Alpha-1-B glycoprotein: Difference between revisions
m (Bot: HTTP→HTTPS) |
imported>Nat965 (Fixed access-date in references) |
||
| Line 12: | Line 12: | ||
=== Neighborhood === | === Neighborhood === | ||
A1BG is located on the negative DNA strand of [[chromosome 19]] from 58,858,172 – 58,864,865.<ref name="A1BG alpha-1-B glycoprotein">{{cite web|title=A1BG alpha-1-B glycoprotein|url=https://www.ncbi.nlm.nih.gov/gene/1|accessdate= | A1BG is located on the negative DNA strand of [[chromosome 19]] from 58,858,172 – 58,864,865.<ref name="A1BG alpha-1-B glycoprotein">{{cite web|title=A1BG alpha-1-B glycoprotein|url=https://www.ncbi.nlm.nih.gov/gene/1|accessdate=May 10, 2013}}</ref> Additionally, A1BG is located directly adjacent to the ZSCAN22 gene (58,838,385-58,853,712)) on the positive DNA strand, as well as the [[ZNF837]] (58,878,990 - 58,892,389, complement) and ZNF497 (58865723 - 58,874,214, complement) genes on the negative strand.<ref name="A1BG alpha-1-B glycoprotein"/> | ||
=== Expression === | === Expression === | ||
| Line 24: | Line 24: | ||
=== mRNA structure === | === mRNA structure === | ||
The gene contains 20 distinct [[introns]].<ref name=AceView>{{cite web|title=AceView: A1BG|url=https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/av.cgi?db=human&term=a1bg&submit=Go|accessdate= | The gene contains 20 distinct [[introns]].<ref name=AceView>{{cite web|title=AceView: A1BG|url=https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/av.cgi?db=human&term=a1bg&submit=Go|accessdate=May 11, 2013}}</ref> [[Transcription (genetics)|Transcription]] produces 15 different [[mRNAs]], 10 [[alternatively spliced]] variants and 5 unspliced forms.<ref name="AceView"/> There are 4 probable alternative [[Promoter (genetics)|promoters]], 4 non overlapping alternative last [[exons]] and 7 validated alternative [[polyadenylation]] sites.<ref name="AceView"/> The mRNAs appear to differ by truncation of the 5' end, truncation of the 3' end, presence or absence of 4 cassette exons, overlapping exons with different boundaries, splicing versus retention of 3 introns.<ref name="AceView"/> | ||
== Protein == | == Protein == | ||
| Line 34: | Line 34: | ||
=== Post-translational modifications === | === Post-translational modifications === | ||
The NetAcet 1.0 program calculated that the first five amino acid residues serve as an N-acetylation site.<ref name="pmid15539450">{{cite journal |vauthors=Kiemer L, Bendtsen JD, Blom N | title = NetAcet: prediction of N-terminal acetylation sites | journal = Bioinformatics | volume = 21 | issue = 7 | pages = 1269–70 |date=April 2005 | pmid = 15539450 | doi = 10.1093/bioinformatics/bti130 | url = http://www.cbs.dtu.dk/services/NetAcet/ }}</ref> The NetGlycate 1.0 program predicted that the lysines located at residue 78, 114, and 227 serve as glycation points.<ref name="pmid16762979">{{cite journal |vauthors=Johansen MB, Kiemer L, Brunak S | title = Analysis and prediction of mammalian protein glycation | journal = Glycobiology | volume = 16 | issue = 9 | pages = 844–53 |date=September 2006 | pmid = 16762979 | doi = 10.1093/glycob/cwl009 | url = http://www.cbs.dtu.dk/services/NetGlycate/ }}</ref> The NetNES 1.1 program predicted the leucine at residue 47 to be a nuclear export signal.<ref name="pmid15314210">{{cite journal |vauthors=la Cour T, Kiemer L, Mølgaard A, Gupta R, Skriver K, Brunak S | title = Analysis and prediction of leucine-rich nuclear export signals | journal = Protein Eng. Des. Sel. | volume = 17 | issue = 6 | pages = 527–36 |date=June 2004 | pmid = 15314210 | doi = 10.1093/protein/gzh062 | url = http://www.cbs.dtu.dk/services/NetNES/|accessdate= | The NetAcet 1.0 program calculated that the first five amino acid residues serve as an N-acetylation site.<ref name="pmid15539450">{{cite journal |vauthors=Kiemer L, Bendtsen JD, Blom N | title = NetAcet: prediction of N-terminal acetylation sites | journal = Bioinformatics | volume = 21 | issue = 7 | pages = 1269–70 |date=April 2005 | pmid = 15539450 | doi = 10.1093/bioinformatics/bti130 | url = http://www.cbs.dtu.dk/services/NetAcet/ }}</ref> The NetGlycate 1.0 program predicted that the lysines located at residue 78, 114, and 227 serve as glycation points.<ref name="pmid16762979">{{cite journal |vauthors=Johansen MB, Kiemer L, Brunak S | title = Analysis and prediction of mammalian protein glycation | journal = Glycobiology | volume = 16 | issue = 9 | pages = 844–53 |date=September 2006 | pmid = 16762979 | doi = 10.1093/glycob/cwl009 | url = http://www.cbs.dtu.dk/services/NetGlycate/ }}</ref> The NetNES 1.1 program predicted the leucine at residue 47 to be a nuclear export signal.<ref name="pmid15314210">{{cite journal |vauthors=la Cour T, Kiemer L, Mølgaard A, Gupta R, Skriver K, Brunak S | title = Analysis and prediction of leucine-rich nuclear export signals | journal = Protein Eng. Des. Sel. | volume = 17 | issue = 6 | pages = 527–36 |date=June 2004 | pmid = 15314210 | doi = 10.1093/protein/gzh062 | url = http://www.cbs.dtu.dk/services/NetNES/|accessdate=May 10, 2013 }}</ref> The NetNGlyc 1.0 program predicted four N-glycosylation sites - two of which are highly conserved internally repeated sequences.<ref>{{cite web|last=Gupta|first=R.|title=Prediction of N-glycosylation sites in human proteins|url=http://www.cbs.dtu.dk/services/NetNGlyc/|accessdate=May 10, 2013}}</ref><ref name="pmid1591615">{{cite journal |vauthors=Higgins DG, Bleasby AJ, Fuchs R | title = CLUSTAL V: improved software for multiple sequence alignment | journal = Comput. Appl. Biosci. | volume = 8 | issue = 2 | pages = 189–91 |date=April 1992 | pmid = 1591615 | doi = 10.1093/bioinformatics/8.2.189}}</ref> The NetCGlyc1.0 program predicted that none of the tryptophan residues serve as C-mannosylation sites.<ref>{{cite journal|last=Julenius|first=Karin|title=NetCGlyc1.0: Prediction of mammalian C-mannosylation sites|journal=Glycobiology|year=2007|volume=17|pages=868–876|url=http://www.cbs.dtu.dk/services/NetCGlyc/|accessdate=May 10, 2013|doi=10.1093/glycob/cwm050|pmid=17494086}}</ref> | ||
=== Protein interactions === | === Protein interactions === | ||
| Line 44: | Line 44: | ||
=== Orthologs === | === Orthologs === | ||
In addition to the table below, alpha-1B glycoprotein is also conserved in the [[white-cheeked crested gibbon]], [[baboon]], [[bolivian squirrel monkey]], [[sheep]], [[dog]], [[wild boar]], [[Treeshrew|Chinese tree shrew]], [[Chinese hamster]], [[black flying fox]], [[rabbit]], [[guinea pig]], [[giant panda]], [[cow]], [[rat]], and the [[naked mole-rat]].<ref name="Blast results for A1BG protein">{{cite web|title=NCBI Blast results for A1BG protein sequence|url=http://blast.ncbi.nlm.nih.gov/Blast.cgi#478530307|accessdate= | In addition to the table below, alpha-1B glycoprotein is also conserved in the [[white-cheeked crested gibbon]], [[baboon]], [[bolivian squirrel monkey]], [[sheep]], [[dog]], [[wild boar]], [[Treeshrew|Chinese tree shrew]], [[Chinese hamster]], [[black flying fox]], [[rabbit]], [[guinea pig]], [[giant panda]], [[cow]], [[rat]], and the [[naked mole-rat]].<ref name="Blast results for A1BG protein">{{cite web|title=NCBI Blast results for A1BG protein sequence|url=http://blast.ncbi.nlm.nih.gov/Blast.cgi#478530307|accessdate=May 11, 2013}}</ref> Additionally, it is very likely that A1BG is further conserved throughout the mammalian [[clade]]. | ||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 50: | Line 50: | ||
! [[Genus]] [[species]] !! Organism common name !! Divergence from humans (MYA) <ref name="Time Tree">{{cite web | title = Time Tree| url =http://www.timetree.org/| accessdate = }}</ref> !! NCBI protein accession number !! Sequence identity !! Protein length !! Common gene name | ! [[Genus]] [[species]] !! Organism common name !! Divergence from humans (MYA) <ref name="Time Tree">{{cite web | title = Time Tree| url =http://www.timetree.org/| accessdate = }}</ref> !! NCBI protein accession number !! Sequence identity !! Protein length !! Common gene name | ||
|- | |- | ||
| ''[[Homo sapiens]]''<ref>{{cite web|title=alpha-1-B glycoprotein [Homo sapiens]|url=https://www.ncbi.nlm.nih.gov/protein/119592981?report=genbank&log$=prottop&blast_rank=2&RID=SVXAZ6UZ01R|accessdate= | | ''[[Homo sapiens]]''<ref>{{cite web|title=alpha-1-B glycoprotein [Homo sapiens]|url=https://www.ncbi.nlm.nih.gov/protein/119592981?report=genbank&log$=prottop&blast_rank=2&RID=SVXAZ6UZ01R|accessdate=May 11, 2013}}</ref> || Humans || -- || NP_570602 || 100% || 495 || A1BG | ||
|- | |- | ||
| ''[[Pan troglodytes]]''<ref name="NCBI Reference Sequence: XP_001146669.1">{{cite web|title=PREDICTED: alpha-1B-glycoprotein isoform 4 [Pan troglodytes]|url=https://www.ncbi.nlm.nih.gov/protein/114679419?report=genbank&log$=prottop&blast_rank=6&RID=SVXAZ6UZ01R|publisher=NCBI|accessdate= | | ''[[Pan troglodytes]]''<ref name="NCBI Reference Sequence: XP_001146669.1">{{cite web|title=PREDICTED: alpha-1B-glycoprotein isoform 4 [Pan troglodytes]|url=https://www.ncbi.nlm.nih.gov/protein/114679419?report=genbank&log$=prottop&blast_rank=6&RID=SVXAZ6UZ01R|publisher=NCBI|accessdate=May 10, 2013}}</ref> || Common chimp || 6.2 || XP_001146669 || 97.0% || 501 || PREDICTED: Alpha-1B-glycoprotein isoform 4 | ||
|- | |- | ||
| ''[[Pan paniscus]]''<ref name="NCBI Reference Sequence: XP_003816677.1">{{cite web|title=PREDICTED: alpha-1B-glycoprotein [Pan paniscus]|url=https://www.ncbi.nlm.nih.gov/protein/397491452?report=genbank&log$=prottop&blast_rank=7&RID=SVXAZ6UZ01R|accessdate= | | ''[[Pan paniscus]]''<ref name="NCBI Reference Sequence: XP_003816677.1">{{cite web|title=PREDICTED: alpha-1B-glycoprotein [Pan paniscus]|url=https://www.ncbi.nlm.nih.gov/protein/397491452?report=genbank&log$=prottop&blast_rank=7&RID=SVXAZ6UZ01R|accessdate=May 11, 2013}}</ref> || Bonobo || 6.3 || XP_003816677 || 97.0% || 499 || A1BG | ||
|- | |- | ||
| ''[[Gorilla gorilla gorilla]]'' <ref name="NCBI Reference Sequence: XP_004061652.1">{{cite web|title=PREDICTED: alpha-1B-glycoprotein|url=https://www.ncbi.nlm.nih.gov/protein/426390528?report=genbank&log$=prottop&blast_rank=39&RID=SVXAZ6UZ01R|accessdate= | | ''[[Gorilla gorilla gorilla]]'' <ref name="NCBI Reference Sequence: XP_004061652.1">{{cite web|title=PREDICTED: alpha-1B-glycoprotein|url=https://www.ncbi.nlm.nih.gov/protein/426390528?report=genbank&log$=prottop&blast_rank=39&RID=SVXAZ6UZ01R|accessdate=May 10, 2013}}</ref> || Gorilla || 8.8 || XP_004061652 || 95.0% || 275 || PREDICTED: alpha-1B-glycoprotein | ||
|- | |- | ||
| ''[[Pongo abelii]]''<ref name="NCBI Reference Sequence: XP_002829953.1">{{cite web|title=Send to: PREDICTED: alpha-1B-glycoprotein isoform 1 [Pongo abelii]|url=https://www.ncbi.nlm.nih.gov/protein/297706253?report=genbank&log$=prottop&blast_rank=8&RID=SVXAZ6UZ01R|accessdate= | | ''[[Pongo abelii]]''<ref name="NCBI Reference Sequence: XP_002829953.1">{{cite web|title=Send to: PREDICTED: alpha-1B-glycoprotein isoform 1 [Pongo abelii]|url=https://www.ncbi.nlm.nih.gov/protein/297706253?report=genbank&log$=prottop&blast_rank=8&RID=SVXAZ6UZ01R|accessdate=May 11, 2013}}</ref> ||Sumatran Organutan|| 15.7 || XP_002829953 || 95.0% || 495 || alpha-1B-glycoprotein isoform 1 | ||
|- | |- | ||
| ''[[Macaca mulatta]]''<ref name="GenBank: EHH30490.1">{{cite web|title=hypothetical protein EGK_11172, partial [Macaca mulatta]|url=https://www.ncbi.nlm.nih.gov/protein/355703999?report=genbank&log$=prottop&blast_rank=9&RID=SVXAZ6UZ01R|accessdate= | | ''[[Macaca mulatta]]''<ref name="GenBank: EHH30490.1">{{cite web|title=hypothetical protein EGK_11172, partial [Macaca mulatta]|url=https://www.ncbi.nlm.nih.gov/protein/355703999?report=genbank&log$=prottop&blast_rank=9&RID=SVXAZ6UZ01R|accessdate=May 11, 2013}}</ref> || Rhesus monkey || 29.0 || XM_001101821 || 88.0% || 351 || hypothetical protein EGK_11172, partial | ||
|- | |- | ||
| ''[[Callithrix jacchus]]''<ref name="NCBI Reference Sequence: XP_002762619.2">{{cite web|title=PREDICTED: alpha-1B-glycoprotein [Callithrix jacchus]|url=https://www.ncbi.nlm.nih.gov/protein/390479473?report=genbank&log$=prottop&blast_rank=12&RID=SVXAZ6UZ01R|accessdate= | | ''[[Callithrix jacchus]]''<ref name="NCBI Reference Sequence: XP_002762619.2">{{cite web|title=PREDICTED: alpha-1B-glycoprotein [Callithrix jacchus]|url=https://www.ncbi.nlm.nih.gov/protein/390479473?report=genbank&log$=prottop&blast_rank=12&RID=SVXAZ6UZ01R|accessdate=May 11, 2013}}</ref> || Marmoset || 42.6 || XP_002762619 || 83.0% || 500 || A1BG | ||
|- | |- | ||
| ''[[Mus musculus]]'' <ref name="NCBI Reference Sequence: NP_001074536.1">{{cite web|title=alpha-1B-glycoprotein precursor [Mus musculus]|url=https://www.ncbi.nlm.nih.gov/protein/124486702?report=genbank&log$=prottop&blast_rank=37&RID=SVXAZ6UZ01R|accessdate= | | ''[[Mus musculus]]'' <ref name="NCBI Reference Sequence: NP_001074536.1">{{cite web|title=alpha-1B-glycoprotein precursor [Mus musculus]|url=https://www.ncbi.nlm.nih.gov/protein/124486702?report=genbank&log$=prottop&blast_rank=37&RID=SVXAZ6UZ01R|accessdate=May 11, 2013}}</ref> || Mouse || 91.0 || NP_001074536 || 44.0% || 512 || alpha-1B-glycoprotein precursor | ||
|- | |- | ||
| ''[[Felis catus]]''<ref name="NCBI Reference Sequence: XP_003997399.1">{{cite web|title=PREDICTED: alpha-1B-glycoprotein [Felis catus]|url=https://www.ncbi.nlm.nih.gov/protein/410982098?report=genbank&log$=prottop&blast_rank=24&RID=SVXAZ6UZ01R|accessdate= | | ''[[Felis catus]]''<ref name="NCBI Reference Sequence: XP_003997399.1">{{cite web|title=PREDICTED: alpha-1B-glycoprotein [Felis catus]|url=https://www.ncbi.nlm.nih.gov/protein/410982098?report=genbank&log$=prottop&blast_rank=24&RID=SVXAZ6UZ01R|accessdate=May 11, 2013}}</ref> || Cat || 94.2 || XP_003997399 || 62.0% || 481 || PREDICTED: alpha-1B-glycoprotein | ||
|- | |- | ||
| ''[[Equus caballus]]''<ref name="NCBI Reference Sequence: XP_001495344.3">{{cite web|title=PREDICTED: alpha-1B-glycoprotein-like|url=https://www.ncbi.nlm.nih.gov/protein/338710440?report=genbank&log$=prottop&blast_rank=33&RID=SVXAZ6UZ01R|accessdate= | | ''[[Equus caballus]]''<ref name="NCBI Reference Sequence: XP_001495344.3">{{cite web|title=PREDICTED: alpha-1B-glycoprotein-like|url=https://www.ncbi.nlm.nih.gov/protein/338710440?report=genbank&log$=prottop&blast_rank=33&RID=SVXAZ6UZ01R|accessdate=May 11, 2013}}</ref> || Horse || 97.4 || XP_001495344 || 58.0% || 568 || PREDICTED: alpha-1B-glycoprotein-like | ||
|- | |- | ||
| ''[[Loxodonta africana]]'' <ref name="NCBI Reference Sequence: XP_003406722.1">{{cite web|title=PREDICTED: alpha-1B-glycoprotein-like [Loxodonta africana]|url=https://www.ncbi.nlm.nih.gov/protein/344269773?report=genbank&log$=prottop&blast_rank=28&RID=SVXAZ6UZ01R}}</ref> || African bush elephant || 104.7 || XP_003406722 || 61.0% || 520 || PREDICTED: alpha-1B-glycoprotein-like | | ''[[Loxodonta africana]]'' <ref name="NCBI Reference Sequence: XP_003406722.1">{{cite web|title=PREDICTED: alpha-1B-glycoprotein-like [Loxodonta africana]|url=https://www.ncbi.nlm.nih.gov/protein/344269773?report=genbank&log$=prottop&blast_rank=28&RID=SVXAZ6UZ01R}}</ref> || African bush elephant || 104.7 || XP_003406722 || 61.0% || 520 || PREDICTED: alpha-1B-glycoprotein-like | ||
| Line 76: | Line 76: | ||
=== Paralogs === | === Paralogs === | ||
No [[paralogs]] have been found for alpha-1B glycoprotein.<ref name="Gene Cards">{{cite web|title=A1BG Gene|url= | No [[paralogs]] have been found for alpha-1B glycoprotein.<ref name="Gene Cards">{{cite web|title=A1BG Gene|url=https://www.genecards.org/cgi-bin/carddisp.pl?gene=A1BG&search=a1bg#publications|publisher=Weissman Institute of Science|accessdate=May 10, 2013}}</ref> | ||
=== Homologous domains === | === Homologous domains === | ||
An initial NCBI Blast alignment of alpha-1B glycoprotein illustrates that the protein is mainly composed of three [[immunoglobulin]] domains.<ref>{{cite web|title=NCBI conserved domain search|url=https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?SEQUENCE=NP_570602.2&FULL|accessdate= | An initial NCBI Blast alignment of alpha-1B glycoprotein illustrates that the protein is mainly composed of three [[immunoglobulin]] domains.<ref>{{cite web|title=NCBI conserved domain search|url=https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?SEQUENCE=NP_570602.2&FULL|accessdate=May 10, 2013}}</ref> There is a large segment of amino acids from position 297 to 400 that is not shown to be an immunoglobulin domain. However, a NCBI BLAST alignment of just the amino acids from 297-400 does illustrate that the latter sequence is indeed a fourth immunoglobulin domain.<ref>{{cite web|title=NCBI Blast: Protein Sequence|url=http://blast.ncbi.nlm.nih.gov/Blast.cgi|accessdate=May 10, 2013}}</ref> Ultimately, alpha-1B glycoprotein seems to be primarily composed of four immunoglobulin domains. | ||
== Clinical significance == | == Clinical significance == | ||
Revision as of 20:36, 31 May 2018
| VALUE_ERROR (nil) | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Aliases | |||||||
| External IDs | GeneCards: [1] | ||||||
| Orthologs | |||||||
| Species | Human | Mouse | |||||
| Entrez |
|
| |||||
| Ensembl |
|
| |||||
| UniProt |
|
| |||||
| RefSeq (mRNA) |
|
| |||||
| RefSeq (protein) |
|
| |||||
| Location (UCSC) | n/a | n/a | |||||
| PubMed search | n/a | n/a | |||||
| Wikidata | |||||||
| |||||||
Alpha-1-B glycoprotein is a 54.3 kDa protein in humans that is encoded by the A1BG gene. [1] The protein encoded by this gene is a plasma glycoprotein of unknown function. The protein shows sequence similarity to the variable regions of some immunoglobulin supergene family member proteins. Patients who have pancreatic ductal adenocarcinoma show an overexpression of A1BG in pancreatic juice.[2]
Gene
Neighborhood
A1BG is located on the negative DNA strand of chromosome 19 from 58,858,172 – 58,864,865.[3] Additionally, A1BG is located directly adjacent to the ZSCAN22 gene (58,838,385-58,853,712)) on the positive DNA strand, as well as the ZNF837 (58,878,990 - 58,892,389, complement) and ZNF497 (58865723 - 58,874,214, complement) genes on the negative strand.[3]
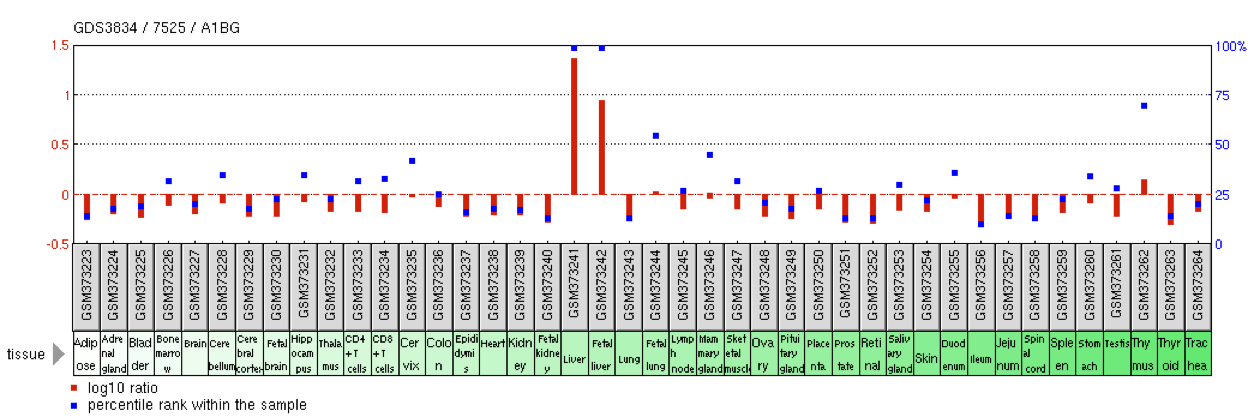
Expression
A1BG is expressed at high levels in the adult and fetal liver.[4] Additionally, the mammary gland shows roughly half as much expression as the liver.[4] Trace amounts of A1BG expression can be found in the blood, brain, lung, lymph node, ovary, testis, pancreas, and pancreas.[4] Liver tumors exhibit elevated levels of A1BG transcripts.[4]
mRNA
mRNA structure
The gene contains 20 distinct introns.[5] Transcription produces 15 different mRNAs, 10 alternatively spliced variants and 5 unspliced forms.[5] There are 4 probable alternative promoters, 4 non overlapping alternative last exons and 7 validated alternative polyadenylation sites.[5] The mRNAs appear to differ by truncation of the 5' end, truncation of the 3' end, presence or absence of 4 cassette exons, overlapping exons with different boundaries, splicing versus retention of 3 introns.[5]
Protein
Properties
The San Diego Super Computer's Statistical Analysis of Protein (SAPS) program determined that alpha-1B glycoprotein has 495 amino acids residues, an isoelectric point of 5.47, and a molecular mass of 54.3 kDa. Additionally, it suggested that no transmembrane domains exist in alpha-1B glycoprotein.[6] According to NCBI, the amino acid sequence MLVVFLLLWGVTWGPVTEA is a signal peptide on the N-terminus of the protein that might function as an endoplasmic reticulum import signal.[6]
Post-translational modifications
The NetAcet 1.0 program calculated that the first five amino acid residues serve as an N-acetylation site.[7] The NetGlycate 1.0 program predicted that the lysines located at residue 78, 114, and 227 serve as glycation points.[8] The NetNES 1.1 program predicted the leucine at residue 47 to be a nuclear export signal.[9] The NetNGlyc 1.0 program predicted four N-glycosylation sites - two of which are highly conserved internally repeated sequences.[10][11] The NetCGlyc1.0 program predicted that none of the tryptophan residues serve as C-mannosylation sites.[12]
Protein interactions
A study by Udby et al. showed that Cysteine-rich secretory protein 3 is a ligand of alpha-1B glycoprotein in human plasma and they suggest that the A1BG-CRISP-3 complex displays a similar function in protecting the circulation from a potentially harmful effect of free CRISP-3.[13]
Homology
Orthologs
In addition to the table below, alpha-1B glycoprotein is also conserved in the white-cheeked crested gibbon, baboon, bolivian squirrel monkey, sheep, dog, wild boar, Chinese tree shrew, Chinese hamster, black flying fox, rabbit, guinea pig, giant panda, cow, rat, and the naked mole-rat.[14] Additionally, it is very likely that A1BG is further conserved throughout the mammalian clade.
| Genus species | Organism common name | Divergence from humans (MYA) [15] | NCBI protein accession number | Sequence identity | Protein length | Common gene name |
|---|---|---|---|---|---|---|
| Homo sapiens[16] | Humans | -- | NP_570602 | 100% | 495 | A1BG |
| Pan troglodytes[17] | Common chimp | 6.2 | XP_001146669 | 97.0% | 501 | PREDICTED: Alpha-1B-glycoprotein isoform 4 |
| Pan paniscus[18] | Bonobo | 6.3 | XP_003816677 | 97.0% | 499 | A1BG |
| Gorilla gorilla gorilla [19] | Gorilla | 8.8 | XP_004061652 | 95.0% | 275 | PREDICTED: alpha-1B-glycoprotein |
| Pongo abelii[20] | Sumatran Organutan | 15.7 | XP_002829953 | 95.0% | 495 | alpha-1B-glycoprotein isoform 1 |
| Macaca mulatta[21] | Rhesus monkey | 29.0 | XM_001101821 | 88.0% | 351 | hypothetical protein EGK_11172, partial |
| Callithrix jacchus[22] | Marmoset | 42.6 | XP_002762619 | 83.0% | 500 | A1BG |
| Mus musculus [23] | Mouse | 91.0 | NP_001074536 | 44.0% | 512 | alpha-1B-glycoprotein precursor |
| Felis catus[24] | Cat | 94.2 | XP_003997399 | 62.0% | 481 | PREDICTED: alpha-1B-glycoprotein |
| Equus caballus[25] | Horse | 97.4 | XP_001495344 | 58.0% | 568 | PREDICTED: alpha-1B-glycoprotein-like |
| Loxodonta africana [26] | African bush elephant | 104.7 | XP_003406722 | 61.0% | 520 | PREDICTED: alpha-1B-glycoprotein-like |
Paralogs
No paralogs have been found for alpha-1B glycoprotein.[27]
Homologous domains
An initial NCBI Blast alignment of alpha-1B glycoprotein illustrates that the protein is mainly composed of three immunoglobulin domains.[28] There is a large segment of amino acids from position 297 to 400 that is not shown to be an immunoglobulin domain. However, a NCBI BLAST alignment of just the amino acids from 297-400 does illustrate that the latter sequence is indeed a fourth immunoglobulin domain.[29] Ultimately, alpha-1B glycoprotein seems to be primarily composed of four immunoglobulin domains.
Clinical significance
Steroid-resistant nephrotic syndrome
The alpha-1-glycoprotein is upregulated 11-fold in the urine of patients who have steroid resistant nephrotic syndrome.[30] A1BG was present in 7/19 patients with SRNS and was absent from all patients with steroid sensitive nephrotic syndrome.[30] The 13.8 kDa A1BG fragment had a high discriminatory power for steroid resistance in pediatric nephrotic syndrome, but is only present in a subset of patients.[30]
References
- ↑ "Entrez Gene: Alpha-1-B glycoprotein". Retrieved 2012-11-09.
- ↑ Tian M, Cui YZ, Song GH, Zong MJ, Zhou XY, Chen Y, Han JX (2008). "Proteomic analysis identifies MMP-9, DJ-1 and A1BG as overexpressed proteins in pancreatic juice from pancreatic ductal adenocarcinoma patients". BMC Cancer. 8: 241. doi:10.1186/1471-2407-8-241. PMC 2528014. PMID 18706098.
- ↑ 3.0 3.1 "A1BG alpha-1-B glycoprotein". Retrieved May 10, 2013.
- ↑ 4.0 4.1 4.2 4.3 "EST Profile - Hs.529161". UniGene. National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2013-05-11.
- ↑ 5.0 5.1 5.2 5.3 "AceView: A1BG". Retrieved May 11, 2013.
- ↑ 6.0 6.1 Brendel V, Bucher P, Nourbakhsh IR, Blaisdell BE, Karlin S (March 1992). "Methods and algorithms for statistical analysis of protein sequences". Proc. Natl. Acad. Sci. U.S.A. 89 (6): 2002–6. doi:10.1073/pnas.89.6.2002. PMC 48584. PMID 1549558.
- ↑ Kiemer L, Bendtsen JD, Blom N (April 2005). "NetAcet: prediction of N-terminal acetylation sites". Bioinformatics. 21 (7): 1269–70. doi:10.1093/bioinformatics/bti130. PMID 15539450.
- ↑ Johansen MB, Kiemer L, Brunak S (September 2006). "Analysis and prediction of mammalian protein glycation". Glycobiology. 16 (9): 844–53. doi:10.1093/glycob/cwl009. PMID 16762979.
- ↑ la Cour T, Kiemer L, Mølgaard A, Gupta R, Skriver K, Brunak S (June 2004). "Analysis and prediction of leucine-rich nuclear export signals". Protein Eng. Des. Sel. 17 (6): 527–36. doi:10.1093/protein/gzh062. PMID 15314210. Retrieved May 10, 2013.
- ↑ Gupta, R. "Prediction of N-glycosylation sites in human proteins". Retrieved May 10, 2013.
- ↑ Higgins DG, Bleasby AJ, Fuchs R (April 1992). "CLUSTAL V: improved software for multiple sequence alignment". Comput. Appl. Biosci. 8 (2): 189–91. doi:10.1093/bioinformatics/8.2.189. PMID 1591615.
- ↑ Julenius, Karin (2007). "NetCGlyc1.0: Prediction of mammalian C-mannosylation sites". Glycobiology. 17: 868–876. doi:10.1093/glycob/cwm050. PMID 17494086. Retrieved May 10, 2013.
- ↑ Udby L, Sørensen OE, Pass J, Johnsen AH, Behrendt N, Borregaard N, Kjeldsen L (October 2004). "Cysteine-rich secretory protein 3 is a ligand of alpha1B-glycoprotein in human plasma". Biochemistry. 43 (40): 12877–86. doi:10.1021/bi048823e. PMID 15461460.
- ↑ "NCBI Blast results for A1BG protein sequence". Retrieved May 11, 2013.
- ↑ "Time Tree".
- ↑ "alpha-1-B glycoprotein [Homo sapiens]". Retrieved May 11, 2013.
- ↑ "PREDICTED: alpha-1B-glycoprotein isoform 4 [Pan troglodytes]". NCBI. Retrieved May 10, 2013.
- ↑ "PREDICTED: alpha-1B-glycoprotein [Pan paniscus]". Retrieved May 11, 2013.
- ↑ "PREDICTED: alpha-1B-glycoprotein". Retrieved May 10, 2013.
- ↑ "Send to: PREDICTED: alpha-1B-glycoprotein isoform 1 [Pongo abelii]". Retrieved May 11, 2013.
- ↑ "hypothetical protein EGK_11172, partial [Macaca mulatta]". Retrieved May 11, 2013.
- ↑ "PREDICTED: alpha-1B-glycoprotein [Callithrix jacchus]". Retrieved May 11, 2013.
- ↑ "alpha-1B-glycoprotein precursor [Mus musculus]". Retrieved May 11, 2013.
- ↑ "PREDICTED: alpha-1B-glycoprotein [Felis catus]". Retrieved May 11, 2013.
- ↑ "PREDICTED: alpha-1B-glycoprotein-like". Retrieved May 11, 2013.
- ↑ "PREDICTED: alpha-1B-glycoprotein-like [Loxodonta africana]".
- ↑ "A1BG Gene". Weissman Institute of Science. Retrieved May 10, 2013.
- ↑ "NCBI conserved domain search". Retrieved May 10, 2013.
- ↑ "NCBI Blast: Protein Sequence". Retrieved May 10, 2013.
- ↑ 30.0 30.1 30.2 Piyaphanee N, Ma Q, Kremen O, Czech K, Greis K, Mitsnefes M, Devarajan P, Bennett MR (June 2011). "Discovery and initial validation of α 1-B glycoprotein fragmentation as a differential urinary biomarker in pediatric steroid-resistant nephrotic syndrome". Proteomics: Clinical Applications. 5 (5–6): 334–42. doi:10.1002/prca.201000110. PMID 21591266.
External links
- Human A1BG genome location and A1BG gene details page in the UCSC Genome Browser.