Phylogenetic structure of the prokaryotic domain: the primary kingdoms". Proc Natl Acad Sci U S A. 74 (11): 5088–5090. PMID 270744.
- ↑ Woese C, Kandler O, Wheelis M (1990). "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya". Proc Natl Acad Sci U S A. 87 (12): 4576–4579. PMID 2112744.
- ↑ Schopf J (1994). "Disparate rates, differing fates: tempo and mode of evolution changed from the Precambrian to the Phanerozoic". Proc Natl Acad Sci U S A. 91 (15): 6735–42. PMID 8041691.
- ↑ DeLong E, Pace N (2001). "Environmental diversity of bacteria and archaea". Syst Biol. 50 (4): 470–78. PMID 12116647.
- ↑ Brown JR, Doolittle WF (1997). "Archaea and the prokaryote-to-eukaryote transition". Microbiol. Mol. Biol. Rev. 61 (4): 456–502. PMID 9409149.
- ↑ Di Giulio M (2003). "The universal ancestor and the ancestor of bacteria were hyperthermophiles". J Mol Evol. 57 (6): 721–30. PMID 14745541.
- ↑ Battistuzzi F, Feijao A, Hedges S. "A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land". BMC Evol Biol. 4: 44. PMID 15535883.
- ↑ Poole A, Penny D (2007). "Evaluating hypotheses for the origin of eukaryotes". Bioessays. 29 (1): 74–84. PMID 17187354.
- ↑ Dyall S, Brown M, Johnson P (2004). "Ancient invasions: from endosymbionts to organelles". Science. 304 (5668): 253&ndash, 7. PMID 15073369.
- ↑ Lang B, Gray M, Burger G. "Mitochondrial genome evolution and the origin of eukaryotes". Annu Rev Genet. 33: 351–97. PMID 10690412.
- ↑ McFadden G (1999). "Endosymbiosis and evolution of the plant cell". Curr Opin Plant Biol. 2 (6): 513–9. PMID 10607659.
- ↑ Schulz H, Jorgensen B. "Big bacteria". Annu Rev Microbiol. 55: 105&ndash, 37. PMID 11544351.
- ↑ Robertson J, Gomersall M, Gill P. (1975). "Mycoplasma hominis: growth, reproduction, and isolation of small viable cells". J Bacteriol. 124 (2): 1007&ndash, 18. PMID 1102522.
- ↑ Fritz I, Strömpl C, Abraham W (2004). "Phylogenetic relationships of the genera Stella, Labrys and Angulomicrobium within the 'Alphaproteobacteria' and description of Angulomicrobium amanitiforme sp. nov". Int J Syst Evol Microbiol. 54 (Pt 3): 651–7. PMID 15143003.
- ↑ Cabeen M, Jacobs-Wagner C (2005). "Bacterial cell shape". Nat Rev Microbiol. 3 (8): 601&ndash, 10. PMID 16012516.
- ↑ Young K (2006). "The selective value of bacterial shape". Microbiol Mol Biol Rev. 70 (3): 660&ndash, 703. PMID 16959965.
- ↑ Douwes K, Schmalzbauer E, Linde H, Reisberger E, Fleischer K, Lehn N, Landthaler M, Vogt T (2003). "Branched filaments no fungus, ovoid bodies no bacteria: Two unusual cases of mycetoma". J Am Acad Dermatol. 49 (2 Suppl Case Reports): S170&ndash, 3. PMID 12894113.
- ↑ Donlan R (2002). "Biofilms: microbial life on surfaces". Emerg Infect Dis. 8 (9): 881–90. PMID 12194761.
- ↑ Branda S, Vik S, Friedman L, Kolter R (2005). "Biofilms: the matrix revisited". Trends Microbiol. 13 (1): 20–26. PMID 15639628.
- ↑ 39.0 39.1 Davey M, O'toole G (2000). "Microbial biofilms: from ecology to molecular genetics". Microbiol Mol Biol Rev. 64 (4): 847–67. PMID 11104821.
- ↑ Donlan RM, Costerton JW (2002). "Biofilms: survival mechanisms of clinically relevant microorganisms". Clin Microbiol Rev. 15 (2): 167–93. PMID 11932229.
- ↑ Shimkets L. "Intercellular signaling during fruiting-body development of Myxococcus xanthus". Annu Rev Microbiol. 53: 525–49. PMID 10547700.
- ↑ Kaiser D. "Signaling in myxobacteria". Annu Rev Microbiol. 58: 75–98. PMID 15487930.
- ↑ Berg JM, Tymoczko JL Stryer L (2002). Molecular Cell Biology (5th ed. ed.). WH Freeman. ISBN 0-7167-4955-6.
- ↑ Gitai Z (2005). "The new bacterial cell biology: moving parts and subcellular architecture". Cell. 120 (5): 577–86. PMID 15766522.
- ↑ Shih YL, Rothfield L (2006). "The bacterial cytoskeleton". Microbiol. Mol. Biol. Rev. 70 (3): 729–54. PMID 16959967.
- ↑ Harold F (1972). "Conservation and transformation of energy by bacterial membranes". Bacteriol Rev. 36 (2): 172–230. PMID 4261111.
- ↑ Thanbichler M, Wang S, Shapiro L (2005). "The bacterial nucleoid: a highly organized and dynamic structure". J Cell Biochem. 96 (3): 506–21. PMID 15988757.
- ↑ Poehlsgaard J, Douthwaite S (2005). "The bacterial ribosome as a target for antibiotics". Nat Rev Microbiol. 3 (11): 870–81. PMID 16261170.
- ↑ Fuerst J (2005). "Intracellular compartmentation in planctomycetes". Annu Rev Microbiol. 59: 299–328. PMID 15910279.
- ↑ Yeo M, Chater K (2005). "The interplay of glycogen metabolism and differentiation provides an insight into the developmental biology of Streptomyces coelicolor". Microbiology. 151 (Pt 3): 855–61. PMID 15758231.
- ↑ Shiba T, Tsutsumi K, Ishige K, Noguchi T (2000). "Inorganic polyphosphate and polyphosphate kinase: their novel biological functions and applications". Biochemistry (Mosc). 65 (3): 315–23. PMID 10739474.
- ↑ Brune DC. (1995). "Isolation and characterization of sulfur globule proteins from Chromatium vinosum and Thiocapsa roseopersicina". Arch Microbiol. 163 (6): 391–99. PMID 7575095.
- ↑ Kadouri D, Jurkevitch E, Okon Y, Castro-Sowinski S. (2005). "Ecological and agricultural significance of bacterial polyhydroxyalkanoates". Crit Rev Microbiol. 31 (2): 55–67. PMID 15986831.
- ↑ Walsby A (1994). "Gas vesicles". Microbiol Rev. 58 (1): 94–144. PMID 8177173.
- ↑ van Heijenoort J (2001). "Formation of the glycan chains in the synthesis of bacterial peptidoglycan". Glycobiology. 11 (3): 25R&ndash, 36R. PMID 11320055.
- ↑ 56.0 56.1 Koch A (2003). "Bacterial wall as target for attack: past, present, and future research". Clin Microbiol Rev. 16 (4): 673&ndash, 87. PMID 14557293.
- ↑ 57.0 57.1 Gram, HC (1884). "Über die isolierte Färbung der Schizomyceten in Schnitt- und Trockenpräparaten". Fortschr. Med. 2: 185&ndash, 189.
- ↑ Hugenholtz P (2002). "Exploring prokaryotic diversity in the genomic era". Genome Biol. 3 (2): REVIEWS0003. PMID 11864374.
- ↑ Walsh F, Amyes S (2004). "Microbiology and drug resistance mechanisms of fully resistant pathogens". Curr Opin Microbiol. 7 (5): 439–44. PMID 15451497.
- ↑ Engelhardt H, Peters J (1998). "Structural research on surface layers: a focus on stability, surface layer homology domains, and surface layer-cell wall interactions". J Struct Biol. 124 (2&ndash, 3): 276–302. PMID 10049812.
- ↑ Beveridge T, Pouwels P, Sára M, Kotiranta A, Lounatmaa K, Kari K, Kerosuo E, Haapasalo M, Egelseer E, Schocher I, Sleytr U, Morelli L, Callegari M, Nomellini J, Bingle W, Smit J, Leibovitz E, Lemaire M, Miras I, Salamitou S, Béguin P, Ohayon H, Gounon P, Matuschek M, Koval S (1997). "Functions of S-layers". FEMS Microbiol Rev. 20 (1&ndash, 2): 99&ndash, 149. PMID 9276929.
- ↑ Kojima S, Blair D. "The bacterial flagellar motor: structure and function of a complex molecular machine". Int Rev Cytol. 233: 93&ndash, 134. PMID 15037363.
- ↑ Beachey E (1981). "Bacterial adherence: adhesin-receptor interactions mediating the attachment of bacteria to mucosal surface". J Infect Dis. 143 (3): 325&ndash, 45. PMID 7014727.
- ↑ Silverman P (1997). "Towards a structural biology of bacterial conjugation". Mol Microbiol. 23 (3): 423&ndash, 9. PMID 9044277.
- ↑ Stokes R, Norris-Jones R, Brooks D, Beveridge T, Doxsee D, Thorson L (2004). "The glycan-rich outer layer of the cell wall of Mycobacterium tuberculosis acts as an antiphagocytic capsule limiting the association of the bacterium with macrophages". Infect Immun. 72 (10): 5676&ndash, 86. PMID 15385466.
- ↑ Daffé M, Etienne G (1999). "The capsule of Mycobacterium tuberculosis and its implications for pathogenicity". Tuber Lung Dis. 79 (3): 153&ndash, 69. PMID 10656114.
- ↑ Finlay B, Falkow S (1997). "Common themes in microbial pathogenicity revisited". Microbiol Mol Biol Rev. 61 (2): 136&ndash, 69. PMID 9184008.
- ↑ Nicholson W, Munakata N, Horneck G, Melosh H, Setlow P (2000). "Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments". Microbiol Mol Biol Rev. 64 (3): 548&ndash, 72. PMID 10974126.
- ↑ Siunov A, Nikitin D, Suzina N, Dmitriev V, Kuzmin N, Duda V. "Phylogenetic status of Anaerobacter polyendosporus, an anaerobic, polysporogenic bacterium" (PDF). Int J Syst Bacteriol. 49 Pt 3: 1119&ndash, 24. PMID 10425769.
- ↑ Nicholson W, Fajardo-Cavazos P, Rebeil R, Slieman T, Riesenman P, Law J, Xue Y (2002). "Bacterial endospores and their significance in stress resistance". Antonie Van Leeuwenhoek. 81 (1&ndash, 4): 27&ndash, 32. PMID 12448702.
- ↑ Vreeland R, Rosenzweig W, Powers D (2000). "Isolation of a 250 million-year-old halotolerant bacterium from a primary salt crystal". Nature. 407 (6806): 897&ndash, 900. PMID 11057666.
- ↑ Cano R, Borucki M (1995). "Revival and identification of bacterial spores in 25- to 40-million-year-old Dominican amber". Science. 268 (5213): 1060&ndash, 4. PMID 7538699.
- ↑ Nicholson W, Schuerger A, Setlow P (2005). "The solar UV environment and bacterial spore UV resistance: considerations for Earth-to-Mars transport by natural processes and human spaceflight". Mutat Res. 571 (1&ndash, 2): 249&ndash, 64. PMID 15748651.
- ↑ Hatheway C (1990). "Toxigenic clostridia". Clin Microbiol Rev. 3 (1): 66&ndash, 98. PMID 2404569.
- ↑ Nealson K (1999). "Post-Viking microbiology: new approaches, new data, new insights". Orig Life Evol Biosph. 29 (1): 73–93. PMID 11536899.
- ↑ Xu J (2006). "Microbial ecology in the age of genomics and metagenomics: concepts, tools, and recent advances". Mol Ecol. 15 (7): 1713–31. PMID 16689892.
- ↑ Zillig W (1991). "Comparative biochemistry of Archaea and Bacteria". Curr Opin Genet Dev. 1 (4): 544–51. PMID 1822288.
- ↑ Hellingwerf K, Crielaard W, Hoff W, Matthijs H, Mur L, van Rotterdam B (1994). "Photobiology of bacteria". Antonie Van Leeuwenhoek. 65 (4): 331&ndash, 47. PMID 7832590.
- ↑ Zumft W (1997). "Cell biology and molecular basis of denitrification". Microbiol Mol Biol Rev. 61 (4): 533&ndash, 616. PMID 9409151.
- ↑ Drake H, Daniel S, Küsel K, Matthies C, Kuhner C, Braus-Stromeyer S (1997). "Acetogenic bacteria: what are the in situ consequences of their diverse metabolic versatilities?". Biofactors. 6 (1): 13&ndash, 24. PMID 9233536.
- ↑ Morel, FMM (1998). "The chemical cycle and bioaccumulation of mercury". Annual Review of Ecological Systems. 29: 543&mdash, 566. Unknown parameter
|coauthors=ignored (help) - ↑ Dalton H (2005). "The Leeuwenhoek Lecture 2000 the natural and unnatural history of methane-oxidizing bacteria". Philos Trans R Soc Lond B Biol Sci. 360 (1458): 1207&ndash, 22. PMID 16147517.
- ↑ Zehr J, Jenkins B, Short S, Steward G (2003). "Nitrogenase gene diversity and microbial community structure: a cross-system comparison". Environ Microbiol. 5 (7): 539&ndash, 54. PMID 12823187.
- ↑ Koch A (2002). "Control of the bacterial cell cycle by cytoplasmic growth". Crit Rev Microbiol. 28 (1): 61&ndash, 77. PMID 12003041.
- ↑ Eagon R. "Pseudomonas natriegens, a marine bacterium with a generation time of less than 10 minutes". J Bacteriol. 83: 736&ndash, 7. PMID 13888946.
- ↑ Stewart EJ, Madden R, Paul G, Taddei F (2005). "Aging and death in an organism that reproduces by morphologically symmetric division". PLoS Biol. 3 (2): e45. PMID 15685293.
- ↑ 87.0 87.1 87.2 Thomson R, Bertram H (2001). "Laboratory diagnosis of central nervous system infections". Infect Dis Clin North Am. 15 (4): 1047&ndash, 71. PMID 11780267.
- ↑ Paerl H, Fulton R, Moisander P, Dyble J. "Harmful freshwater algal blooms, with an emphasis on cyanobacteria". ScientificWorldJournal. 1: 76&ndash, 113. PMID 12805693.
- ↑ Challis G, Hopwood D. "Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species". Proc Natl Acad Sci U S A. 100 Suppl 2: 14555&ndash, 61. PMID 12970466.
- ↑ Kooijman S, Auger P, Poggiale J, Kooi B (2003). "Quantitative steps in symbiogenesis and the evolution of homeostasis". Biol Rev Camb Philos Soc. 78 (3): 435&ndash, 63. PMID 14558592.
- ↑ Prats C, López D, Giró A, Ferrer J, Valls J (2006). "Individual-based modelling of bacterial cultures to study the microscopic causes of the lag phase". J Theor Biol. 241 (4): 939&ndash, 53. PMID 16524598.
- ↑ Hecker M, Völker U. "General stress response of Bacillus subtilis and other bacteria". Adv Microb Physiol. 44: 35&ndash, 91. PMID 11407115.
- ↑ Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar H, Moran N, Hattori M (2006). "The 160-kilobase genome of the bacterial endosymbiont Carsonella". Science. 314 (5797): 267. PMID 17038615.
- ↑ Pradella S, Hans A, Spröer C, Reichenbach H, Gerth K, Beyer S (2002). "Characterisation, genome size and genetic manipulation of the myxobacterium Sorangium cellulosum So ce56". Arch Microbiol. 178 (6): 484–92. PMID 12420170.
- ↑ Hinnebusch J, Tilly K (1993). "Linear plasmids and chromosomes in bacteria". Mol Microbiol. 10 (5): 917–22. PMID 7934868.
- ↑ Belfort M, Reaban ME, Coetzee T, Dalgaard JZ (1995). "Prokaryotic introns and inteins: a panoply of form and function". J. Bacteriol. 177 (14): 3897–903. PMID 7608058.
- ↑ Brüssow H, Canchaya C, Hardt W (2004). "Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion". Microbiol Mol Biol Rev. 68 (3): 560–602. PMID 15353570.
- ↑ Denamur E, Matic I (2006). "Evolution of mutation rates in bacteria". Mol Microbiol. 60 (4): 820&ndash, 7. PMID 16677295.
- ↑ Wright B (2004). "Stress-directed adaptive mutations and evolution". Mol Microbiol. 52 (3): 643&ndash, 50. PMID 15101972.
- ↑ Davison J (1999). "Genetic exchange between bacteria in the environment". Plasmid. 42 (2): 73&ndash, 91. PMID 10489325.
- ↑ Hastings P, Rosenberg S, Slack A (2004). "Antibiotic-induced lateral transfer of antibiotic resistance". Trends Microbiol. 12 (9): 401&ndash, 4. PMID 15337159.
- ↑ 102.0 102.1 102.2 Bardy S, Ng S, Jarrell K (2003). "Prokaryotic motility structures". Microbiology. 149 (Pt 2): 295&ndash, 304. PMID 12624192.
- ↑ Merz A, So M, Sheetz M (2000). "Pilus retraction powers bacterial twitching motility". Nature. 407 (6800): 98–102. PMID 10993081.
- ↑ Wu M, Roberts J, Kim S, Koch D, DeLisa M (2006). "Collective bacterial dynamics revealed using a three-dimensional population-scale defocused particle tracking technique". Appl Environ Microbiol. 72 (7): 4987&ndash, 94. PMID 16820497.
- ↑ Lux R, Shi W (2004). "Chemotaxis-guided movements in bacteria". Crit Rev Oral Biol Med. 15 (4): 207–20. PMID 15284186.
- ↑ Frankel R, Bazylinski D, Johnson M, Taylor B (1997). "Magneto-aerotaxis in marine coccoid bacteria". Biophys J. 73 (2): 994&ndash, 1000. PMID 9251816.
- ↑ Kaiser D. "Signaling in myxobacteria". Annu Rev Microbiol. 58: 75–98. PMID 15487930.
- ↑ Goldberg MB (2001). "Actin-based motility of intracellular microbial pathogens". Microbiol Mol Biol Rev. 65 (4): 595–626. PMID 11729265.
- ↑ Boucher Y, Douady CJ, Papke RT, Walsh DA, Boudreau ME, Nesbo CL, Case RJ, Doolittle WF (2003). "Lateral gene transfer and the origins of prokaryotic groups". Annu Rev Genet. 37: 283–328. PMID 14616063. Text " http://arjournals.annualreviews.org/doi/abs/10.1146/annurev.genet.37.050503.084247 " ignored (help)
- ↑ Olsen G, Woese C, Overbeek R (1994). "The winds of (evolutionary) change: breathing new life into microbiology". J Bacteriol. 176 (1): 1&ndash, 6. PMID 8282683.
- ↑ IJSEM - Home
- ↑ Bergey's Manual Trust
- ↑ Woese C, Kandler O, Wheelis M (1990). "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya". Proc Natl Acad Sci U S A. 87 (12): 4576&ndash, 9. PMID 2112744.
- ↑ Gupta R (2000). "The natural evolutionary relationships among prokaryotes". Crit Rev Microbiol. 26 (2): 111–31. PMID 10890353.
- ↑ Doolittle RF (2005). "Evolutionary aspects of whole-genome biology". Curr Opin Struct Biol. 15 (3): 248–253. PMID 11837318.
- ↑ Cavalier-Smith T (2002). "The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification". Int J Syst Evol Microbiol. 52 (Pt 1): 7–76. PMID 11837318.
- ↑ Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P (2006). "Toward automatic reconstruction of a highly resolved tree of life". Science. 311 (5765): 1283–7. PMID 16513982.
- ↑ Woods G, Walker D (1996). "Detection of infection or infectious agents by use of cytologic and histologic stains". Clin Microbiol Rev. 9 (3): 382&ndash, 404. PMID 8809467.
- ↑ Weinstein M (1994). "Clinical importance of blood cultures". Clin Lab Med. 14 (1): 9&ndash, 16. PMID 8181237.
- ↑ Louie M, Louie L, Simor AE (2000). "The role of DNA amplification technology in the diagnosis of infectious diseases". CMAJ. 163 (3): 301–309. PMID 10951731.
- ↑ Oliver J. "The viable but nonculturable state in bacteria". J Microbiol. 43 Spec No: 93–100. PMID 15765062.
- ↑ Curtis T, Sloan W, Scannell J (2002). "Estimating prokaryotic diversity and its limits". Proc Natl Acad Sci U S A. 99 (16): 10494–9. PMID 12097644.
- ↑ Schloss P, Handelsman J (2004). "Status of the microbial census". Microbiol Mol Biol Rev. 68 (4): 686–91. PMID 15590780.
- ↑ Stams A, de Bok F, Plugge C, van Eekert M, Dolfing J, Schraa G (2006). "Exocellular electron transfer in anaerobic microbial communities". Environ Microbiol. 8 (3): 371&ndash, 82. PMID 16478444.
- ↑ Barea J, Pozo M, Azcón R, Azcón-Aguilar C (2005). "Microbial co-operation in the rhizosphere". J Exp Bot. 56 (417): 1761&ndash, 78. PMID 15911555.
- ↑ O'Hara A, Shanahan F (2006). "The gut flora as a forgotten organ". EMBO Rep. 7 (7): 688&ndash, 93. PMID 16819463.
- ↑ Zoetendal E, Vaughan E, de Vos W (2006). "A microbial world within us". Mol Microbiol. 59 (6): 1639&ndash, 50. PMID 16553872.
- ↑ Gorbach S (1990). "Lactic acid bacteria and human health". Ann Med. 22 (1): 37&ndash, 41. PMID 2109988.
- ↑ Salminen S, Gueimonde M, Isolauri E (2005). "Probiotics that modify disease risk". J Nutr. 135 (5): 1294&ndash, 8. PMID 15867327.
- ↑ Fish D. "Optimal antimicrobial therapy for sepsis". Am J Health Syst Pharm. 59 Suppl 1: S13&ndash, 9. PMID 11885408.
- ↑ Belland R, Ouellette S, Gieffers J, Byrne G (2004). "Chlamydia pneumoniae and atherosclerosis". Cell Microbiol. 6 (2): 117&ndash, 27. PMID 14706098.
- ↑ Heise E. "Diseases associated with immunosuppression". Environ Health Perspect. 43: 9&ndash, 19. PMID 7037390.
- ↑ Saiman, L. "Microbiology of early CF lung disease". Paediatr Respir Rev.volume=5 Suppl A: S367&ndash, 369. Unknown parameter
|yar=ignored (help) PMID 14980298 - ↑ Yonath A, Bashan A (2004). "Ribosomal crystallography: initiation, peptide bond formation, and amino acid polymerization are hampered by antibiotics". Annu Rev Microbiol. 58: 233&ndash, 51. PMID 15487937.
- ↑ Khachatourians G (1998). "Agricultural use of antibiotics and the evolution and transfer of antibiotic-resistant bacteria". CMAJ. 159 (9): 1129–36. PMID 9835883.
- ↑ Johnson M, Lucey J (2006). "Major technological advances and trends in cheese". J Dairy Sci. 89 (4): 1174–8. PMID 16537950.
- ↑ Hagedorn S, Kaphammer B (1994). "Microbial biocatalysis in the generation of flavor and fragrance chemicals". Annu. Rev. Microbiol. 48: 773–800. doi:10.1146/annurev.mi.48.100194.004013. PMID 7826026.
- ↑ Cohen Y (2002). "Bioremediation of oil by marine microbial mats". Int Microbiol. 5 (4): 189&ndash, 93. PMID 12497184.
- ↑ Neves LC, Miyamura TT, Moraes DA, Penna TC, Converti A (2006). "Biofiltration methods for the removal of phenolic residues". Appl. Biochem. Biotechnol. 129-132: 130–52. PMID 16915636.
- ↑ Liese A, Filho M (1999). "Production of fine chemicals using biocatalysis". Curr Opin Biotechnol. 10 (6): 595&ndash, 603. PMID 10600695.
- ↑ Aronson AI, Shai Y (2001). "Why Bacillus thuringiensis insecticidal toxins are so effective: unique features of their mode of action". FEMS Microbiol. Lett. 195 (1): 1–8. PMID 11166987.
- ↑ Bozsik A (2006). "Susceptibility of adult Coccinella septempunctata (Coleoptera: Coccinellidae) to insecticides with different modes of action". Pest Manag Sci. 62 (7): 651–4. PMID 16649191.
- ↑ Chattopadhyay A, Bhatnagar N, Bhatnagar R (2004). "Bacterial insecticidal toxins". Crit Rev Microbiol. 30 (1): 33–54. PMID 15116762.
- ↑ Serres M, Gopal S, Nahum L, Liang P, Gaasterland T, Riley M (2001). "A functional update of the Escherichia coli K-12 genome". Genome Biol. 2 (9): RESEARCH0035. PMID 11574054.
- ↑ Almaas E, Kovács B, Vicsek T, Oltvai Z, Barabási A (2004). "Global organization of metabolic fluxes in the bacterium Escherichia coli". Nature. 427 (6977): 839–43. PMID 14985762.
- ↑ Reed JL, Vo TD, Schilling CH, Palsson BO (2003). "An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR)". Genome Biol. 4 (9): R54. doi:10.1186/gb-2003-4-9-r54. PMID 12952533.
- ↑ Walsh G (2005). "Therapeutic insulins and their large-scale manufacture". Appl Microbiol Biotechnol. 67 (2): 151&ndash, 9. PMID 15580495.
- ↑ Graumann K, Premstaller A (2006). "Manufacturing of recombinant therapeutic proteins in microbial systems". Biotechnol J. 1 (2): 164&ndash, 86. PMID 16892246.
Further reading
- Alcamo IE (2001). Fundamentals of microbiology. Boston: Jones and Bartlett. ISBN 0-7637-1067-9.
- Atlas RM (1995). Principles of microbiology. St. Louis: Mosby. ISBN 0-8016-7790-4.
- Martinko JM, Madigan MT (2005). Brock Biology of Microorganisms (11th ed. ed.). Englewood Cliffs, N.J: Prentice Hall. ISBN 0-13-144329-1.
- Holt JC, Bergey DH (1994). Bergey's manual of determinative bacteriology (9th ed. ed.). Baltimore: Williams & Wilkins. ISBN 0-683-00603-7.
- Hugenholtz P, Goebel BM, Pace NR (1998). "Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity". J Bacteriol. 180 (18): 4765&ndash, 74. PMID 9733676.
- Funke BR, Tortora GJ, Case CL (2004). Microbiology: an introduction (8th ed, ed.). San Francisco: Benjamin Cummings. ISBN 0-8053-7614-3.
External links
- Bacterial Nomenclature Up-To-Date from DSMZ
- The largest bacteria
- Tree of Life: Eubacteria
- Videos of bacteria swimming and tumbling, use of optical tweezers and other videos.
- Planet of the Bacteria by Stephen Jay Gould
- On-line text book on bacteriology
- Animated guide to bacterial cell structure.
Template:Link FA Template:Link FA Template:Link FA Template:Link FA
af:Bakterie ar:بكتيريا an:Eubacteria ay:Jisk'a laq'u az:Bakteriya bn:ব্যাক্টেরিয়া zh-min-nan:Sè-khún br:Bakteri bg:Бактерии ca:Bacteri cs:Bakterie cy:Bacteria da:Bakterie de:Bakterien et:Bakterid el:Βακτήριο eo:Bakterioj eu:Bakterio fa:باکتری fo:Bakteria ga:Baictéar gl:Bacteria ko:세균 hi:जीवाणु hr:Bakterije io:Bakterio ilo:Bacteria id:Bakteri ia:Bacterio is:Gerlar it:Bacteria he:חיידקים kn:ಬ್ಯಾಕ್ಟೀರಿಯ kaa:Bakteriya ka:ბაქტერიები ku:Bakterî la:Bacterium lv:Baktērija lb:Bakterien lt:Bakterijos hu:Baktériumok mk:Бактерија mr:जीवाणू ms:Bakteria nl:Bacterie no:Bakterier nn:Bakterie nrm:Bactéthie oc:Bactèri nds:Bakterien qu:Añaki simple:Bacteria sl:Bakterije sr:Бактерија sh:Bakterija su:Baktéri fi:Bakteerit sv:Bakterier tl:Bakterya ta:கோலுரு நுண்ணுயிர் te:బాక్టీరియా th:แบคทีเรีย tg:Бактерия uk:Бактерії wa:Bactereye yi:באקטעריע sk:Baktérie