Ebola causes
|
Ebola Microchapters |
|
Diagnosis |
|---|
|
Treatment |
|
Postmortem Care |
|
Case Studies |
|
Ebola causes On the Web |
|
American Roentgen Ray Society Images of Ebola causes |
| style="background:#Template:Taxobox colour;"|Ebola virus | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| style="background:#Template:Taxobox colour;" | Virus classification | ||||||||
| ||||||||
| Type species | ||||||||
| Zaïre Ebolavirus | ||||||||
| Species | ||||||||
|
Reston Ebolavirus |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]
Overview
Ebola infection is caused by a virus that belongs to the family Filoviridae. Four viral subtypes have been reported to cause clinical illness in humans: Bundibugyo ebolavirus, Sudan ebolavirus, Tai Forest ebolavirus, and Zaire ebolavirus. The human disease has so far been limited to parts of Africa. A very small number of people in the United States and in the Philippines were infected with the fifth type of the virus, known as Reston ebolavirus, and did not develop any signs of disease.
Taxonomy
Viruses; ssRNA viruses; ssRNA negative-strand viruses; Mononegavirales; Filoviridae; Ebolavirus[1]
- Ebolavirus
- Bundibugyo ebolavirus
- Reston ebolavirus
- Reston ebolavirus - Reston
- Reston ebolavirus - Reston (1989)
- Reston ebolavirus - Siena/Philippine-92
- Sudan ebolavirus
- Sudan ebolavirus - Boniface (1976)
- Sudan ebolavirus - Maleo (1979)
- Sudan ebolavirus - Nakisamata
- Sudan ebolavirus - Uganda (2000)
- Tai Forest ebolavirus
- Tai Forest virus - Côte d’Ivoire, Côte d’Ivoire, 1994
- Zaire ebolavirus
- Ebola virus - Mayinga, Zaire, 1976
- Zaire ebolavirus - Eckron (Zaire, 1976)
- Zaire ebolavirus - Gabon (1994-1997)
- Zaire ebolavirus - Zaire (1995)
- Unclassified Ebolavirus
- Ebola virus Yambio0401
- Ebola virus Yambio0402
- Ebola virus Yambio0403
- Ebola virus sp.
Biology
Ebola infection is caused by a virus that belongs to the family Filoviridae. Four viral subtypes have been reported to cause clinical illness in humans: Bundibugyo ebolavirus, Sudan ebolavirus, Tai Forest ebolavirus, and Zaire ebolavirus. The human disease has so far been limited to parts of Africa. A very small number of people in the United States and in the Philippines were infected with the fifth type of the virus, known as Reston ebolavirus, and did not develop any signs of disease. The virus can be passed to humans from infected animals and animal materials. Ebola can also be spread among humans by close contact with infected body fluids or by infected needles.
Structure
Size and Shape
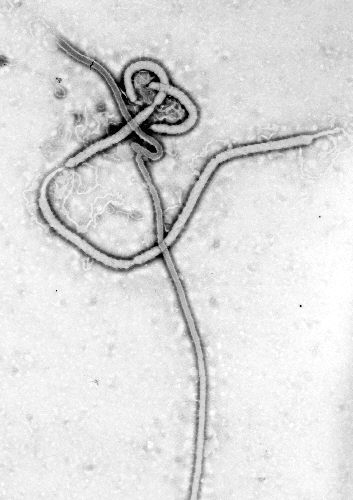
Electron micrographs of members of Ebola virus show them to have the characteristic thread-like structure of a filovirus.[2] EBOV VP30 is around 288 amino acids long.[2] The virions are tubular and variable in shape and may appear as a "U", "6", coiled, circular, or branched shape, however, laboratory purification techniques, such as centrifugation, may contribute to the various shapes.[2] Virions are generally 80 nm in diameter.[2] They are variable in length, and can be up to 1400 nm long. On average, however, the length of a typical Ebola virus is closer to 1000 nm. In the center of the virion is a structure called nucleocapsid, which is formed by the helically wound viral genomic RNA complexed with the proteins NP, VP35, VP30 and L. It has a diameter of 40 – 50 nm and contains a central channel of 20–30 nm in diameter. Virally encoded glycoprotein (GP) spikes 10 nm long and 10 nm apart are present on the outer viral envelope of the virion, which is derived from the host cell membrane. Between envelope and nucleocapsid, in the so-called matrix space, the viral proteins VP40 and VP24 are located.
Genome
Each virion contains one minor molecule of linear, single-stranded, negative-sense RNA, totaling 18959 to 18961 nucleotides in length. The 3′ terminus is not polyadenylated and the 5′ end is not capped. It was found that 472 nucleotides from the 3' end and 731 nucleotides from the 5' end were sufficient for replication.[2] It codes for seven structural proteins and one non-structural protein. The gene order is 3′ - leader - NP - VP35 - VP40 - GP/sGP - VP30 - VP24 - L - trailer - 5′; with the leader and trailer being non-transcribed regions which carry important signals to control transcription, replication and packaging of the viral genomes into new virions. The genomic material by itself is not infectious, because viral proteins, among them the RNA-dependent RNA polymerase, are necessary to transcribe the viral genome into mRNAs, as well as for replication of the viral genome.
Life Cycle
- Virus attaches to host receptors through the GP (glycoprotein) surface peplomer and is endocytosed into vesicles in the host cell.
- Fusion of virus membrane with the vesicle membrane occurs; nucleocapsid is released into the cytoplasm.
- The encapsidated, negative-sense genomic ssRNA is used as a template for the synthesis (3' - 5') of polyadenylated, monocistronic mRNAs.
- Translation of the mRNA into viral proteins occurs using the host cell's machinery.
- Post-translational processing of viral proteins occurs. GP0 (glycoprotein precursor) is cleaved to GP1 and GP2, which are heavily glycosylated. These two molecules assemble, first into heterodimers, and then into trimers to give the surface peplomers. SGP (secreted glycoprotein) precursor is cleaved to SGP and delta peptide, both of which are released from the cell.
- As viral protein levels rise, a switch occurs from translation to replication. Using the negative-sense genomic RNA as a template, a complementary +ssRNA is synthesized; this is then used as a template for the synthesis of new genomic (-)ssRNA, which is rapidly encapsidated.
- The newly-formed nucleocapsides and envelope proteins associate at the host cell's plasma membrane; budding occurs, and the virions are released
Scientists have identified four types of the Ebola virus. Three have been reported to cause disease in humans: Ebola-Zaire virus, Ebola-Sudan virus, and Ebola-Ivory Coast virus. The human disease has so far been limited to parts of Africa. A very small number of people in the United States who were infected with the fourth type of the virus, known as Ebola Reston, did not develop any signs of disease. The disease can be passed to humans from infected animals and animal materials. Ebola can also be spread between humans by close contact with infected bodily fluids or through infected needles in the hospital.
References
- ↑ "Taxonomy browser (Ebolavirus)".
- ↑ 2.0 2.1 2.2 2.3 2.4 Klenk, Hans-Dieter (2004). Ebola and Marburg Viruses, Molecular and Cellular Biology. Wymondham, Norfolk: Horizon Bioscience. ISBN 0954523237. Unknown parameter
|coauthors=ignored (help)