DNA microarray
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]
Overview
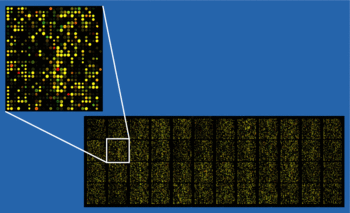
A DNA microarray (also commonly known as gene or genome chip, DNA chip, or gene array) is a collection of microscopic DNA spots, commonly representing single genes, arrayed on a solid surface by covalent attachment to chemically suitable matrices. DNA arrays are different from other types of microarray only in that they either measure DNA or use DNA as part of its detection system. Qualitative or quantitative measurements with DNA microarrays utilize the selective nature of DNA-DNA or DNA-RNA hybridization under high-stringency conditions and fluorophore-based detection. DNA arrays are commonly used for expression profiling, i.e., monitoring expression levels of thousands of genes simultaneously, or for comparative genomic hybridization.
Introduction
Arrays of DNA can either be spatially arranged, as in the commonly known gene or genome chip, DNA chip, or gene array, or can be specific DNA sequences tagged or labelled such that they can be independently identified in solution. The traditional solid-phase array is a collection of microscopic DNA spots attached to a solid surface, such as glass, plastic or silicon chip. The affixed DNA segments are known as probes (although some sources will use different nomenclature such as reporters), thousands of which can be placed in known locations on a single DNA microarray. Microarray technology evolved from Southern blotting, whereby fragmented DNA is attached to a substrate and then probed with a known gene or fragment.
Applications of these arrays include:
- mRNA or gene expression profiling - Monitoring expression levels for thousands of genes simultaneously is relevant to many areas of biology and medicine, such as studying treatments, disease, and developmental stages. For example, microarrays can be used to identify disease genes by comparing gene expression in diseased and normal cells (reference?).
- comparative genomic hybridization (Array CGH) - Assessing large genomic rearrangements.
- SNP detection arrays - Looking for Single nucleotide polymorphism in the genome of populations.
- Chromatin immunoprecipitation (chIP) studies - Determining protein binding site occupancy throughout the genome, employing ChIP-on-chip technology.
Fabrication
Microarrays can be fabricated using a variety of technologies, including printing with fine-pointed pins onto glass slides, photolithography using pre-made masks, photolithography using dynamic micromirror devices, ink-jet printing, [1] or electrochemistry on microelectrode arrays.
DNA microarrays can be used to detect RNAs that may or may not be translated into active proteins. Scientists refer to this kind of analysis as "expression analysis" or expression profiling. Since there can be tens of thousands of distinct probes on an array, each microarray experiment can accomplish the equivalent number of genetic tests in parallel. Arrays have therefore dramatically accelerated many types of investigations.
The use of a collection of distinct DNAs in arrays for expression profiling was first described in 1987, and the arrayed DNAs were used to identify genes whose expression is modulated by interferon. [2] These early gene arrays were made by spotting cDNAs onto filter paper with a pin-spotting device. The use of miniaturized microarrays for gene expression profiling was first published in 1995[3] (Science) and the first complete eukaryotic genome (Saccharomyces cerevisiae) on a microarray was published in 1997 (Science).
Spotted microarrays
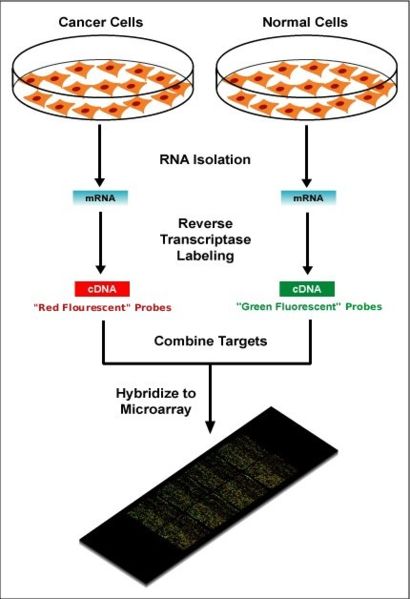
In spotted microarrays (or two-channel or two-colour microarrays), the probes are oligonucleotides, cDNA or small fragments of PCR products that correspond to mRNAs and are spotted onto the microarray surface. This type of array is typically hybridized with cDNA from two samples to be compared (e.g. diseased tissue versus healthy tissue) that are labeled with two different fluorophores. Fluorescent dyes commonly used for cDNA labelling include Cy3, which has a fluorescence emission wavelength of 570 nm (corresponding to the green part of the light spectrum), and Cy5 with a fluorescence emission wavelength of 670 nm (corresponding to the red part of the light spectrum). The two Cy-labelled cDNA samples are mixed and hybridized to a single microarray that is then scanned in a microarray scanner to visualize fluorescence of the two fluorophores after excitation with a laser beam of a defined wavelength. Relative intensities of each fluorophore may then be used in ratio-based analysis to identify up-regulated and down-regulated genes. Absolute levels of gene expression cannot be determined in the two-colour array, but relative differences in expression among different spots (=genes) can be estimated with some oligonucleotide arrays. Examples of providers for such microarrays includes Agilent with their Dual-Mode platform, Eppendorf with their DualChip platform, and TeleChem International with ArrayIt.
Oligonucleotide microarrays

In oligonucleotide microarrays (or single-channel microarrays), the probes are designed to match parts of the sequence of known or predicted mRNAs. There are commercially available designs that cover complete genomes from companies such as GE Healthcare, Affymetrix or Agilent. These microarrays give estimations of the absolute value of gene expression and therefore the comparison of two conditions requires the use of two separate microarrays.
Oligonucleotide Arrays can be either produced by piezoelectric deposition with full length oligonucleotides or in-situ synthesis.
Long Oligonucleotide Arrays are composed of 60-mers, or 50-mers and are produced by ink-jet printing on a silica substrate. Short Oligonucleotide Arrays are composed of 25-mer or 30-mer and are produced by photolithographic synthesis (Affymetrix) on a silica substrate or piezoelectric deposition (GE Healthcare) on an acrylamide matrix. More recently, Maskless Array Synthesis from NimbleGen Systems has combined flexibility with large numbers of probes. Arrays can contain up to 390,000 spots, from a custom array design. New array formats are being developed to study specific pathways or disease states for a systems biology approach.
Oligonucleotide microarrays often contain control probes designed to hybridize with RNA spike-ins. The degree of hybridization between the spike-ins and the control probes is used to normalize the hybridization measurements for the target probes.
Genotyping microarrays
DNA microarrays can also be used to read the sequence of a genome in particular positions.
SNP microarrays are a particular type of DNA microarrays that are used to identify genetic variation in individuals and across populations. Short oligonucleotide arrays can be used to identify the single nucleotide polymorphisms (SNPs) that are thought to be responsible for genetic variation and the source of susceptibility to genetically caused diseases. Generally termed genotyping applications, DNA microarrays may be used in this fashion for forensic applications, rapidly discovering or measuring genetic predisposition to disease, or identifying DNA-based drug candidates.
These SNP microarrays are also being used to profile somatic mutations in cancer, specifically loss of heterozygosity events and amplifications and deletions of regions of DNA. Amplifications and deletions can also be detected using comparative genomic hybridization, or aCGH, in conjunction with microarrays, but may be limited in detecting novel Copy Number Polymorphisms, or CNPs, by probe coverage.
Resequencing arrays have also been developed to sequence portions of the genome in individuals. These arrays may be used to evaluate germline mutations in individuals, or somatic mutations in cancers.
Genome tiling arrays include overlapping oligonucleotides designed to blanket an entire genomic region of interest. Many companies have successfully designed tiling arrays that cover whole human chromosomes.
Microarrays and bioinformatics
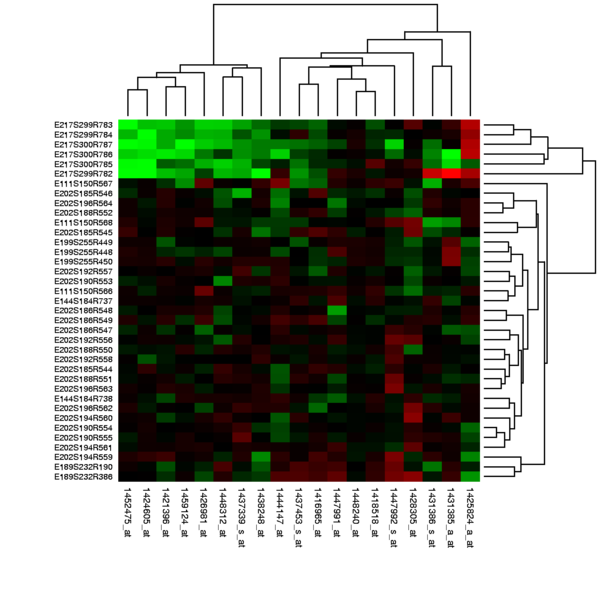
Experimental Design
Due to the biological complexity of gene expression, the considerations of experimental design that are discussed in the expression profiling article are of critical importance if statistically and biologically valid conclusions are to be drawn from the data.
- There are three main elements to consider when designing a microarray experiment. First, replication of the biological samples is essential for drawing conclusions from the experiment. Second, technical replicates (two RNA samples obtained from each experimental unit) help to ensure precision and allow for testing differences within treatment groups. The technical replicates may be two independent RNA extractions or two aliquots of the same extraction. Third, spots of each cDNA clone or oligonucleotide are present at least as duplicates on the microarray slide, to provide a measure of technical precision in each hybridization. It is critical that information about the sample preparation and handling is discussed in order to help identify the independent units in the experiment as well as to avoid inflated estimates of significance [4]
Standardization
The lack of standardization in arrays presents an interoperability problem in bioinformatics, which hinders the exchange of array data. Various grass-roots open-source projects are attempting to facilitate the exchange and analysis of data produced with non-proprietary chips.
- The "Minimum Information About a Microarray Experiment" (MIAME) checklist helps define the level of detail that should exist and is being adopted by many journals as a requirement for the submission of papers incorporating microarray results. MIAME describes the minimum required information for complying experiments, but not its format. Thus, as of 2007, whilst many formats can support the MIAME requirements there is no format which permits verification of complete semantic compliance.
- The "MicroArray Quality Control (MAQC) Project" is being conducted by the FDA to develop standards and quality control metrics which will eventually allow the use of MicroArray data in drug discovery, clinical practice and regulatory decision-making. [5]
- The MicroArray and Gene Expression (MAGE) group is working on the standardization of the representation of gene expression data and relevant annotations.
Statistical analysis
The analysis of DNA microarrays poses a large number of statistical problems, including the normalization of the data. There are dozens of proposed normalization methods in the published literature; as in many other cases where authorities disagree, a sound conservative approach is to try a number of popular normalization methods and compare the conclusions reached: how sensitive are the main conclusions to the method chosen?
From a hypothesis-testing perspective, the large number of genes present on a single array means that the experimenter must take into account a multiple testing problem: even if the statistical P-value assigned to a given gene indicates that it is extremely unlikely that differential expression of this gene was due to random rather than treatment effects, the very high number of genes on an array makes it likely that differential expression of some genes represent false positives or false negatives. Statistical methods tailored to microarray analyses have recently become available that assess statistical power based on the variation present in the data and the number of experimental replicates, and can help minimize type I and type II errors in the analyses.[6]
A basic difference between microarray data analysis and much traditional biomedical research is the dimensionality of the data. A large clinical study might collect 100 data items per patient for thousands of patients. A medium-size microarray study will obtain many thousands of numbers per sample for perhaps a hundred samples. Many analysis techniques treat each sample as a single point in a space with thousands of dimensions, then attempt by various techniques to reduce the dimensionality of the data to something humans can visualize.
Relation between probe and gene
The relation between a probe and the mRNA that it is expected to detect is problematic. On the one hand, some mRNAs may cross-hybridize probes in the array that are supposed to detect another mRNA. On the other hand, probes that are designed to detect the mRNA of a particular gene may be relying on genomic EST information that is incorrectly associated with that gene.
Public databases of microarray data
| Database | Microarray Experiment Sets | Sample Profiles | as of Date |
| Gene Expression Omnibus - NCBI | 5366 | 134669 | April 1, 2007 |
| Stanford Microarray database | 12742 | ? | April 1, 2007 |
| UPenn RAD database | ~100 | ~2500 | Sept. 1, 2007 |
| UNC Microarray database | ~31 | 2093 | April 1, 2007 |
| MUSC database | ~45 | 555 | April 1, 2007 |
| ArrayExpress at EBI | 1643 | 136 | April 1, 2007 |
| caArray at NCI | 41 | 1741 | November 15, 2006 |
- For a directory of Microarray Databases, see: Template:Dmoz
- See also the Microarray databases page in Wikipedia
Online microarray data analysis programs and tools
Several Open Directory Project categories list online microarray data analysis programs and tools:
- Template:Dmoz
- Template:Dmoz
- Template:Dmoz
- Template:Dmoz
- Bioconductor: open source and open development software project for the analysis and comprehension of genomic data
- Genevestigator : Web-based database and analysis tool to study gene expression across large sets of tissues, developmental stages, drugs, stimuli, and genetic modifications.
Notable microarray related articles
- For microarray companies, see: Template:Dmoz
References
- ↑ http://genomebiology.com/2004/5/8/R58
- ↑ Kulesh, D, Clive, DR, Zarlenga, DS, Greene, J. (1987). Identification of interferon-modulated proliferation-related cDNA sequences. Proc. Natl. Acad. Sci. USA. Dec;84(23):8453-7.
- ↑ Schena M, Shalon D, Davis RW, Brown PO. (1995). Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. Oct 20; 270 (5235): 467-70.
- ↑ http://www.vmrf.org/research-websites/gcf/Forms/Churchill.pdf.
- ↑ http://www.fda.gov/nctr/science/centers/toxicoinformatics/maqc/
- ↑ Wei C, Li J, Bumgarner RE. (2004). "Sample size for detectng differentially expressed genes in microarray experiments". BMC Genomics. 5: 87. PMID 15533245.
External links
- Many important links can be found at the Open Directory Project
- Microarray data processing using Self-Organizing Maps tutorial: Part 1 Part 2
ar:مصفوفة دنا صغيرة de:Microarray el:Μικροσυστοιχίες γονιδίων it:Microarray he:מערכי DNA nl:DNA-microarray sv:Microarrays ur:ڈی این اے خورد منظومہ