Homology (biology)
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [2]
Overview
In biology, homology refers to the general and quite ancient observation of similarity of form seen in the biological world of animals or plants.[1] It is the name given to the anatomical correspondences between different species that biologists and paleontologists have noted and studied for centuries.
More specifically, in evolutionary biology, homology has come to mean any similarity between characters that is due to their shared ancestry. There are examples in different branches of biology. Anatomical structures that perform the same function in different biological species and evolved from the same structure in some ancestor species are homologous. In genetics, homology can be observed in DNA sequences that code for proteins (genes) and in noncoding DNA. For protein coding genes, one can compare translated amino-acid sequences of different genes. Genes that share a high sequence identity or similarity support the hypothesis that they share a common ancestor and are therefore homologous. Sequence homology may also indicate common function. Homologous chromosomes are non-identical chromosomes that can pair (synapse) during meiosis, and are believed to share common ancestry.
The word homologous derives from the ancient Greek ομολογειν, 'to agree'.
Homology of structures in evolution
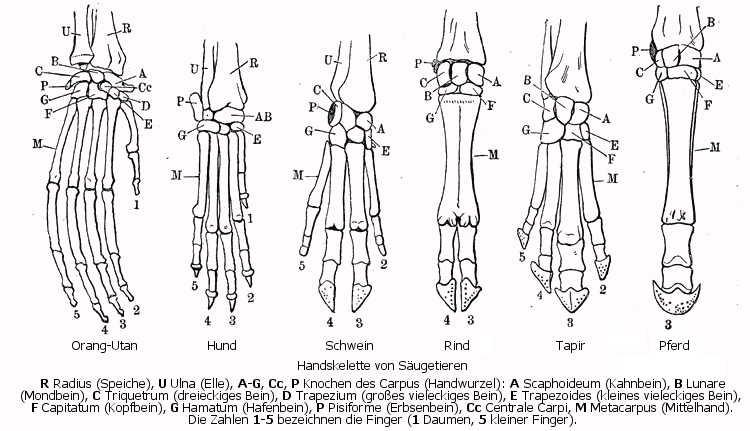
Shared ancestry can be evolutionary or developmental. Evolutionary ancestry means that structures evolved from some structure in a common ancestor; for example, the wings of bats and the arms of humans are homologous in this sense. Developmental ancestry means that structures arose from the same tissue in embryonal development; the ovaries of female humans and the testicles of male humans are homologous in this sense.
Homology is different from analogy. The wings of a maple seed and the wings of an albatross are analogous but not homologous (they both allow the organism to travel on the wind, but they didn't both develop from the same structure). This is called homoplasy. But structures can be homologous and analogous. The wings of a bat and a bird are homologous, in that they both developed from the pectoral fins of fish. They are also analogous, in that the forelimbs of the ancestors of birds and of bats developed into organs of a similar new function independently. Thus evolution can be initially divergent, giving rise to homologous structures, and subsequently convergent, causing the structures to become analogous again.
It has recently been discovered that structures previously considered analogous but not homologous are actually homologous. Sean Carroll's books (see notes, below) contain elegant descriptions of these new discoveries.
Homology of sequences in genetics
Homology among proteins and DNA is often concluded on the basis of sequence similarity, especially in bioinformatics. For example, in general, if two or more genes have highly similar DNA sequences, it is likely that they are homologous. But sequence similarity may arise from different ancestors: short sequences may be similar by chance, and sequences may be similar because both were selected to bind to a particular protein, such as a transcription factor. Such sequences are similar but not homologous. Sequence regions that are homologous are also called conserved. This is not to be confused with conservation in amino acid sequences in which the amino acid at a specific position has changed but the physio-chemical properties of the amino acid remain unchanged.
The phrase "percent homology" is sometimes used but is incorrect. "Percent identity" or "percent similarity" should be used to quantify the similarity between the biomolecule sequences. For two naturally occurring sequences, percent identity is a factual measurement, whereas homology is a hypothesis supported by evidence. One can, however, refer to partial homology where a fraction of the sequences compared (are presumed to) share descent, while the rest does not. For example, partial homology may result from a gene fusion event.
Many algorithms exist to cluster protein sequences into sequence families, which are sets of mutually homologous sequences. (See sequence clustering and sequence alignment.)
Homologous sequences are of two types: orthologous and paralogous. [3]
Orthology
Homologous sequences are orthologous if they were separated by a speciation event: when a species diverges into two separate species, the divergent copies of a single gene in the resulting species are said to be orthologous. Orthologs, or orthologous genes, are genes in different species that are similar to each other because they originated from a common ancestor. The term "ortholog" was coined in 1970.
The strongest evidence that two similar genes are orthologous is the result of a phylogenetic analysis of the gene lineage. Genes that are found within one clade are orthologs, descended from a common ancestor. Orthologs often, but not always, have the same function.
Orthologous sequences provide useful information in taxonomic classification studies of organisms. The pattern of genetic divergence can be used to trace the relatedness of organisms. Two organisms that are very closely related are likely to display very similar DNA sequences between two orthologs. Conversely, an organism that is further removed evolutionarily from another organism is likely to display a greater divergence in the sequence of the orthologs being studied.
Paralogy
Homologous sequences are paralogous if they were separated by a gene duplication event: if a gene in an organism is duplicated to occupy two different positions in the same genome, then the two copies are paralogous.
A set of sequences that are paralogous are called paralogs of each other. Paralogs typically have the same or similar function, but sometimes do not: due to lack of the original selective pressure upon one copy of the duplicated gene, this copy is free to mutate and acquire new functions.
Paralogous sequences provide useful insight to the way genomes evolve. The genes encoding myoglobin and hemoglobin are considered to be ancient paralogs. Similarly, the four known classes of hemoglobins (hemoglobin A, hemoglobin A2, hemoglobin S, and hemoglobin F) are paralogs of each other. While each of these genes serves the same basic function of oxygen transport, they have already diverged slightly in function: fetal hemoglobin (hemoglobin F) has a higher affinity for oxygen than adult hemoglobin.
Another example can be found in rodents such as rats and mice. Rodents have a pair of paralogous insulin genes, although it is unclear if any divergence in function has occurred.
Paralogous genes often belong to the same species, but this is not necessary: for example, the hemoglobin gene of humans and the myoglobin gene of chimpanzees are paralogs. This is a common problem in bioinformatics: when genomes of different species have been sequenced and homologous genes have been found, one can not immediately conclude that these genes have the same or similar function, as they could be paralogs whose function has diverged.
Ohnology
Ohnologous genes are paralogous genes that have originated by a process of whole-genome duplication (WGD). The name was first given in honour of Susumu Ohno by Ken Wolfe.[2] Ohnologs are interesting for evolutionary analysis because they all have been diverging for the same length of time since their common origin.
Xenology
Homologs resulting from horizontal gene transfer between two organisms are termed xenologs. Xenologs can have different functions, if the new environment is vastly different for the horizontally moving gene. In general, though, xenologs typically have similar function in both organisms.[3]
Gametology
Gametology denotes the relationship between homologous genes on nonrecombining, opposite sex chromosomes. Gametologs result from the origination of genetic sex determination and barriers to recombination between sex chromosomes. Examples of gametologs include CHDW and CHDZ in birds.
Homologous chromosome sets
Homologous chromosomes are non-identical chromosomes that can pair (synapse) during meiosis.[4] Except for the sex chromosomes, homologous chromosomes share significant sequence similarity across their entire length, typically contain the same sequence of genes, and pair up to allow for proper disjunction during meiosis. The chromosomes can also undergo cross-over at this stage. There may be some variations between genes on homologs giving rise to alternate forms or alleles. Sex chromosomes have a shorter region of sequence similarity. Based on the sequence similarity and our knowledge of biology, it is believed that they are paralogous.
Sources
- Fitch, W.M. 2000. Homology: a personal view on some of the problems. Trends Genet. 16(5):227-31.
- Dewey, C and Pachter L. "Evolution at the Nucleotide Level: The Problem of Whole-Genome Multiple Alignment" Human Molecular Genetics 15:R51-R56.
- Haeckel, Е. Generelle Morphologie der Organismen. Bd 1-2. Вerlin, 1866.
- Gegenbaur, G. Vergleichende Anatomie der Wirbelthiere... Leipzig, 1898.
- Owen, R. On the archetype and homologies of the vertebrate skeleton. London, 1847.
- DePinna, M.C.C. 1991. "Concepts and tests of homology in the cladistic paradigm." Cladistics 7: 367-394.
- Mindell, D.P. and A. Meyer. 2001. "Homology evolving." Trends in Ecology and Evolution 16:434-440.
- Carroll, S. Endless Forms Most Beautiful, New York, 2005
- Carroll, S. The Making of the Fittest New York, 2006
References
- ↑ http://www.natcenscied.org/icons/icon3homology.html icons of evolution
- ↑ Ken Wolfe (2000) Robustness—it's not where you think it is. Nature Genetics May;25(1):3-4. [1]
- ↑ NCBI Phylogenetics Factsheet
- ↑ RC King and WD Stansfield (1997). A Dictionary of Genetics (5th Edition ed.). Oxford University Press.
See also
| Wikimedia Commons has media related to Homology. |
External links
ca:Homologia (Biologia) de:Homologie (Biologie) nl:Homoloog (genetica) no:Homologi (biologi) simple:Orthology sr:Хомологија (биологија) sv:Homologi