Salmonella: Difference between revisions
Joao Silva (talk | contribs) No edit summary |
Joao Silva (talk | contribs) |
||
| Line 19: | Line 19: | ||
|} | |} | ||
''Salmonella'' is a [[ | ''Salmonella'' is a [[gram-negative]], facultative [[intracellular]], [[anaerobic]], non-spore-forming [[bacillum]]. It measures 2 to 3 by 0.4 to 0.6 μm. [[Salmonella]] is a non-[[lactose]] fermenting bacterium. It reduces [[nitrates]], produces acid on glucose fermentation and does not produce cytochrome oxidase.<ref>{{cite book | last = Mandell | first = Gerald | title = Mandell, Douglas, and Bennett's principles and practice of infectious diseases | publisher = Churchill Livingstone/Elsevier | location = Philadelphia, PA | year = 2010 | isbn = 0443068399 }}</ref> Due to the presence of [[flagella]], almost all [[salmonella]] species are motile. 1% of the bacteria is able to ferment [[lactose]], which may be responsible for its non-detection in some culture media. | ||
For the isolation of [[salmonella]] in culture | For the isolation of [[salmonella]] in culture, freshly passed stool are preferred. Common media for the growth of [[salmonella]] include: [[MacConkey agar]], deoxycholate agar, and xylose-lysine-deoxycholate agar.<ref name="PerezCavalli2003">{{cite journal|last1=Perez|first1=J. M.|last2=Cavalli|first2=P.|last3=Roure|first3=C.|last4=Renac|first4=R.|last5=Gille|first5=Y.|last6=Freydiere|first6=A. M.|title=Comparison of Four Chromogenic Media and Hektoen Agar for Detection and Presumptive Identification of Salmonella Strains in Human Stools|journal=Journal of Clinical Microbiology|volume=41|issue=3|year=2003|pages=1130–1134|issn=0095-1137|doi=10.1128/JCM.41.3.1130-1134.2003}}</ref> | ||
When the sample has a low number of [[bacteria]], special enrichment broths, such as the selenite-based enrichment broth, may be used to | When the sample has a low number of [[bacteria]], special enrichment broths, such as the selenite-based enrichment broth, may be used to raise the number of [[bacteria]].<ref name="PerezCavalli2003">{{cite journal|last1=Perez|first1=J. M.|last2=Cavalli|first2=P.|last3=Roure|first3=C.|last4=Renac|first4=R.|last5=Gille|first5=Y.|last6=Freydiere|first6=A. M.|title=Comparison of Four Chromogenic Media and Hektoen Agar for Detection and Presumptive Identification of Salmonella Strains in Human Stools|journal=Journal of Clinical Microbiology|volume=41|issue=3|year=2003|pages=1130–1134|issn=0095-1137|doi=10.1128/JCM.41.3.1130-1134.2003}}</ref> | ||
===Infectious Cycle=== | ===Infectious Cycle=== | ||
[[Salmonella enterica]] enters the body through the mouth, by ingestion of contaminated food and water. For the [[bacteria]] to cause disease, an [[inoculum]] of about 50 000 [[ | [[Salmonella enterica]] enters the body through the mouth, by [[ingestion]] of contaminated food and water. For the [[bacteria]] to cause disease, an [[inoculum]] of about 50 000 [[pathogens]] is often required. Once in the [[intestine]], the [[bacteria]] will first infect the apical [[epithelium]]. [[Salmonella]] will then initiate bacterial mechanisms that allow invasion of the host cells, inducing [[inflammatory]] changes, such as:<ref name="pmid534385">{{cite journal| author=McGovern VJ, Slavutin LJ| title=Pathology of salmonella colitis. | journal=Am J Surg Pathol | year= 1979 | volume= 3 | issue= 6 | pages= 483-90 | pmid=534385 | doi= | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=534385 }} </ref><ref name="pmid4630603">{{cite journal| author=Giannella RA, Formal SB, Dammin GJ, Collins H| title=Pathogenesis of salmonellosis. Studies of fluid secretion, mucosal invasion, and morphologic reaction in the rabbit ileum. | journal=J Clin Invest | year= 1973 | volume= 52 | issue= 2 | pages= 441-53 | pmid=4630603 | doi=10.1172/JCI107201 | pmc=PMC302274 | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=4630603 }} </ref><ref name="pmid3551701">{{cite journal| author=Clarke RC, Gyles CL| title=Virulence of wild and mutant strains of Salmonella typhimurium in ligated intestinal segments of calves, pigs, and rabbits. | journal=Am J Vet Res | year= 1987 | volume= 48 | issue= 3 | pages= 504-10 | pmid=3551701 | doi= | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=3551701 }} </ref><ref name="pmid2919285">{{cite journal| author=Finlay BB, Heffron F, Falkow S| title=Epithelial cell surfaces induce Salmonella proteins required for bacterial adherence and invasion. | journal=Science | year= 1989 | volume= 243 | issue= 4893 | pages= 940-3 | pmid=2919285 | doi= | pmc= | url=http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=2919285 }} </ref> | ||
*Diffuse and focal infiltration of [[PMN]] | *Diffuse and focal infiltration of [[PMN]] | ||
*[[Crypt abscess]] | *[[Crypt abscess]]es | ||
*[[Necrosis]] of the [[epithelium]] | *[[Necrosis]] of the [[epithelium]] | ||
*Fluid secretion | *Fluid secretion | ||
Revision as of 23:00, 22 August 2014
|
Salmonellosis Microchapters |
|
Diagnosis |
|---|
|
Treatment |
|
Case Studies |
|
Salmonella On the Web |
|
American Roentgen Ray Society Images of Salmonella |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]; Associate Editor(s)-in-Chief: João André Alves Silva, M.D. [2] Jolanta Marszalek, M.D. [3]
Overview
Salmonella is a rod-shaped, facultative intracellular, gram-negative enterobacteria.[1] Salmonella species shows motility, produces hydrogen sulfide, and only 1% is able to ferment lactose.[2] It may be isolated from the stool of infected patients, and grown in culture media, such as MacConkey agar and deoxycholate agar. Salmonella enters the body through contaminated food or water, and invades the intestinal epithelial cells, causing inflammation.
This bacterium may be classified into 2 different species: Salmonella enterica and Salmonella bongori. Salmonella enterica is divided in 6 different subspecies, of which I, contains most pathogenic serotypes for humans. Salmonella may be serogrouped into more than 2500 serovars with polyvalent antisera, according to the capsular antigen, polysaccharide O antigens, and flagellar antigens. The bacteria show tropism for the epithelial cells of the gastrointestinal tract, macrophages, dendritic cells, and neutrophils. Different serotypes of Salmonella may have different natural reservoirs, some have humans as their only natural reservoir (serotypes Sendai and Typhi), while others (serotype Dublin) may infect humans and cattle.
Taxonomy
Cellular organism; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae[3]
Biology
 |
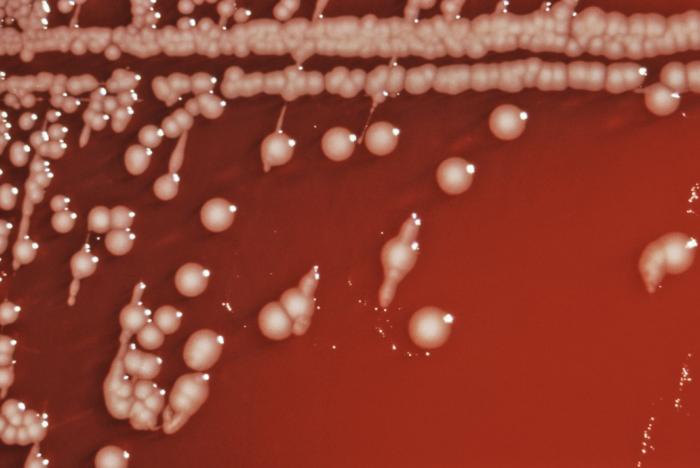 |
Salmonella is a gram-negative, facultative intracellular, anaerobic, non-spore-forming bacillum. It measures 2 to 3 by 0.4 to 0.6 μm. Salmonella is a non-lactose fermenting bacterium. It reduces nitrates, produces acid on glucose fermentation and does not produce cytochrome oxidase.[5] Due to the presence of flagella, almost all salmonella species are motile. 1% of the bacteria is able to ferment lactose, which may be responsible for its non-detection in some culture media.
For the isolation of salmonella in culture, freshly passed stool are preferred. Common media for the growth of salmonella include: MacConkey agar, deoxycholate agar, and xylose-lysine-deoxycholate agar.[6]
When the sample has a low number of bacteria, special enrichment broths, such as the selenite-based enrichment broth, may be used to raise the number of bacteria.[6]
Infectious Cycle
Salmonella enterica enters the body through the mouth, by ingestion of contaminated food and water. For the bacteria to cause disease, an inoculum of about 50 000 pathogens is often required. Once in the intestine, the bacteria will first infect the apical epithelium. Salmonella will then initiate bacterial mechanisms that allow invasion of the host cells, inducing inflammatory changes, such as:[7][8][9][10]
- Diffuse and focal infiltration of PMN
- Crypt abscesses
- Necrosis of the epithelium
- Fluid secretion
- Edema
Different serovars will have different preferable intestinal locations. An example is the enterocolitis at the terminal ileum, cecum, and proximal colon caused by serovar Typhimurium. Intestinal disease is marked by neutrophil migration to the intestinal epithelium. This recruitment is done by the secretion of interleukin-8, induced by Salmonella.[11]
Classification
Before 1983 Salmonella were classified in different species. However, genome studies have shown high levels of DNA similarities among the different types of salmonella, which lead to the actual classification of salmonella in 2 different species:
Salmonella enterica
- Contains six subspecies - I, II, IIIa, IIIb, IV, and VI:[12][13]
- I - enterica
- II - salamae
- III - arizonae
- IIIb - diarizonae
- IV - houtenae
- VI - indica
- Subspecies I contains most pathogenic serotypes for humans (99.5%)
- Subspecies IIIa and IIIb, formerly belonging to the genus Arizonae, are responsible for rare human infections
Salmonella bongori
- Formerly subspecies V[14]
Serovars
Salmonella subspecies may be serogrouped, into more than 2500 serovars with polyvalent antisera. For this division the following bacterial structures are considered:[15]
- Capsular antigen
- Polysaccharide O antigens
- Flagellar antigens
Different salmonella serotypes may also be distinguished in cultures according to their different metabolism of sugars.[16]
Tropism
In vitro Salmonella are able to interact with different cell types. However, in vivo, the bacteria were found to enter only certain human cells, namely:[17]
- Epithelial cells of the gastrointestinal tract - initially infected by Salmonella
- Macrophages - where Salmonella survives and replicates
- Dendritic cells - relevant for dissemination of bacteria in initial stages of infection, yet, they are not considered adequate reservoirs for Salmonella at latter stages
- Neutrophils - interaction is based on immune response against the bacteria, hence might not be considered tropism
Natural Reservoir
Different salmonella serovars may have different natural reservoirs. Common types of serovars of salmonella enterica that infect the human gastrointestinal tract include serovars Sendai, Typhi, and Paratyphi. Humans are their only natural reservoir. Other serotypes, such as serotype Dublin, have cattle as their natural reservoir, also being able to cause human infection.[18][19]
Related Chapters
- 1984 Rajneeshee bioterror attack
- List of foodborne illness outbreaks
- Food Testing Strips
References
- ↑ Ryan KJ, Ray CG (editors) (2004). Sherris Medical Microbiology (4th ed. ed.). McGraw Hill. ISBN 0-8385-8529-9.
- ↑ Giannella RA (1996). "Salmonella". In Baron S et al (eds.). Baron's Medical Microbiology (4th ed. ed.). Univ of Texas Medical Branch. ISBN 0-9631172-1-1.
- ↑ "Salmonella (Taxonomy)".
- ↑ 4.0 4.1 "Public Health Image Library (PHIL), Centers for Disease Control and Prevention".
- ↑ Mandell, Gerald (2010). Mandell, Douglas, and Bennett's principles and practice of infectious diseases. Philadelphia, PA: Churchill Livingstone/Elsevier. ISBN 0443068399.
- ↑ 6.0 6.1 Perez, J. M.; Cavalli, P.; Roure, C.; Renac, R.; Gille, Y.; Freydiere, A. M. (2003). "Comparison of Four Chromogenic Media and Hektoen Agar for Detection and Presumptive Identification of Salmonella Strains in Human Stools". Journal of Clinical Microbiology. 41 (3): 1130–1134. doi:10.1128/JCM.41.3.1130-1134.2003. ISSN 0095-1137.
- ↑ McGovern VJ, Slavutin LJ (1979). "Pathology of salmonella colitis". Am J Surg Pathol. 3 (6): 483–90. PMID 534385.
- ↑ Giannella RA, Formal SB, Dammin GJ, Collins H (1973). "Pathogenesis of salmonellosis. Studies of fluid secretion, mucosal invasion, and morphologic reaction in the rabbit ileum". J Clin Invest. 52 (2): 441–53. doi:10.1172/JCI107201. PMC 302274. PMID 4630603.
- ↑ Clarke RC, Gyles CL (1987). "Virulence of wild and mutant strains of Salmonella typhimurium in ligated intestinal segments of calves, pigs, and rabbits". Am J Vet Res. 48 (3): 504–10. PMID 3551701.
- ↑ Finlay BB, Heffron F, Falkow S (1989). "Epithelial cell surfaces induce Salmonella proteins required for bacterial adherence and invasion". Science. 243 (4893): 940–3. PMID 2919285.
- ↑ McCormick BA, Colgan SP, Delp-Archer C, Miller SI, Madara JL (1993). "Salmonella typhimurium attachment to human intestinal epithelial monolayers: transcellular signalling to subepithelial neutrophils". J Cell Biol. 123 (4): 895–907. PMC 2200157. PMID 8227148.
- ↑ "The type species of the genus Salmonella Lignieres 1900 is Salmonella enterica (ex Kauffmann and Edwards 1952) Le Minor and Popoff 1987, with the type strain LT2T, and conservation of the epithet enterica in Salmonella enterica over all earlier epithets that may be applied to this species. Opinion 80". Int J Syst Evol Microbiol. 55 (Pt 1): 519–20. 2005. PMID 15653929.
- ↑ Tindall BJ; Grimont PAD, Garrity GM; Euzéby JP (2005). "Nomenclature and taxonomy of the genus Salmonella". Int J Syst Evol Microbiol. 55: 521&ndash, 524. PMID 15653930.
- ↑ Popoff MY, Bockemühl J, Gheesling LL (2004). "Supplement 2002 (no. 46) to the Kauffmann-White scheme". Res Microbiol. 155 (7): 568–70. doi:10.1016/j.resmic.2004.04.005. PMID 15313257.
- ↑ Murray, Patrick (2013). Medical microbiology. Philadelphia: Elsevier/Saunders. ISBN 0323086926.
- ↑ Mandell, Gerald (2010). Mandell, Douglas, and Bennett's principles and practice of infectious diseases. Philadelphia, PA: Churchill Livingstone/Elsevier. ISBN 0443068399.
- ↑ Santos RL, Bäumler AJ (2004). "Cell tropism of Salmonella enterica". Int J Med Microbiol. 294 (4): 225–33. doi:10.1016/j.ijmm.2004.06.029. PMID 15532980.
- ↑ Mandell, Gerald (2010). Mandell, Douglas, and Bennett's principles and practice of infectious diseases. Philadelphia, PA: Churchill Livingstone/Elsevier. ISBN 0443068399.
- ↑ "Salmonella enterica Serotypes and Food Commodities, United States, 1998–2008".
- CS1 maint: Extra text: authors list
- CS1 maint: Extra text
- CS1 maint: Extra text: editors list
- CS1 maint: Multiple names: authors list
- CS1 maint: PMC format
- Bacterial diseases
- Gram negative bacteria
- Enterobacteria
- Foodborne illnesses
- Zoonoses
- Disease
- Infectious disease
- Emergency medicine
- Intensive care medicine
- Rat carried diseases