Nucleosol
|
WikiDoc Resources for Nucleosol |
|
Articles |
|---|
|
Most recent articles on Nucleosol |
|
Media |
|
Evidence Based Medicine |
|
Clinical Trials |
|
Ongoing Trials on Nucleosol at Clinical Trials.gov Clinical Trials on Nucleosol at Google
|
|
Guidelines / Policies / Govt |
|
US National Guidelines Clearinghouse on Nucleosol
|
|
Books |
|
News |
|
Commentary |
|
Definitions |
|
Patient Resources / Community |
|
Patient resources on Nucleosol Discussion groups on Nucleosol Directions to Hospitals Treating Nucleosol Risk calculators and risk factors for Nucleosol
|
|
Healthcare Provider Resources |
|
Causes & Risk Factors for Nucleosol |
|
Continuing Medical Education (CME) |
|
International |
|
|
|
Business |
|
Experimental / Informatics |
Editor-In-Chief: Henry A. Hoff
Overview
The structural and functional unit of all known living organisms is the cell. Eukaryotic cells of which humans are composed contain membrane-bound compartments. The most important among these is the cell nucleus. It is the "control center" for each cell that has at least one. Separating the nucleus from the cellular cytoplasm is the double-membraned structure of the nuclear envelope.
Inside the nuclear membrane, including attached to the inner layer of this membrane, are a variety of substances serving special purposes. Depending on conditions, for a time, some of these compose the nucleosol, some compose the nucleohyaloplasm, and still others compose the nucleoplasm. Those substances and the special conditions present, even if temporary, in water can be the nucleosol. The nucleus cannot succeed in its efforts to control the cell without the special properties of the nucleosol.
Introduction
Generally, in chemistry a fluid suspension of a colloidal solid in a liquid is referred to as a sol. Whereas, a solution is a liquid mixture of a minor component (the solute) distributed uniformly within the major component (the solvent). In the nucleosol the liquid is water and the solvent is also water.
Using the water (data page) and the atomic radii of the elements (data page), a water molecule is approximately 120 x 187 pm in maximum diameters (average 154 pm). For its small size a water molecule (H2O), 18 Da, at about 38°C, under 1 atm of pressure (about 101325 kgm-1s-2), in large enough numbers to produce a liquid exhibits a high viscosity, short-range order, clustering, and miscibility.
The molecule is polar enough to produce and maintain ions when in a liquid. Liquid water can also dissolve organic compounds such as carbohydrates, fatty acids, amino acids, nucleobases, nucleosides, nucleotides, and peptides. Diffusion of molecules, ions, or particles in liquid water is depended upon by most living organisms. In cell biology, diffusion is the main form of transport for the smaller necessary materials such as amino acids within cells.[1] Metabolism and respiration rely in part upon diffusion in addition to bulk or active processes. For example, in the alveoli of mammalian lungs, due to differences in partial pressures across the alveolar-capillary membrane, oxygen diffuses into the blood (another water-based liquid) and carbon dioxide diffuses out.
Diffusion in liquid water depends on the lateral speed of the water molecule within liquid water. The lateral speed of a water molecule within liquid water appears to be unknown but can be estimated.Template:Wiktionarypar
A plasm is a formative or formed material; i.e., something molded. Usually a formed material keeps it shape once the mold is removed. Should something be glassy or transparent it can be said to be hyaloid. Whether a fluid is molded or liquid is often a matter of viscosity and whether a fluid is transparent, translucent, or opaque is often a matter of absorption.
Before any internal molding exists within the nucleus or when any portion of the existing nucleoskeleton is dissolved and reconstructed elsewhere, genes within the euchromatin are transcribed, mRNAs are synthesized, and the mers that are to become new portions of the nucleoskeleton become part of the nucleosol: actins (microfilaments), lamins (intermediate filaments), and tubulins (microtubules).
In addition to internal molding by the nucleoskeleton there are local increases in viscosity due to macromolecular crowding that help to compose the nucleohyaloplasm and the nucleoplasm of which it is a part.
Water
Water, whether a gas (water vapor) or liquid, is an amorphous substance.
| File:Wiktionary-logo-en-v2.svg | Look up amorphous in Wiktionary, the free dictionary. |
The chemical bonding within many amorphous substances can produce short-range order while there is long-range disorder.
| File:Wiktionary-logo-en-v2.svg | Look up disorder in Wiktionary, the free dictionary. |
The short-range order is often a symmetrical arrangement of polyhedra. The long-range disorder can be approached with the disordered arrangement of space-filling polyhedra. These polyhedra are bonded together in a solid and undergo bond breaking through the transitions from solid to fluid. A model based on the configuron is an approach to understanding the viscosity changes that occur with changes in temperature.
In the kinetic theory of gases (such as water vapor) there is a correlation between average energy (Eav) and temperature (T):
Eav = 3/2 kT with Eav = 1/2 mvav2.
⇒ vav2 = 3kT/m.
For a water molecule m = 18 Da, or 2.9913 x 10-23 gmolecule-1.
⇒ vrms = √3kT/m = √3·1.382x10-23 J/K·306 K/(0.018 kg/6.022x1023) = 651 ms-1 at 33°C. vrms is the root mean square velocity.
However, water in the nucleosol is a liquid, not a gas. For example, vrms (O2) in air at 25°C is 482.1 ms-1.
Enthalpy of vaporization for water
The enthalpy of vaporization for water at 25°C is 44 kJmol-1. The enthalpy of vaporization can be viewed as the energy required to overcome the intermolecular interactions in water. The molecules in liquid water are held together by relatively strong hydrogen bonds. But care must be taken, however, when using enthalpies of vaporization to measure the strength of intermolecular forces, as these forces may persist to an extent in the gas phase (as is the case with hydrogen fluoride), and so the calculated value of the bond strength will be too low. With water the enthalpy of vaporization is larger than the available kinetic energy suggested by the ideal gas law:
Eav = 3/2 kT with Eav = 1/2 mvav2.
The lateral speed of a water molecule estimated using the enthalpy of vaporization is ~2,210 ms-1. For water vapor at 25°C, by the ideal gas law, vrms = 643 ms-1.
Enthalpy of flow for liquid water
As a liquid much of the available kinetic energy is expressed through additional degrees of freedom. Some of this energy is in the form of intermolecular bonds. These bonds are a resistance to flow.
Water has a resistance to flow that is considered relatively "thin", having a lower viscosity than other liquids such as vegetable oil. At 25°C, water has a nominal viscosity of 1.0 × 10-3 Pa∙s and motor oil has a nominal apparent viscosity of 250 × 10-3 Pa∙s.[2] Because of its density of <math>\rho</math> = 1 g/cm3 (varies slightly with temperature), and its dynamic viscosity near 1 mPa·s, the viscosity values of water are, to rough precision, all powers of ten:
Dynamic viscosity:
Kinematic viscosity:
- <math>{\nu}</math> = 1 cSt = 10-2 stokes = 1 mm²/s
<math>{\nu}</math> = <math>{\mu}</math>/<math>{\rho}</math>, where <math>{\rho}</math> is the density (kg/m3).
In water
- viscosity is independent of pressure (except at very high pressure); and
- viscosity tends to fall as temperature increases: from 1.79 cP to 0.28 cP in the temperature range from 0°C to 100°C; see temperature dependence of liquid viscosity for more details.
The viscosity of water is 8.94 × 10−4 Pa·s or 8.94 × 10−3 dyn·s/cm2 or 10−1 cP at about 25 °C.
As a function of temperature T (K):
μ(Pa·s) = A × 10B/(T−C)
where A=2.414 × 10−5 Pa·s ; B = 247.8 K ; and C = 140 K.
Whereas, for a gas such as air, the viscosity is much lower and also depends mostly on temperature. At 15.0 °C, the viscosity of air is 1.78 × 10−5 kg/(m·s) or 1.78 × 10−4 cP. One can get the viscosity of air as a function of temperature from the Gas Viscosity Calculator.
Viscous flow, which results from viscosity, in amorphous materials such as water is a thermally activated process:[3]
- <math>{\mu_L} = A_L \cdot e^{Q_L/RT},</math>
where QL is the activation energy in the liquid state, T is temperature, R is the molar gas constant and AL is approximately a constant.
With
- QL ≥ Hm,
where Hm is the enthalpy of motion of the broken hydrogen bonds. Hm ~ 1/2 mvav2. Here, the energy of motion for flow suggests a lateral speed of ~1590 ms-1.
Because of its small molecular size and ability to quickly rearrange, it cannot be made amorphous without resorting to specialized hyperquenching techniques. These produce amorphous ice, which has a quenching rate in the range of metallic glasses.[4]
The higher the temperature of an amorphous material the higher the configuron concentration. The higher the configuron concentration the lower the viscosity. As configurons form percolating clusters, an amorphous solid can transition to a liquid. This clustering facilitates viscous flow. Thermodynamic parameters of configurons can be found from viscosity-temperature relationships.[4]
Short-range order
Like a liquid an amorphous solid has a topologically disordered distribution of particles but elastic properties of an isotropic solid. The symmetry similarity of both liquid and solid phases cannot explain the qualitative differences in their behavior.
Due to chemical bonding characteristics amorphous solids such as glasses do possess a high degree of short-range order with respect to local atomic polyhedra.[5] The amorphous structure of glassy silica has no long range order but shows local ordering with respect to the tetrahedral arrangement of oxygen atoms around silicon atoms.
The most ubiquitous, and perhaps simplest, example of a hydrogen bond is found between water molecules. This intermolecular hydrogen bonding is responsible for the high boiling point of water (100 °C) due to the high number of hydrogen bonds each molecule can have relative to its low molecular mass, and the great strength of these bonds. The length of hydrogen bonds depends on bond strength, temperature, and pressure. The bond strength itself is dependent on temperature, pressure, bond angle, and environment (usually characterized by local dielectric constant). The typical length of a hydrogen bond (H-bond) in water is 1.97 Å (197 pm).
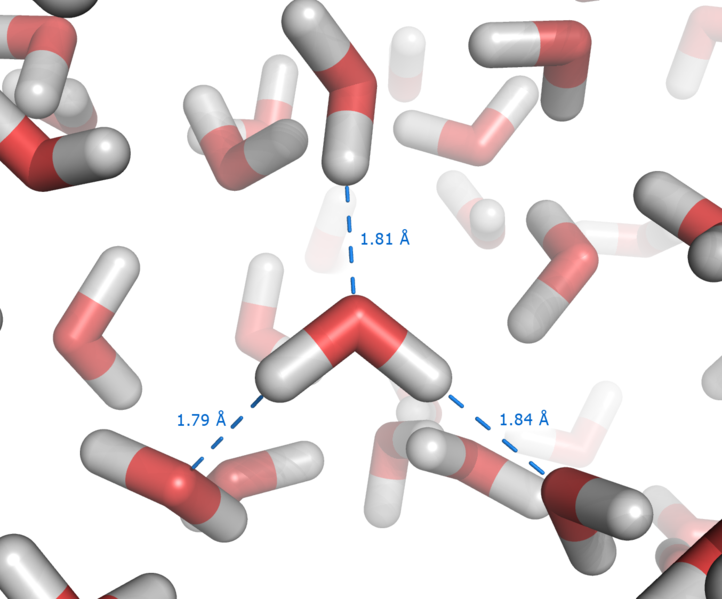
Every water molecule can be H-bonded with up to four other molecules, as shown in the figure (two through its two lone pairs, and two through its two hydrogen atoms.) Water has a very high boiling point, melting point and viscosity compared to other similar substances not conjoined by hydrogen bonds.
The exact number of hydrogen bonds in which a molecule in liquid water participates fluctuates with time and depends on the temperature. From liquid water simulations at 25 °C, each water molecule may participate in an average of 3.59 hydrogen bonds.[6] At 100 °C, this number apparently decreases to 3.24 due to the increased molecular motion and decreased density, while at 0 °C, the average number of hydrogen bonds increases to 3.69.[6] A more recent study found a much smaller number of hydrogen bonds: 2.357 at 25 °C.[7] The differences may be due to the use of a different method for defining and counting the hydrogen bonds.
Where the bond strengths are more equivalent, one might instead find the atoms of two interacting water molecules partitioned into two polyatomic ions of opposite charge, specifically hydroxide (OH−) and hydronium (H3O+). (Hydronium ions are also known as 'hydroxonium' ions.)
- H-O− H3O+
Indeed, in pure water under conditions of standard temperature and pressure, this latter formulation is applicable only rarely; on average about one in every 5.5 × 108 molecules gives up a proton to another water molecule, in accordance with the value of the dissociation constant for water under such conditions. It is a crucial part of the uniqueness of water.
It can be that a single hydrogen atom participates in two hydrogen bonds, rather than one. This type of bonding is called "bifurcated". It was suggested that a bifurcated hydrogen atom is an essential step in water reorientation.[8]
Water clusters
In chemistry a water cluster is a discrete hydrogen bonded assembly or cluster of molecules of water [9]. These clusters have been found experimentally or predicted in silico in various forms of water; in ice, in crystal lattices and in bulk liquid water, the simplest one being the water dimer (W2). Water manifests itself as clusters. Water clusters are also implicated in the stabilization of certain supramolecular structures. Supramolecular forces include hydrogen bonding that result in molecular self-assembly, folding, and mechanically-interlocked molecular architectures.[10]
In silico, cyclic water clusters (H2O)n are found with n = 3 to 6. Many isomeric forms seem to exist for the hexamer: from ring, book, bag, cage, to prism shape with nearly identical energy. Two cage-like isomers exist for heptamers and octamers: either cyclic or in the shape of a cube. Even larger clusters are predicted: the fullerene-like cluster W28 is called bucky water and even for a 280 water molecule monster icosahedral network (with each water molecule coordinated to 4 others) there is found a local energy minimum.
When water is trapped in a liquid helium environment the hexamer occurs as a cyclic planar assembly but in the gas-phase it is the cage form and in an organic host (water trapped in the crystal lattice of an organic compound) the hexamer occurs as a conformation reminiscent of a cyclohexane chair. Cubic configurations for clusters in the range W8-W10 have been found.
Another heptamer (cyclic twisted nonplanar) is the conformation for water as a hydrate in a crystal structure.[11]
Recent research has revealed that liquid water at ambient conditions has two distinct structures, either a very disordered or a very tetrahedral (clusters) at any temperature, which are spatially separated.[12] As temperature increases the very tetrahedral structure concentration decreases and the very disordered increases.
Glass transition temperature of water
The glass transition temperature for water is about 136 K or -137°C. Factors in the formation of amorphous ice include ingredients that form a heterogenous mixture with water (such as is used in the production of ice cream), pressure (which may convert one form into another), and cryoprotectants that lower its freezing point and increase viscosity. Melting low-density amorphous ice (LDA) between 140 and 210 K through its transition temperature shows that it is more viscous than normal water.[13] LDA has a density of 0.94 g/cm³, less dense than the densest water (1.00 g/cm³ at 277 K), but denser than ordinary ice.
Amorphous ice is used in some scientific experiments, especially in electron cryomicroscopy of biomolecules.[14] The individual molecules can be preserved for imaging in a state close to what they are in liquid water.
Enthalpy of motion for water configurons
A simple estimate of QL can be obtained by using the two temperatures 0°C and 100°C, where µ=1.79 x 10-3 Pa·s at 0°C and 0.28 x 10-3 Pa·s at 100°C, and solving for AL and QL. AL = 7.7 x 10-7 Pa·s and QL = 18 kJmol-1. R=8.314472 JK-1mol-1. Temperature is in K (273.15 + °C). QL includes the energy to break the hydrogen bond and move the configuron, as such HM ≤ QL. Using AL and QL to calculate the viscosity of water and comparing the calculated values to the experimentally determined ones for a range of temperature values shows that there is a systematic deviation at the higher temperatures. As the data for the viscosity of water vapor is available, AV and QV can be estimated: AV ~ 1.2 x 10-4 Pa·s and QV ~ - 6.0 kJmol-1. This added to the calculated configuron contribution
- <math>{\mu_L} = A_L \cdot e^{Q_L/RT} + A_V \cdot e^{Q_V/RT},</math>
improves the fit to the liquid water viscosity data remarkably well, suggesting that like other gas molecules mixed into water, water vapor can also be.[15]
But, here again the enthalpy of motion for water configurons suggests an average lateral speed of 1410 ms-1 and the enthalpy of dissolved water molecules suggests ~816 ms-1 which is also too high.
Average speed for water molecules
Although a liquid, some idea of the average speed for water molecules can be estimated using the self-diffusion coefficients for water at various temperatures. In the gas phase, <math>\, \lambda</math> is often defined as the diffusional mean free path, by assuming a simple approximate relation is exact:
<math>D = \frac{1}{2}\lambda \,v</math>,
where <math>\, v</math> is the root mean square (rms) speed of the gas molecules:
<math>v = \sqrt {{3\, k\, T}\over{m}}</math>
where
<math>\, m</math> is the mass of the diffusing species. This approximate equation becomes exact when used to define the diffusional mean free path. If the diffusional mean free path were nominally the same number of µm as vrms, then, with the apparent self-diffusion coefficients (ADCs) ranging from 0.58 x 10-9 to 1.23 x 10-9 m2s-1[16], an approximate vrms would range from 34-50 µms-1 for λ from 34-50 µm. So if in liquid water λ is much greater, then vrms may be much less.
The self-diffusion coefficient, D, for pure liquid water has been measured for temperatures between 2°C and 225°C at pressures up to 1.75 kbar.[17]
The average speeds of translation for various small molecules having the same kinetic energy as a water molecule, when water is at 500 nms-1, m = 18 Da, are Pi at 200 nms-1, m = 96 Da, adenosine (Ado) at 100 nms-1, m = 267 Da, and EC 3.1.3.5 at 6 nms-1, m = 140 kDa (homodimer), for the reaction:
Ado + Pi <=> H2O + AMP,
although EC 3.1.3.5 (70 kDa) may not occur within the nucleosol. But EC 3.1.3.5 NT5C1A cytosolic 5'-nucleotidase 1A, m = 41 kDa[18] can, with 82 kDa as a homodimer.
But, if the average speed of lateral translation for a water molecule were on the order of 500 nms-1, an average cell could not operate ~64,000 RNA polymerase II transcription units by diffusion of ATP, CTP, GTP, and UTP to the RNA polymerase II transcription locations to produce, e.g., the mRNA of dystrophin, at the rate needed of ~42 NTPs per second. For a lateral speed of ~37 µms-1 of a water molecule, an average cell could operate ~14,400 transcription units by diffusion, ~75 µms-1, ~28,800 transcription units, and ~167 µms-1, ~64,000 transcription units. This in turn suggests λs of ~32, ~15, and ~6.9 µm.
Signal transduction also depends on the lateral speed of a water molecule (diffusion) to succeed. With completion times on the order of a few minutes, this would mean the speed of a water molecule is on the order of 15 µms-1 and not much faster due to benefits from concentration. Increased lateral speed of a water molecule much above ~35 µms-1 may make signal transduction occur too quickly.
The diffusion of solutes and macromolecules in aqueous cellular compartments such as the nucleosol within the nucleoplasm and the nucleus is required for numerous processes including metabolism, signaling and protein-protein interactions.[19] Diffusion of small molecules through the cytosol is about fourfold slower than in pure water, due mostly to collisions with large numbers of macromolecules.[19] Recent measurements have indicated unexpectedly high mobilities (µ) of small solutes and macromolecules.[19]
By the Einstein relation,
<math>\mu = \frac{q}{k\, T}D</math>,
where
- <math>\, q</math> is the charge of the species,
- <math>\, k</math> is the Boltzmann constant,
- <math>\, T</math> is the gas temperature;
thus, approximately, for water, solutes and macromolecules, mobilities are directly proportional to vrms:
<math>\mu = \frac{q}{2k\, T}\lambda \,v_{\rm rms}</math>.
Miscible molecules
Miscible molecules such as O2, CO2, N2, and NH3 occur in any bodily fluid. These molecules are mixed into the liquid, but not turned into ions. Water contains only 1/20 parts O2. N2 mixes into the bloodstream and body fats.
Inorganic ions
The inorganic ions in the nucleosol should be very similar in type and concentration to the cytosol as each sol is free to interdiffuse with the other.
| Ion | Concentration in cytosol (millimolar) | Concentration in blood (millimolar) |
|---|---|---|
| Potassium (K+) | 139 | 4 |
| Sodium (Na+) | 12 | 145 |
| Magnesium (Mg2+) | 0.8 | 1.5 |
| Calcium (Ca2+) | <0.0002 | 1.8 |
| Chloride (Cl−) | 4 | 116 |
| Bicarbonate (HCO3−) | 12 | 29 |
| Amino acids in proteins | 138 | 9 |
Relative to the outside of a cell, the concentration of Ca2+ is low.[21] In addition to sodium and potassium ions the nucleosol also contains Mg2+[22]. Some of these magnesium ions are associated with incoming ribonucleoside triphosphate (NTP) as they enter the catalytic center for transcription by RNA polymerase (RNAP) II.[22] The remaining typical ions found in any cytosol include chloride and bicarbonate.[20]
Intranuclear posttranscriptional modifications such as mRNA editing convert cytidine to uridine within some mRNA.[23] This conversion by enzyme EC 3.5.4.5 though infrequent releases ammonia[24] or produces ammonium (NH4+) in solution. This enzyme is Zn2+ dependent. The zinc ion in the active site plays a central role in the proposed catalytic mechanism, activating a water molecule to form a hydroxide ion (OH-) that performs a nucleophilic attack on the substrate.[25]
Cells also maintain an intracellular iron ion (Fe2+) homeostasis.[26] Cu2+ serves as a cofactor.[27] Iron homeostasis involves interconversions of Fe2+ with Fe3+.
When a nucleotide is incorporated into a growing DNA or RNA strand by a polymerase, pyrophosphate (PPi) is released. The pyrophosphate anion has the structure P2O74−, and is an acid anhydride of phosphate. It is unstable in aqueous solution and in the absence of enzymic catalysis hydrolyzes extremely slowly into inorganic phosphate HPO42− (orthophosphate, Pi) in all but highly acidic media.[28]
Enzyme EC 3.6.1.1 catalyzes the hydrolysis of PPi to Pi:
PPi + H2O <=> 2 Pi.
The enzyme is Mg2+ binding, occurs in the cytosol, has a 33 kDa form, and no NLS. The enzymes of EC 3.6.1.1, in general, exist as homooligomers.
| Ion | Concentration in cytosol | Concentration in blood |
|---|---|---|
| Hydrogen (H+) | 35 - 45 nmol/l (nM), (pH > 7.45; H+ < 35) | 35 - 45 nmol/l (nM), (pH < 7.35; H+ >45) |
| Lithium (Li+) | ? | 1.5-2.5 x 10-8 g/cm3 (total lithium, whole blood) |
| Ammonium (NH4+) | ? | ? |
| Vanadium (V2+) | ? | ? |
| Manganese (Mn2+) | ? | 0.76 to 1.46 µg/L (plasma, women), 0.4 to 2.12 µg/L, 1.04 average µg/L (serum, men), 3.93 to 17.1 µg/L (whole blood) [29] |
| Iron (Fe2+) | ? | 400-450mg/L (total iron) [30] |
| Cobalt (Co2+) | ? | ? |
| Nickel (Ni2+) | ? | ? |
| Copper (Cu2+) | ? | 10 to 25 µmol/L (serum) [31] |
| Zinc (Zn2+), usually bound | ? | about 10 to 15 μmol/L (plasma) [32] |
| Molybdenum (Mo2+) | ? | ~5 nmol/L (total molybdenum, average) [33] |
| Cadmium (Cd2+) | ? | 1-5 x 10-9 g/cm3 (total cadmium, whole blood) |
| Tin (Sn2+) | ? | 0-4 x 10-7 g/cm3 (total tin, whole blood), 0-1 x 10-7 g/cm3 (total tin, plasma or serum) |
| Lead (Pb2+) | ? | 1-5 x 10-7 g/cm3 (total lead, whole blood), 1-7.8 x 10-8 g/cm3 (total lead, plasma or serum) |
| Boron (B3+) | ? | ? |
| Chromium (Cr3+) | ? | 2 to 3 nmol/L (total chromium) [34] |
| Iron (Fe3+) | ? | ? |
| Arsenic (As3+) | ? | 2-62 x 10-9 g/cm3 (whole blood, total arsenic) |
| Silicon (Si4+) | ? | 21 µmol/L (serum, total silicon) [35] |
| Hydroxide (OH−) | ? | ? |
| Fluoride (F−) | ? | 1-4.5 x 10-7 g/cm3 (whole blood and plasma or serum) |
| Cyanide (CN−) | ? | 4 x 10-9 g/cm3 (plasma or serum, non-smokers) |
| Bromide (Br−) | ? | 7-10 x 10-9 g/cm3 (plasma or serum) |
| Iodine (I−) | ? | 2.4-3.2 x 10-8 g/cm3 (whole blood), 4.5-14.5 x 10-8 g/cm3 (plasma or serum) |
| Orthophosphate, Pi (HPO42−) | 75 milliequivalents/L[36] | 0.8 to 1.5 mM (arterial blood)[37], 4 milliequivalents/L (ISF)[36] |
| Orthophosphate, Pi (H2PO4−) | ? | 1 milliequivalents/L (ISF) [38] |
| Pyrophosphate, PPi (P2O74−) | ? | 300-700 pmol/µg protein (plasma, normal human children) [39] |
Unless otherwise noted, minor ion concentrations are from the List of human blood components.
Carbohydrates
Of the carbohydrates, monosaccharides and oligosaccharides are water soluble. Polysaccharides on the other hand tend to be insoluble in water. As to alcohols, there are two opposing solubility trends: the tendency of the polar OH to promote solubility in water, and of the carbon chain to resist it. Thus, methanol, ethanol, and propanol are miscible in water because the hydroxyl group wins out over the short carbon chain. Butanol, with a four-carbon chain, is moderately soluble because of a balance between the two trends. Alcohols of five or more carbons (pentanol and higher) are effectively insoluble in water because of the hydrocarbon chain's dominance.
Fatty acids
Short chain carboxylic acids such as formic acid and acetic acid are miscible with water and dissociate to form reasonably strong acids (pKa 3.77 and 4.76, respectively). Longer-chain fatty acids do not show a great change in pKa. Nonanoic acid, for example, has a pKa of 4.96. However, as the chain length increases the solubility of the fatty acids in water decreases very rapidly, so that the longer-chain fatty acids have very little effect on the pH of a solution.
When the body uses stored fat as a source of energy, glycerol and fatty acids are released into the bloodstream. The glycerol component can be converted to glucose by the liver and provides energy for cellular metabolism.
Amino acids
The average mass range for amino acids: 75 - 222 Da. By comparison a water molecule is 18 Da. In addition to the proteinogenic standard amino acids, there are a number of other amino acids (aa) involved in the synthesis of the proteinogenic aa: citrulline (Cit), cystathionine (Cth), homocysteine (Hcy), ornithine (Orn), sarcosine (Sar) and taurine (Tau), for example. As Tau does not contain a carboxyl group it is not an aa, but since in its place it does contain a sulfonate group, it may be called an amino sulfonic acid.
Nucleobases
Purine (Pur) 120 Da is not a protein. The purines are the most widely distributed naturally occurring nitrogen-containing heterocycle.[40] The purine nucleobases include adenine (A) 135 Da, hypoxanthine (Hx) 136 Da, guanine (G) 151 Da, and xanthine (Xan) 152 Da. The pyrimidines include pyrimidine (Pyr) 80 Da, also a heterocycle and naturally occurring, cytosine (C) 111 Da, uracil (U) 112 Da, thymine (T) 126 Da, and queuine (Q) 275 Da.
Nucleosides
Nucleosides are glycosylamines, a nucleobase linked to a ribose or deoxyribose ring. Examples include purines: adenosine (Ado) 267 Da, guanosine (Guo) 283 Da, and inosine (Ino) 268 Da, and pyrimidines: cytidine (Cyd) 243 Da, thymidine (Thd) 242 Da, uridine (Urd) 244 Da, and queuosine (Quo) 409 Da. When the nucleobase is attached to deoxyribose, a 'd' is placed in front of the abbreviation, e.g., dCyd is deoxycytidine 227 Da and the molar mass decreases by one oxygen from Cyd.
Nucleotides
Nucleotides such as orotidine 5'-monophosphate (OMP) range in size from 176 Da (OMP) to 523 Da (GTP). The purine nucleotides involved in RNA or DNA synthesis include: inosine monophosphate (IMP), adenosine triphosphate (ATP), and guanosine triphosphate (GTP). The pyrimidine nucleotides involved include OMP, cytidine triphosphate (CTP), uridine triphosphate (UTP), and thymidine triphosphate (TTP) for DNA in place of UTP. Although rare, higher phosphates do occur such as adenosine tetraphosphate (Ap4) 587 Da. The deoxyribonucleotides have a 'd' in front, like dCTP, except for the thymidine deoxyribonucleotides.
Cofactors
Many cofactors are involved in the synthesis of amino acids and nucleotides. They range in size from ascorbic acid (ASA) 176 Da and biotin (BIO) 244 Da, which are vitamins, to nicotinamide adenine dinucleotide phosphate (NADP) 744 Da and flavin adenine dinucleotide (FAD) 785 Da.
One of the coenzymes essential for the synthesis of amino acids is nicotinamide adenine dinucleotide (NAD) 663 Da. Besides assembling NAD+ de novo from simple amino acid precursors, cells also salvage preformed compounds containing nicotinamide. The three natural compounds containing the nicotinamide ring and used in these salvage metabolic pathways are nicotinic acid (Na), nicotinamide (Nam) and nicotinamide riboside (NR).[41] These compounds are also produced within cells, when the nicotinamide group is released from NAD+ in ADP-ribose transfer reactions. Indeed, the enzymes involved in these salvage pathways appear to be concentrated in the cell nucleus, which may compensate for the high level of reactions that consume NAD+ in this organelle.[42] Nicotinamide mononucleotide adenylyl transferase 1 (NMNAT1) (EC 2.7.7.1) catalyzes a key step of NAD synthesis.[43] It has a nuclear localization signal (NLS).[43] NMNAT1 may be a substrate for nuclear kinases.[43]
Peptides
Peptides are short polymers formed from the linking, in a defined order, of α-amino acids. Proteins are polypeptide molecules (or consist of multiple polypeptide subunits). The distinction is that peptides are short and polypeptides/proteins are long. The digestion of dietary proteins produces dipeptides which are absorbed more rapidly than aa. A dipeptide is a molecule consisting of two amino acids joined by a single peptide bond. Examples of dipeptides include carnosine (Car) 244 Da, of the amino acids β-alanine (β-Ala) and histidine (His), homocarnosine (Hcn) 258 Da consisting of γ-aminobutyric acid (GABA) and His, and anserine (Ans) 240 Da.
Oligopeptides
Some tripeptides and tetrapeptides are synthesized in humans. Oligopeptides can range up to 40 aa (9 kDa) generally.
Small proteins (polypeptides)
Due to the size limitation of the nuclear pore, these polypeptides would range from 9 kDa to <70 kDa and not need or have a NLS. For example, emerin 18 kDa (no NLS) mediates inner nuclear membrane anchorage to the nuclear lamina, regulates the flux of beta-catenin into the nucleus, and interacts with nuclear actin.[44][45][46]
On the other hand, LEMD1 (20.3 kDa) is involved in the glutamine (Gln) metabolic process[47] and has a NLS.[48]
Many of the polypeptides are enzymes including peptidases and kinases.
Proteases
Carnosinase occurs as EC 3.4.13.3 (Xaa-His dipeptidase) with Zn2+ as cofactor, 3.4.13.18 (cytosol nonspecific dipeptidase) with Zn2+ as cofactor and Mn2+ activation, and 3.4.13.20 (beta-Ala-His dipeptidase), activated by Cd2+ and citrate, catalyzing the reaction
Car + H2O <=> His + β-Ala.
It is intracellular to the cytosol and can occur in 14 kDa, 35 kDa, and 44 kDa sizes, often forming a homodimer. As a nonspecific dipeptidase, it degrades a number of dipeptides including Car[49], Ans and Hcn[49] as EC 3.4.13.3 and EC 3.4.13.20 per the reaction:
Hcn + H2O <=> γ-aminobutyric acid (GABA) + His.
Oligopeptides can be degraded by aminopeptidases such as EC 3.4.11.6 19-68 kDa forms (intracellular to the cytosol) with Zn2+ as cofactor and activation by Cl- per the reactions:
oligopeptide (n) + H2O <=> Lys + oligopeptide (n-1)
oligopeptide (n) + H2O <=> Arg + oligopeptide (n-1).
Synthases
Enzymes EC 2.3.1.37 (cofactor: pyridoxal phosphate, PLP) aminolevulinate, delta-, synthase 1 (ALAS1) and aminolevulinate, delta-, synthase 2 (ALAS2) anabolically synthesize glycine (Gly) from the amino acid 5-amino-4-oxovaleric acid (ALA) in the two-step reaction:
5-aminolevulinate (C5H9NO3) (ALA) + CO2 <=> 2-amino-3-oxoadipate (C6H9NO5)
+
2-amino-3-oxoadipate + CoA (C21H36N7O16P3S) <=> succinyl-CoA (C25H40N7O19P3S) + Gly
=
5-aminolevulinate + CoA + CO2 <=> succinyl-CoA + Gly
The mRNA for ALAS1 is 82 kDa, the intracellular precursor is a homodimer of 71 kDa, and the mitochondrial mature protein is 65 kDa. But, ALAS1 also occurs in a 30 kDa form.[50]
CTP synthase EC 6.3.4.2 is the final step in the de novo synthesis of CTP from UTP. As a monomer 67 kDa or dimer it is inactive because three monomers contribute to ligand binding at the active site.[51] The active form is a homotetramer (a dimer of dimers), with no NLS, intracellular to the cytosol,[51][52] for the following reactions.
UTP + Gln + ATP + H2O <=> CTP + Glu + ADP + Pi
ATP + UTP + NH3 <=> ADP + Pi + CTP
The reactions
2 ATP + HCO3- + NH3 <=> 2 ADP + Pi + carbamoyl phosphate (multistep)[53]
Gln + H2O <=> Glu + NH3[54]
2 ADP + Pi + Glu + carbamoyl phosphate <=> 2 ATP + Gln + HCO3- + H2O[55]
are catalyzed by EC 6.3.5.5 carbamoyl-phosphate synthase II (CAD). It has no NLS, occurs as a homohexamer, uses Zn2+ as a cofactor and is intracellular to the nucleus.[56] It does occur in a 22 kDa form.[57]
Kinases
EC 3.6.1.5 (cofactor: Ca2+) catalyzes the following reactions:
AMP + Pi <=> ADP + H2O
AMP + 2 Pi <=> ATP + 2 H2O
ADP + Pi <=> ATP + H2O
CMP + Pi <=> CDP + H2O
CMP + 2 Pi <=> CTP + 2 H2O
CDP + Pi <=> CTP + H2O
GMP + Pi <=> GDP + H2O
GMP + 2 Pi <=> GTP + 2 H2O
GDP + Pi <=> GTP + H2O
UMP + Pi <=> UDP + H2O
UMP + 2 Pi <=> UTP + 2 H2O
UDP + Pi <=> UTP + H2O
Ca2+ or Mg2+ can serve as activating ions. ENTPD1 (CD39) is a 56 kDa protein.[58] CD39 associates with RanBPM (RANBP9).[59] RANBP9 (90 kDa)[60] binds Ran, a small GTP binding protein that is essential for the translocation of RNA and proteins through the nuclear pore complex.[61] RanBPM localizes in the nucleus and cytoplasm,[59] but RanBPM has no NLS. CANT1 (EC 3.6.1.6 and 3.6.1.5) 49 kDa[62] catalyzes similar reactions:
a nucleotide + Pi <=> a nucleoside diphosphate + H2O,
also acting on IDP, GDP, UDP and on D-ribose 5-diphosphate.[63] ENTPD7 (EC 3.6.1.-) occurs in the mouse nucleus.[64]
Enzymes 2.4.2.7 adenine phosphoribosyltransferase (APRT) 19.4 kDa (forms a dimer, 38.8 kDa) is intracellular (cytoplasm)[65] and 2.4.2.8 hypoxanthine phosphoribosyltransferase 1 (HPRT1) 24 kDa (forms a tetramer, 96 kDa) is intracellular (cytosolic)[66] catalyze the following reaction:
Adenine + 5-phospho-alpha-D-ribose 1-diphosphate (PRPP) <=> AMP + PPi
Neither APRT nor HPRT1 has a NLS. APRT is a dimer in solution at pH 6.5, but a monomer at pH 8.0, and like HPRT1 needs Mg2+, or Mn2+.[67]
Enzymes EC 3.6.1.5 ATP pyrophosphohydrolase, ADPase[68]/ADP synthase[69] ectonucleoside triphosphate diphosphohydrolase 1 (ENTPD1) 56 kDa catalyzes the conversion of AMP into ADP (see below). Cofactor: Ca2+.[70] Ca2+ or Mg2+ can serve as activating ions. Also acts on ADP, and on other nucleoside triphosphates and diphosphates.[70]
AMP + Pi <=> ADP + H2O
Enzymes EC 2.7.4.6 nucleoside-diphosphate kinases A, B, C, D (NDKA-D) catalyzes the following reaction inside the nucleosol as it is intracellular (nucleus)[71] and each gene is translated as a 7-11 kDa particle[72]. However, these kinases exist either as tetramers (28-44 kDa) in bacteria or hexamers (42-66 kDa).[73] Once the hexamer has formed the particle may be too big to pass through the nuclear pores. NDKA has been shown to mediate transcription, associate with a promoter region of a gene, and be a member of the SET or INHAT complex which can modulate gene expression.[74] Nucleoside-diphosphate kinase does form nuclear and cytoplasmic hexamers.[75] NDKA-D do not have a NLS.
ADP + GTP <=> ATP + GDP
Transaminases
In catabolic transamination, with PLP as a cofactor, EC 2.6.1.2 transfers the amine from glutamic acid (glutamate) (Glu) to alanine (Ala) via a two step reaction:
PLP + Glu <=> pyridoxamine monophosphate (PMP) + α-ketoglutarate (2-oxoglutarate)
+
PMP + pyruvate <=> PLP + Ala
=
pyruvate + Glu <=> Ala + 2-oxoglutarate.
Although this enzyme has several different names, e.g., alanine transaminase, glutamic-pyruvate transaminase (GPT), or alanine aminotransferase, it can occur as a monomer of 55 kDa or homodimer of 101 kDa, and as either a cytosolic (GPT1), or mitochondrial form (GPT2).
Synthetases
The reactions
ATP + Glu + NH3 (or NH4+) <=> ADP + Pi + Gln
ATP + Asp + NH4+ <=> ADP + Pi + Asn
ATP + Asp + NH4+ <=> AMP + PPi + Asn
are catalyzed by the enzyme EC 6.3.1.2 glutamine synthetase (GS), glutamine-ammonia ligase (GLUL). GLUL 42 kDa is intracellular, occurs as a homooctamer,[76] also as 12 kDa and 22 kDa forms,[77] and complexes with phosphate, ADP, and Mn2+.
Histone pools in the nucleosol
Histones H2A, H2B, H3, and H4 are in the nucleosol, and H2B, H3, and H4 are also in the cytosol.[78]
Enzyme activity
EC 2.4.1.101 (UDP-N-acetylglucosaminyl transferase) occurs and is active in the nuclesol.[79]
Briefly, any enzyme that can occur as a monomer small enough to pass through the nuclear pore complex before it becomes a dimer or larger oligomer or polymer can occur and act in the nucleosol, whether it has a NLS or not.
Exonucleosol proteins
Keratin type II cytoskeletal 80 (KRT80) as a monomer is 51 kDa (variant 2) and thereby can diffuse into the nucleosol, variant 3 is 54 kDA, and variant 1 is 47 kDa. KRT80 is an intermediate filament protein that makes up one of the major structural fibers of epithelial cells.[80]
While keratin type I cytoskeletal 13 (KRT13) is usually paired with keratin type II cytoskeletal 4 (KRT4) 64 kDa, KRT13 has a 46 kDa variant 2 and a 39 kDA variant (KRT-13-201).[81] Keratins only assemble as heteropolymers: a type I and a type II protein forming a heterodimer.
Polo-like kinase 1 (PLK1) phosphorylizes KRT4 at serine 157.[82] PLK1 27 kDa occurs in the nucleoplasm and cytosol, is ATP binding, and engages in protein amino acid phosphorylation.[83] PLK1 associates with, binds to, and phosphorylates alpha-, beta- and gamma-tubulins in interphase independent of the microtubule polymerization state.[84]
Intermediate filaments (IFs) are composed of proteins (40-210 kDa) that self-assemble into complex cytoskeletal fibers.[85] Inside an interphase cell, ~5% of the total intermediate filament protein exists in a soluble tetrameric form.[86] Newly synthesized intermediate filament protein is soluble and self-assembly proceeds at concentrations >40-50 µg/ml.[86] Homodimers of IF proteins can form and be added to already forming IFs but cannot assemble into IFs themselves.[86]
History of the nucleosol
While earlier articles or book sections may exist mentioning the term "nucleosol", a book from 1970 online refers to the nucleosol.[87]
Acknowledgements
The content on this page was first contributed by: Henry A. Hoff.
Initial content for this page in some instances came from Wikipedia.
References
- ↑ Maton A, Hopkins J, Johnson S, LaHart D, Warner MQ, Wright JD (1997). Cells Building Blocks of Life. Upper Saddle River, New Jersey: Prentice Hall. pp. 66–67.
- ↑ Raymond A. Serway (1996). Physics for Scientists & Engineers (4th Edition ed.). Saunders College Publishing. ISBN 0-03-005932-1.
- ↑ Ojovan MI, Lee WE (2004). "Viscosity of network liquids within Doremus approach". J Appl Phys. 95 (7): 3803–10. doi:10.1063/1.1647260. Text "month" ignored (help)
- ↑ 4.0 4.1 Ojovan MI (2008). "Configurons: thermodynamic parameters and symmetry changes at glass transition". Entropy. 10: 334–64. doi:10.3390/e10030334. Text "http://www.mdpi.org/entropy/papers/e10030334.pdf " ignored (help); Unknown parameter
|month=ignored (help) - ↑ Salmon PS (2002). "Amorphous materials: Order within disorder". Nature Materials. 1 (2): 87–8. doi:10.1038/nmat737.
- ↑ 6.0 6.1 Jorgensen WL, Madura JD (1985). "Temperature and size dependence for Monte Carlo simulations of TIP4P water". Mol Phys. 56 (6): 1381. doi:10.1080/00268978500103111.
- ↑ Zielkiewicz J (2005). "Structural properties of water: Comparison of the SPC, SPCE, TIP4P, and TIP5P models of water". J Chem Phys. 123: 104501. doi:10.1063/1.2018637.
- ↑ Laage D, Hynes JT (2006). "A Molecular Jump Mechanism for Water Reorientation". Science. 311: 832. doi:10.1126/science.1122154.
- ↑ Ludwig R (2001). "Water: From Clusters to the Bulk". Angew Chem Int Ed. 40 (10): 1808–27. doi:10.1002/1521-3773(20010518)40:10<1808::AID-ANIE1808>3.0.CO;2-1.
- ↑ Oshovsky GV, Reinhoudt DN, Verboom W (2007). "Supramolecular Chemistry in Water". Angew Chem Int Ed. 46 (14): 2366–93. doi:10.1002/anie.200602815.
- ↑ Mir MH, Vittal JJ (2007). "Phase Transition Accompanied by Transformation of an Elusive Discrete Cyclic Water Heptamer to a Bicyclic (H2O)7 Cluster". Angew Chem Int Ed. 46: 5925–28. doi:10.1002/anie.200701779.
- ↑ Huang C, Wikfeldt KT, Tokushima T, Nordlund D, Harada Y, Bergmann U, Niebuhr M, Weiss TM, Horikawa Y, Leetmaa M, Ljungberg MP, Takahashi O, Lenz A, Ojamäe L, Lyubartsev AP, Shin S, Pettersson LGM, Nilsson A (2009). "The Inhomogeneous Structure of Water at Ambient Conditions". Proc Nat Acad Sci (USA). doi:10.1073/pnas.0904743106. Unknown parameter
|month=ignored (help) - ↑ "Liquid water in the domain of cubic crystalline ice Ic".
- ↑ Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowell AW, Schultz P (1988). "Cryo-electron microscopy of vitrified specimens". Q Rev Biophys. 21: 129–228.
- ↑ Hyatt MT, Levinson HS (1968). "Water Vapor, Aqueous Ethyl Alcohol, and Heat Activation of Bacillus megaterium
Spore Germination". J Bacteriol. 95 (6): 2090–101. PMID 4970224. Unknown parameter
|month=ignored (help); line feed character in|title=at position 80 (help) - ↑ Gideon P, Thomsen C, Henriksen O (1994). "Increased self-diffusion of brain water in normal aging". J Magn Reson Imaging. 4 (2): 185–8. doi:10.1002/jmri.1880040216. PMID 8180459. Unknown parameter
|month=ignored (help) - ↑ Krynicki K, Green CD, Sawyer DW (1978). "Pressure and temperature dependence of self-diffusion in water". Faraday Discuss Chem Soc. 66: 199–208. doi:10.1039/DC9786600199.
- ↑ "Human Protein Atlas NT5C1A gene information".
- ↑ 19.0 19.1 19.2 Verkman AS (2002). "Solute and macromolecule diffusion in cellular aqueous compartments". Trends Biochem Sci. 27 (1): 27–33. doi:10.1016/S0968-0004(01)02003-5. PMID 11796221. Unknown parameter
|month=ignored (help) - ↑ 20.0 20.1 Lodish, Harvey F. (1999). Molecular cell biology. New York: Scientific American Books. ISBN 0-7167-3136-3. OCLC 174431482.
- ↑ Berridge MJ (1997). "Elementary and global aspects of calcium signalling". J. Physiol. (Lond.). 499 ( Pt 2): 291–306. PMC 1159305. PMID 9080360. Unknown parameter
|month=ignored (help) - ↑ 22.0 22.1 Langelier MF, Baali D, Trinh V, Greenblatt J, Archambault J, Coulombe B (2005). "The highly conserved glutamic acid 791 of Rpb2 is involved in the binding of NTP and Mg(B) in the active center of human RNA polymerase II". Nucleic Acids Res. 33 (8): 2629–39. PMID 15886393. Unknown parameter
|month=ignored (help) - ↑ Ashkenas J (1997). "Gene regulation by mRNA editing". Am J Hum Genet. 60 (2): 278–83. PMID 9012400. Unknown parameter
|month=ignored (help) - ↑ "NiceZyme View of ENZYME: EC 3.5.4.5".
- ↑ "NCBI Conserved Domains: cytidine_deaminase-like Super-family".
- ↑ Mukhopadhyay CK, Attieh ZK, Fox PL (1998). "Role of ceruloplasmin in cellular iron uptake". Science. 279 (5351): 714–7. PMID 9445478. Unknown parameter
|month=ignored (help) - ↑ 1.16.3.1 "NiceZyme View of ENZYME: EC 1.16.3.1" Check
|url=value (help). - ↑ Huebner PWA, Milburn RM (1980). "Hydrolysis of pyrophosphate to orthophosphate promoted by cobalt(III). Evidence for the role of polynuclear species". Inorg Chem. 19 (5): 1267–72. doi:10.1021/ic50207a032. Unknown parameter
|month=ignored (help) - ↑ United States National Research Council, Institute of Medicine (2000). Dietary Reference Intakes for Vitamin A, Vitamin K, Arsenic, Boron, Chromium, Copper, Iodine, Iron, Manganese, Molybdenum, Nickel, Silicon, Vanadium, and Zinc. National Academic Press. pp. 399–400.
- ↑ United States National Research Council, Institute of Medicine (2000). Dietary Reference Intakes for Vitamin A, Vitamin K, Arsenic, Boron, Chromium, Copper, Iodine, Iron, Manganese, Molybdenum, Nickel, Silicon, Vanadium, and Zinc. National Academic Press. pp. 320–6.
- ↑ United States National Research Council, Institute of Medicine (2000). Dietary Reference Intakes for Vitamin A, Vitamin K, Arsenic, Boron, Chromium, Copper, Iodine, Iron, Manganese, Molybdenum, Nickel, Silicon, Vanadium, and Zinc. National Academic Press. p. 227.
- ↑ United States National Research Council, Institute of Medicine (2000). Dietary Reference Intakes for Vitamin A, Vitamin K, Arsenic, Boron, Chromium, Copper, Iodine, Iron, Manganese, Molybdenum, Nickel, Silicon, Vanadium, and Zinc. National Academic Press. p. 445.
- ↑ United States National Research Council, Institute of Medicine (2000). Dietary Reference Intakes for Vitamin A, Vitamin K, Arsenic, Boron, Chromium, Copper, Iodine, Iron, Manganese, Molybdenum, Nickel, Silicon, Vanadium, and Zinc. National Academic Press. p. 421.
- ↑ United States National Research Council, Institute of Medicine (2000). Dietary Reference Intakes for Vitamin A, Vitamin K, Arsenic, Boron, Chromium, Copper, Iodine, Iron, Manganese, Molybdenum, Nickel, Silicon, Vanadium, and Zinc. National Academic Press. p. 202.
- ↑ United States National Research Council, Institute of Medicine (2000). Dietary Reference Intakes for Vitamin A, Vitamin K, Arsenic, Boron, Chromium, Copper, Iodine, Iron, Manganese, Molybdenum, Nickel, Silicon, Vanadium, and Zinc. National Academic Press. p. 529.
- ↑ 36.0 36.1 Kavanaugh MP, Miller DG, Zhang W, Law W, Kozak SL, Kabat D, Miller AD (1994). "Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters". Proc Natl Acad Sci U S A. 91 (15): 7071–5. PMID 8041748. Unknown parameter
|month=ignored (help) - ↑ Walter F. Medical Physiology: A Cellular And Molecular Approaoch. Elsevier/Saunders. p. 849. ISBN 1-4160-2328-3.
- ↑ Schwartz MK. "Phosphate metabolism". McGraw-Hill Encyclopedia of Science & Technology (9th ed.). 13: 343–4.
- ↑ Rutsch F, Vaingankar S, Johnson K, Goldfine I, Maddux B, Schauerte P, Kalhoff H, Sano K, Boisvert WA, Superti-Furga A, Terkeltaub R (2001). "PC-1 nucleoside triphosphate pyrophosphohydrolase deficiency in idiopathic infantile arterial calcification". Am J Pathol. 158 (2): 543–54. PMID 11159191. Unknown parameter
|month=ignored (help) - ↑ Rosemeyer, H. Chemistry & Biodiversity 2004, 1, 361.
- ↑ Tempel W, Rabeh WM, Bogan KL; et al. (2007). "Nicotinamide riboside kinase structures reveal new pathways to NAD+". PLoS Biol. 5 (10): e263. PMID 17914902.
- ↑ Anderson RM, Bitterman KJ, Wood JG; et al. (2002). "Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels". J. Biol. Chem. 277 (21): 18881&ndash, 90. PMID 11884393.
- ↑ 43.0 43.1 43.2 Schweiger M, Hennig K, Lerner F, Niere M, Hirsch-Kauffmann M, Specht T, Weise C, Oei SL, Ziegler M (2001). "Characterization of recombinant human nicotinamide mononucleotide adenylyl transferase (NMNAT), a nuclear enzyme essential for NAD synthesis". FEBS Lett. 492 (1–2): 95–100. PMID 11248244. Unknown parameter
|month=ignored (help) - ↑ "Entrez Gene: EMD emerin".
- ↑ "GENATLAS: GENE Database EMD".
- ↑ "Apropos IpiRecord: IPI00032003".
- ↑ "Apropos IpiRecord: IPI00438351".
- ↑ "Nuclear Protein Database LEMD1".
- ↑ 49.0 49.1 Balion CM, Benson C, Raina PS, Papaioannou A, Patterson C, Ismaila AS (2007). "Brain type carnosinase in dementia: a pilot study". BMC Neurol. 7 (Nov 5): 38. PMID 17983474.
- ↑ "Apropos IpiRecord: IPI00789294 ALAS1, 30 KDA PROTEIN".
- ↑ 51.0 51.1 Kursula P, Flodin S, Ehn M, Hammarstrom M, Schüler H, Nordlund P, Stenmark P (2006). "Structure of the synthetase domain of human CTP synthetase, a target for anticancer therapy". Acta Crystallogr Sect F Struct Biol Cryst Commun. 62 (Pt 7): 613–7. doi:10.1107/S1744309106018136. PMID 16820675. Unknown parameter
|month=ignored (help) - ↑ "GENATLAS : GENE Database CTPS".
- ↑ "KEGG REACTION: R07641".
- ↑ "KEGG REACTION: R00256".
- ↑ "KEGG REACTION: R00575".
- ↑ "GENATLAS : GENE Database CAD".
- ↑ "Apropos IpiRecord: IPI00301263 CAD".
- ↑ "Apropos IpiRecord: IPI00382672".
- ↑ 59.0 59.1 Wu Y, Sun X, Kaczmarek E, Dwyer KM, Bianchi E, Usheva A, Robson SC (2006). "RanBPM associates with CD39 and modulates ecto-nucleotidase activity". Biochem J. 396 (1): 23–30. PMID 16478441. Unknown parameter
|month=ignored (help) - ↑ "GENATLAS: GENE Database RANBP9".
- ↑ "Entrex Gene: RANBP9 RAN binding protein 9".
- ↑ "Apropos IpiRecord: IPI00413533".
- ↑ "NiceZyme View of ENZYME: EC 3.6.1.6".
- ↑ "Apropos IpiRecord: IPI00828276".
- ↑ "GENATLAS : GENE Database APRT".
- ↑ "GENATLAS : GENE Database HPRT1".
- ↑ Phillips CL, Ullman B, Brennan RG, Hill CP (1999). "Crystal structures of adenine phosphoribosyltransferase from Leishmania donovani". EMBO J. 18 (13): 3533–45. doi:10.1093/emboj/18.13.3533. PMID 10393170.
- ↑ Moodie FDL, Baum H, Peter J, Butterworth PJ, Timothy J, Peters TJ (1991). "Purification and characterisation of bovine spleen ADPase". Eur. J. Biochem. 202 (3): 1209–15. PMID 1837267. Unknown parameter
|month=ignored (help) - ↑ Goodman JM (2008). "The gregarious lipid droplet". J Biol Chem. 283 (42): 28005–9. doi:10.1074/jbc.R80004220. PMID 18611863. Unknown parameter
|month=ignored (help) - ↑ 70.0 70.1 "NiceZyme View of ENZYME: EC 3.6.1.5".
- ↑ "GENATLAS : GENE Database NME3".
- ↑ "Apropos IpiRecord: IPI00012048".
- ↑ "NCBI Conserved Domains cd04413: NDPK_I".
- ↑ Curtis CD, Likhite VS, McLeod IX, Yates JR, Narulli AM (2007). "Interaction of the Tumor Metastasis Suppressor Nonmetastatic Protein 23 Homologue H1 and Estrogen Receptor {alpha} Alters Estrogen-Responsive Gene Expression". Cancer Res. 67 (21): 10600–7. doi:1158/0008-5472.CAN-07-0055 Check
|doi=value (help). PMID 17975005. Unknown parameter|month=ignored (help) - ↑ Klouckova I, Hrncirova P, Mechref Y, Arnold RJ, Li TK, McBride WJ, Novotny MV (2006). "Changes in liver protein abundance in inbred alcohol-preferring rats due to chronic alcohol exposure, as measured through a proteomics approach". Proteomics. 6 (10): 3060–74. doi:0.1002/pmic.200500725 Check
|doi=value (help). PMID 16619309. Unknown parameter|month=ignored (help) - ↑ "GENATLAS : GENE Database GLUL".
- ↑ "Apropos IpiRecord: IPI00830929".
- ↑ Tsvetkov S, Ivanova E, Djondjurov L (1989). "The pool of histones in the nucleosol and cytosol of proliferating Friend cells is small, uneven and chasable". Biochem J. 264 (3): 785–91. PMID 2619716. Unknown parameter
|month=ignored (help) - ↑ Marshall S, Okuyama R (2004). "Differential effects of vanadate on UDP-N-acetylglucosaminyl transferase activity derived from cytosol and nucleosol". Biochem Biophys Res Commun. 318 (4): 911–5. PMID 15147958. Unknown parameter
|month=ignored (help) - ↑ Rogers MA, Edler L, Winter H, Langbein L, Beckmann I, Schweizer J (2005). "Characterization of new members of the human type II keratin gene family and a general evaluation of the keratin gene domain on chromosome 12q13.13". J Invest Dermatol. 124 (3): 536–44. PMID 15737194. Unknown parameter
|month=ignored (help) - ↑ "Human Protein Atlas KRT13 gene information".
- ↑ Burkard ME, Maciejowski J, Rodriguez-Bravo V, Repka M, Lowery DM, Clauser KR, Zhang C, Shokat KM, Carr SA, Yaffe MB, Jallepalli PV (2009). "Plk1 self-organization and priming phosphorylation of HsCYK-4 at the spindle midzone regulate the onset of division in human cells". PLoS Biol. 7 (5). doi:10.1371/journal.pbio.1000111. Unknown parameter
|month=ignored (help) - ↑ "Human Protein Atlas PLK1 gene information".
- ↑ Feng Y, Hodge DR, Palmieri G, Chase DL, Longo DL, Ferris DK (1999). "Association of polo-like kinase with alpha-, beta- and gamma-tubulins in a stable complex". Biochem J. 339 (Pt 2): 435–42. PMID 10191277. Unknown parameter
|month=ignored (help) - ↑ Coulombe PA, Fuchs E (1990). "Elucidating the early stages of keratin filament assembly". J Cell Biol. 111 (1): 153–69. PMID 1694855. Unknown parameter
|month=ignored (help) - ↑ 86.0 86.1 86.2 Bachant JB, Klymkowsky MW (1996). "A nontetrameric species is the major soluble form of keratin in Xenopus oocytes and rabbit reticulocyte lysates". J Cell Biol. 132 (1–2): 153–65. PMID 8567720. Unknown parameter
|month=ignored (help) - ↑ Société de chimie biologique (France), Société de chimie biologique (1970). Bulletin de la Société de chimie biologique, Volume 52. Masson. p. 401.
See also
External links
- Discussion of amorphous ice at LSBU's website.
- Glassy Water from Science, on phase diagrams of water (requires registration)
- Structure of amorphous ice
- Water clusters at London South Bank University Link
- CS1 maint: Multiple names: authors list
- CS1 maint: Extra text
- Pages with citations using unnamed parameters
- Pages with citations using unsupported parameters
- CS1 errors: invisible characters
- Pages with URL errors
- CS1 maint: Explicit use of et al.
- CS1 errors: DOI
- Pages with broken file links
- Fluid mechanics
- Fundamental physics concepts
- Viscosity
- Phases of matter
- Optics
- Electromagnetic radiation
- Cluster chemistry
- Water chemistry
- Homogeneous chemical mixtures
- Colloidal chemistry
- Cell anatomy